Abstract
The centrosome, an organelle comprising centrioles and associated pericentriolar material, is the major microtubule organizing center in animal cells. For the cell to form a bipolar mitotic spindle and ensure proper chromosome segregation at the end of each cell cycle, it is paramount that the cell contains two and only two centrosomes. Because the number of centrosomes in the cell is determined by the number of centrioles, cells have evolved elaborate mechanisms to control centriole biogenesis and to tightly coordinate this process with DNA replication. Here we review key proteins involved in centriole assembly, compare two major modes of centriole biogenesis, and discuss the mechanisms that ensure stringency of centriole number.
Keywords: centriole, centrosome, duplication, de novo
INTRODUCTION
The centrosome (from Latin for “central body”) is a tiny dense structure that resides near the geometric center of an interphase cell (Wilson, 1925). Since its first descriptions by Van Beneden and Boveri in 1876, the centrosome has remained a focus of attention for cell biologists given it’s involvement, whether direct or indirect, in many vital cellular processes. During mitosis, centrosomes organize the poles of the mitotic spindle and, if the cell contains more than two centrosomes, the spindle is likely to be multipolar. This inevitably randomizes distribution of chromosomes and ultimately leads to aneuploidy, which is a hallmark of malignant tumors. Obviously, the number of centrosomes present in the cell must be precisely controlled.
Centrosomes are composed of cylindrical structures termed centrioles, embedded in a cloud of electron-dense pericentriolar material (PCM). The walls of centrioles are constructed of nine microtubule blades. In different organisms, these blades contain individual microtubules (e.g., Caenorhabditis elegans), microtubule doublets (e.g., Drosophila melanogaster), or microtubule triplets (higher animals). The size of the centriole varies among species, from ~100×150-nm in C. elegans to 200×500-nm in mammalian cells. However, the cylindrical shape and nine-fold symmetry are well conserved even among evolutionarily-distant organisms. Centrioles are directly responsible for the formation of sensory and primary cilia as well as motile cilia/flagella. For this function, centrioles move to the cell cortex, where they attach to the plasma membrane and convert into basal bodies, the structures that initiate assembly of cilia and flagella. Cilia are indispensible for cell motility, extracellular signaling, and development. The list of diseases associated with ciliary dysfunctions continues to lengthen, underscoring the fact that the centriole is essential in higher eukaryotes (Bisgrove and Yost, 2006; Marshall, 2008). The PCM surrounding the centrioles is responsible for nucleation and organization of mitotic and interphase microtubules. Thus, centrosomes in addition to their role in organizing the mitotic poles influence intracellular trafficking and the architecture of the interphase cell.
Centrosome assembly and organization depend on the centrioles; disruption of the centrioles results in a disassembly of the entire centrosome (Bobinnec et al., 1998; Kuriyama and Borisy, 1983). The centriole is therefore the organizer of the centrosome, and the number of centrioles determines the number of centrosomes.
This review focuses on the mechanisms that govern centriole biogenesis and control the number of centrioles in the cell. We will describe two major modes of centriole formation, review the key proteins involved in the formation process, and discuss the results of some recent studies that bring us closer to an understanding of the numerical control of the centriole.
Ab ovo - centriole propagation via duplication of preexisting centrioles
Centriolar cycle
The major mode of centriole formation in somatic cells is duplication of preexisting centrioles. When a new cell is born during mitosis, it inherits two centrioles, each surrounded by its own cloud of PCM (Fig. 1). These two centrioles are of different ages: one of them was formed during the previous cell cycle (i.e., a daughter centriole), while the other one originated from at least two cell cycles ago (i.e., a mother centriole). The mother and daughter differ in several morphological features. For example, only mother but not daughter centrioles bear external appendages attached to its walls (Fig.1). Functionally, only the mother centriole maintains a cloud of PCM that is able to both nucleate microtubules and organize them into the typical radial array. In contrast, PCM associated with the daughter centriole can nucleate microtubules; however, these microtubules are then rapidly released from the centrosome. As the result, the mother centriole during G1 resides at the focus of the microtubule array, while the daughter moves extensively in the cytoplasm (Piel et al., 2001).
Fig. 1. Organization of the centrosome in a typical animal somatic cells (during G0/G1 phase of the cell cycle).
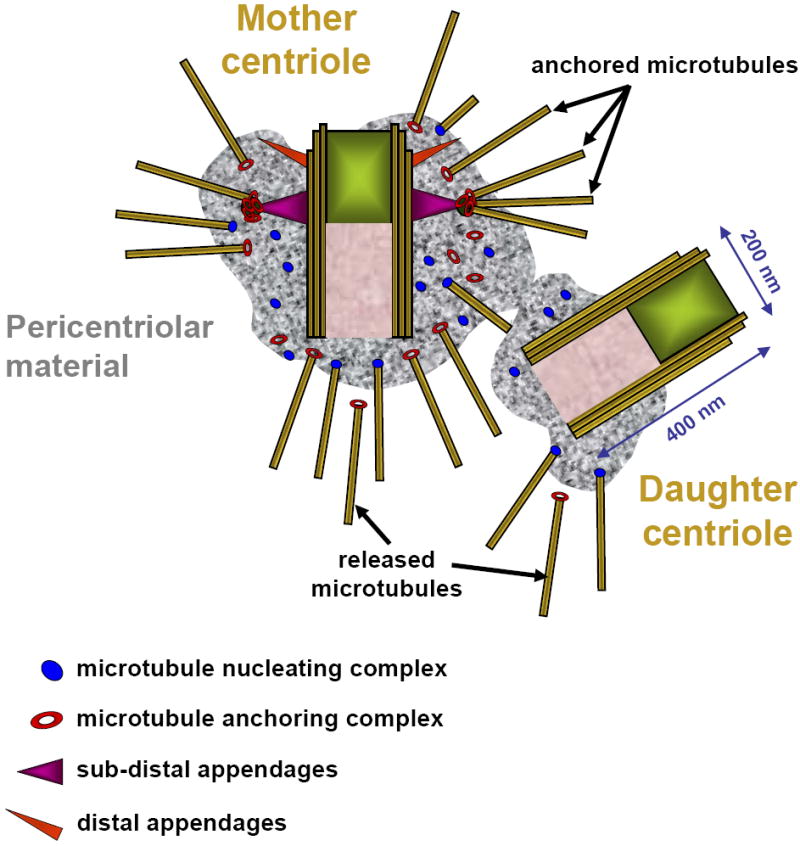
Two centrioles (mother and daughter) are embedded into clouds of the pericentriolar material (PCM). The walls of centrioles are built of nine microtubule blades. In higher animals, each blade is formed by a microtubule triplet. The mother centriole bears two rings of appendages attached to its wall. Subdistal appendages anchor microtubule minus ends, while distal appendages are needed to attach the centriole to the plasma membrane during ciliogenesis. The PCM contains a number of proteinaceous complexes that are responsible for microtubule nucleation and anchoring of microtubule minus ends. Notice that microtubule-anchoring complexes are only present in the PCM organized by the mother centriole. Microtubules nucleated in the PCM of the daughter centrioles are not anchored but released and rapidly escape the centrosome.
As the cell progresses from G1 into S phase, a procentriole, the structure that will ultimately develop into a new daughter centriole, assembles in the vicinity of each existing centriole adjacent to its proximal end (Fig. 2A). Procentrioles and the mother centriole are orthogonally arranged, with the lumen of the procentriole facing the wall of the mother (Chretien et al., 1997; Kuriyama and Borisy, 1981; Vorobjev and Chentsov, 1982). The functional significance of, as well as the molecular mechanisms responsible for, this stringent orientation remain unknown.
Fig. 2. Centriole propagation via duplication of preexisting centrioles.
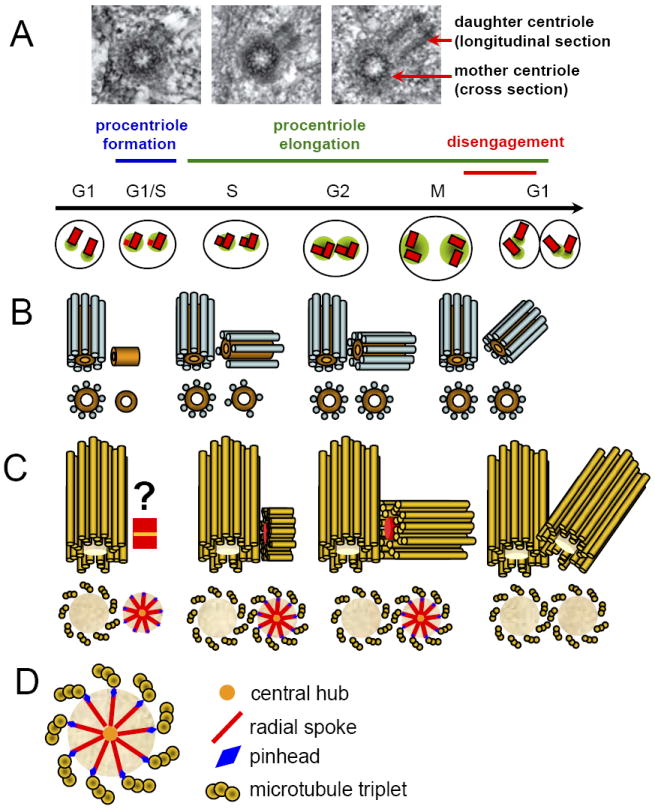
(A) At the beginning of the S phase, a procentriole assembles to the proximal end of the mother centriole. As the cell progresses through S and G2 phases, procentrioles centrioles elongate. At this time and through the first half of mitosis procentriole and the mother centriole maintain orthogonal orientation and the procentriole resides within the PCM cloud organized by the mother. (B) Schematics of procentriole formation in C. elegans. The earliest manifestation of centriole duplication is the formation of an amorphous central tube ~60 nm in length and orthogonal to the wall of the mother. The diameter and the length of the central tube increase as nine individual microtubules assemble around it. The central tube remains permanently embedded into the forming centriole a permanent structure of the centriole. (C) In higher animals, daughter centriole assembly begins with the formation of the ‘cartwheel’ (schematics in (D)). Microtubule blades subsequently assemble around the cartwheel. In some species the cartwheel can be detected inside fully matured centrioles, whereas in other organisms the cartwheel is only present during earlier stages of centriole assembly. (D) The cartwheel comprises nine radial spokes projecting from a central hub. Radial spokes are attached to the microtubules by structures called pinheads. Some studies suggest that the cartwheel serves as the organizer of the centriole symmetry.
Ultrastructural analyses of centrioles, especially more recent studies that used high-pressure freezing for sample fixation, have revealed details of the early stages in procentriole formation. In C. elegans, the earliest detectable structure in the process is an amorphous central tube ~ 60 nm in length oriented orthogonal to the wall of the mother centriole (Pelletier et al., 2006). The diameter and the length of the central tube subsequently increase as microtubules assemble around it (Fig. 2B). The central tube is a permanent structure that lines the inner wall of the centrioles, when mature. However, this tube has not yet been recognized in organisms other than C. elegans. Instead, in Paramecium and Chlamydomonas, the first morphological manifestation of centriole duplication is the appearance of an amorphous generative disc near to the proximal end of the mother centriole (Dippell, 1968; Dutcher, 2007). Whether this disc is functionally analogous to the central tube of C. elegans is not clear.
Another early structure that precedes formation of the typical procentriole is the cartwheel. The cartwheel is present in Drosophila, many protozoa, unicellular algae and in vertebrates. This structure, while possessing the nine-fold symmetry characteristic of the centriole, does not contain microtubules. Instead, the cartwheel comprises nine radial spokes projecting from a central hub (Fig. 2C,D). Several studies suggest that the cartwheel serves as the organizer of the centriole; the positions of the radial spokes, in particular, appears to determine the number and the positions of centriolar microtubules (Hiraki et al., 2007; Nakazawa et al., 2007). However, in Paramecium the cartwheel appears after the formation of the first microtubule blades around the generative disc; and thus, after the diameter and the symmetry of the procentriole would already have been established (Dippell, 1968). In some species the cartwheel can be detected inside all centrioles; however, in other organisms, it disappears from mature centriole (Alvey, 1985).
As the cell progresses through S and G2 phases, newly-formed daughter centrioles elongate until they reach the length of their mothers, usually during mitosis. The elongating daughter remain oriented orthogonal to the mother centriole and reside within the cloud of PCM formed by the mother (Fig. 2). Thus, at this stage, the mother and daughter centrioles are engaged in a structurally rigid complex that is commonly referred to as the ‘diplosome’. Early in mitosis, the distance between the mother and daughter centrioles within the diplosome increases (Chretien et al., 1997). Then, approximately at the onset of anaphase, the mother and daughter centrioles lose their orthogonal relationship (disengage) and the diplosome breaks down. From this point onward, each centriole is surrounded by its own cloud of PCM.
Cell-cycle regulators of centriole duplication
The strict synchronization between S phase and the centriole duplication process was recognized and described decades ago. However, the details of the molecular mechanisms responsible for such synchronization remain obscure. Formation of procentrioles coincides with the rise of Cdk2 activity at the beginning of the S phase, and experiments conducted in Xenopus laevis egg extracts, as well as in vertebrate cells in culture, revealed that Cdk2/cyclin E is necessary for centriole duplication (Hinchcliffe and Sluder, 2001a; Hinchcliffe and Sluder, 2001b). In addition, the Cdk2/cyclin A complex has been shown to be required for centriole reduplication in S phase-arrested CHO cells (Matsumoto et al., 1999; Matsumoto and Maller, 2004; Meraldi et al., 1999). Thus, the activity of Cdk2, whether in complex with cyclin A or cyclin E, appears to be a critical factor in centriole duplication. Three Cdk2 substrates have been proposed to be responsible for the regulation of centriole duplication. The first one, nucleophosmin (NPM/B23), was identified by Okuda and coworkers (2000). This protein is found only in the centrosomes that have unreplicated centrioles. Phosphorylation of nucleophosmin by Cdk2/cyclin E results in nucleophosmin’s removal from the centrosome, coinciding with initiation of procentriole formation. Antibodies that block nucleophosmin phosphorylation prevent centriole duplication (Okuda et al., 2000 ; Tokuyama et al., 2001). It is not known whether nucleophosmin can also be phosphorylated by the Cdk2/cyclin A complex.
Another centrosomal protein phosphorylated by both mitotic and interphase Cdk/cyclin complexes is CP110 (Chen et al., 2002). CP110 expression is cell-cycle regulated; the level of the protein increases at G1/S transition and remains high through the ensuing mitosis. Depletion of CP110 by siRNA prevented centriole reduplication in S-arrested U2-OS cells (Chen et al., 2002; Kleylein-Sohn et al., 2007).
The third Cdk2 substrate at the centrosome is the protein kinase Mps1, a conserved protein present in organisms from yeast to vertebrates. Mps1’s activity is essential in the spindle assembly checkpoint (Stucke et al., 2002). In yeast, Mps1 is necessary for duplication of the spindle pole body (Winey et al., 1991). However, its role in centriole formation in higher eukaryotes remains controversial. Centrosomal accumulation of Mps1 is shown to be indispensable for centriole duplication in mouse and human cells (Fisk et al., 2003; Fisk and Winey, 2001). Further, prevention of orderly degradation of Mps1 during S period has been shown to cause centriole reduplication in human cells (Kasbek et al., 2007). However, work from a different group (Stucke et al., 2002) did not confirm the involvement of Mps1 in centriole duplication.
Although data implicating Cdk2 activity in the regulation of centriole duplication are abundant, several pieces of evidence do not fit into the puzzle. Recent studies revealed that Cdk2, cyclin E1, and cyclin E2 are all dispensable for cell proliferation and centriole duplication in mice (Berthet et al., 2003; Geng et al., 2003). These findings imply that centriole duplication is governed by redundant mechanisms, and that the involvement of Cdk2, cyclin A, and cyclin E in this process, albeit important, may not be direct. It is feasible that Cdk2 activity is needed simply to create cytoplasmic conditions permissive for centriole duplication.
Centrosomal proteins
The process of centriole assembly follows a highly ordered and relatively conserved sequence of molecular events. Recent extensive siRNA screenings, as well as mutational analysis in C. elegans have identified core centrosomal proteins necessary for centriole duplication: SPD-2, ZYG-1, SAS-5, SAS-6, and SAS-4 (Delattre et al., 2006; Leidel et al., 2005; O’Connell et al., 2001; Pelletier et al., 2006). Functional homologues of these proteins are present in other organisms, with notable exception of SAS-5, which has been detected only in C. elegans thus far (reviewed in Strnad and Gonczy, 2008).
The player essential for initiation of centriole assembly in C. elegans is the coiled-coil protein SPD-2 (Kemp et al., 2004; Pelletier et al., 2004). SPD-2 associates with the centrosome throughout the cell cycle, and its level correlates with centrosome’s microtubule-organizing capacity. Depletion of SPD-2 from the cell by siRNA prevents centriole biogenesis, in C. elegans. The same has been shown for Cep192, the mammalian homologue of SPD-2 (Zhu et al., 2008). In contrast, DmSpd2, the Drosophila homologue of SPD-2, does not appear to play an essential role in centriole duplication, at least in somatic cells (Dix and Raff, 2007).
SDP-2 is required for centrosomal recruitment of the kinase ZYG-1. This kinase has functional analogues in humans (Plk4) and Drosophila (Sak), and in all organisms these proteins are indispensable for centriole duplication (Bettencourt-Dias et al., 2005; Bettencourt-Dias et al., 2005; Kleylein-Sohn et al., 2007; Peel et al., 2007). Activation of ZYG-1/Plk4/Sak on the centrosome is required for the sequential recruitment of the three essential coiled-coil proteins, SAS-5, SAS-6 (hsSAS-6 in humans, Bld12p in C. reinhardtii), and SAS-4 (CPAP/CENP-J in humans, DmSAS-4 in Drosophila).
Recent work has also ascertained two new proteins, CP110 and Cep135, as necessary for centriole formation in human cells (Kleylein-Sohn et al., 2007). Depletion of CP110 prevents both centriole formation (Kleylein-Sohn et al., 2007; Spektor et al., 2007) and centriole-to-basal body conversion (Bettencourt-Dias and Carvalho-Santos, 2008; Spektor et al., 2007). Finally, small calcium-binding centriolar proteins, centrins, were initially shown to be necessary for centriole duplication in human cells (Salisbury et al., 2002). However, more recent work (Kleylein-Sohn et al., 2007) co-depletion of two major centrin isoforms, centrin-2 and centrin-3, was not found not to affect centriole formation. The reason for the discrepancy is not clear. It is possible that the residual levels of the centrin proteins after siRNA depletion still allowed centriole duplication in the latter study. Further work is needed to resolve the centrin controversy.
Functions of individual centrosomal proteins during centriole formation
While the list of proteins involved in the formation of new centrioles is well established, significant work remains to be done, to elucidate the specific roles played by each component.
ZYG-1/Sak/Plk44 localizes to the centrosome near the time of centriole duplication. Its centrosomal activity is necessary for initiation of procentriole assembly in all organisms evaluated thus far (Delattre et al., 2006; O’Connell et al., 2001; Pelletier et al., 2006). In C. elegans, ZYG-1 localizes to the centrosome before the formation of the central tube (O’Connell et al., 2001; Pelletier et al., 2006). In human cells accumulation of Plk4 to the parental centrioles also occurs prior to initiation of daughter centriole assembly (Kleylein-Sohn et al., 2007). However, the mechanism of ZYG-1/Sak/Plk4 activation has not been identified, nor have the protein’s in vivo substrate(s) (see Bettencourt-Dias et al., 2005; Kleylein-Sohn et al., 2007; O’Connell et al., 2001). The cytoplasmic level of Plk4 is relatively low during G0 and G1, increases during S and G2, and peaks during mitosis. Plk4 has a short half-life of 2-3 h, presumably due to its extensive degradation via the ubiquitin-proteosome pathway (Winkles and Alberts, 2005). Sak and Plk4 continue to accumulate at the centrosome through S, G2 and mitosis, long after the initiation of formation of procentrioles (Bettencourt-Dias et al., 2005; Kleylein-Sohn et al., 2007). This accumulation of Plk4 could reflect some additional unexplained function of the protein in the centrosome.
The activity of ZYG1 is followed by the centrosomal recruitment of two coiled-coil proteins, SAS-5 and SAS-6. In C. elegans, both proteins are components of the centriolar central tube. In human cells, SAS-5 is not present, while SAS-6 accumulates locally in the proximal portion of centrioles, from their earliest stages until centriole disengagement in late mitosis (Kleylein-Sohn et al., 2007; Strnad et al., 2007). Mutations in Bld12p, the C. reinhardtii homologue of SAS-6, result in the absence of the central hub in the cartwheel, and in the formation of the basal bodies that are structurally defective (Nakazawa et al., 2007). Additionally, in primary spermatocytes of Drosophila, depletion of DmSAS-6 leads to the formation of centrioles with missing microtubule blades and reduced diameter (Rodrigues-Martins et al., 2007).
CEP135 is another essential centriolar protein that has been localized to the proximal portions of the nascent centrioles (Kleylein-Sohn et al., 2007). The Chlamydomonas homologue of CEP135, bld10p, is located at the pinheads of the cartwheel spokes (Hiraki et al., 2007). Mutations in Bld10p result in the formation of cartwheels of smaller and with shorter spokes, formation of centrioles with broken symmetry, and unstable microtubule triplets (Hiraki et al., 2007); for reviews see (Marshall, 2007; Strnad and Gonczy, 2008). Based on their localization to the proximal portions of centrioles, SAS-6/bld12p and CEP135/bls10p are proposed to participate in establishment of the nine-fold symmetry of the cartwheel-containing centrioles.
The addition of microtubules to the wall of centrioles depends on the action of SAS-4 (Pelletier et al., 2006). Nevertheless, how SAS-4 functions at the molecular level is not yet clear. Immuno-electron microscopic analysis of centrioles in C. elegans (Kirkham et al., 2003) localized SAS-4 to the centriole wall, whereas a similar study in human cells (Kleylein-Sohn et al., 2007) mapped CPAP to the centriolar lumen. While total depletion of SAS-4 or CPAP prevents centriole formation, partial depletion of SAS-4 in C. elegans revealed a correlation between centriole size and ability to recruit PCM, and residual levels of SAS-4. It is not clear whether SAS-4 directly dictates the size of PCM, or whether the impaired PCM recruitment is a result of the inability of defective centrioles to properly organize PCM around them. A recent study (Dammermann et al., 2008) suggests that stabilization of SAS-4 on newly formed centrioles in Drosophila depends on γ-tubulin-mediated formation of centriole microtubules.
The role of γ-tubulin as a key player in centriole formation has been documented in several studies (Dammermann et al., 2004; Kleylein-Sohn et al., 2007). Although γ-tubulin has been suggested to play a structural role in the centriole (Fuller et al., 1995; Moudjou et al., 1996), a more likely scenario is that this protein is required during centriole assembly because it nucleates centriolar microtubules. This view is indirectly supported by the demonstration that in Chlamydomonas (O’Toole et al., 2003; Silflow et al., 1999) and Drosophila (Moritz et al., 2000), γ-tubulin accumulates at the proximal region of the centriole and basal body where, together with other proteins, it forms a cap that protects the minus ends of centriolar microtubule blades from depolymerization. In turn, distal tips of nascent and mature centrioles also need to be protected to prevent normally-dynamic microtubule plus ends from both extensive growth and depolymerization. Immuno-electron microscopy has revealed that the distal tips of nascent and mature centrioles are rich in CP110. This protein is recruited to the distal end of daughter centrioles at the earliest stages of their formation. Further, depletion of CP110 via siRNA prevents growth of daughter centrioles. These data suggest that CP110 serves as the protective cap for the centriole microtubules (Kleylein-Sohn et al., 2007). It is interesting to note that, although fusion proteins of GFP and centrin-1 and centrin-2 are widely used to label the distal ends of the centrioles in live-cell studies, the function of centrin proteins at the centriole are still unknown.
Mechanisms controlling the number of procentrioles
The accuracy of centriole duplication in somatic cells is remarkable: only one daughter centriole forms per mother centriole, and each mother duplicates only once per cell cycle. We are just beginning to understand the mechanisms that prevent centriole reduplication, and that control the formation of only one daughter centriole in the vicinity of each radially-symmetrical mother centriole.
One centriole cycle per cell cycle
In most cell types, centriole reduplication does not occur when progression through the cycle is blocked so that the cell remains arrested in S phase. Thus, a mechanism exists that prevents formation of additional procentrioles once the first one has assembled. The nature of this mechanism is beginning to emerge.
Apparent similarity between the processes of centriole duplication and DNA replication (both are semi-conservative) as well as their strict temporal coordination suggest that the reduplication of both DNA and centrioles is prevented by an analogous mechanism. DNA reduplication is known to be prevented by a temporal separation between the formation of the pre-replication complexes (“licensing” event) at the end of mitosis and in early G1, and the firing of replication origins in S phase (Blow and Dutta, 2005; Hook et al., 2007). Whether a similar temporal regulation governs centriole duplication was experimentally tested by Wong and Sterns (Wong and Stearns, 2003). They demonstrated that in hybrid cells obtained by fusion of G1 cells with S cells or G1 cells with G2 cells, centrioles from the G1 cells readily duplicate when the hybrid cell enters S phase. In contrast, already-duplicated centrioles from the G2 cells cannot reduplicate even though they reside within the same cytoplasm. Thus, the block to centriole reduplication is intrinsic to already-duplicated centrioles that are engaged in the diplosome. These elegant experiments lead to the formulation of “block-and-license” hypothesis of centriole duplication (Tsou and Stearns, 2006a; Nigg, 2007). In this view, attachment of the daughter centriole to the mother prevents formation of additional daughters in spite of permisive cytoplasmic conditions that exist throughout S phase. Breakdown of the diplosome during mitosis licenses the centrioles for a new round of duplication; however initiation of duplication cannot occur until cytoplasmic conditions become permissive again in the ensuing S phase. Direct support for this model was obtained from laser ablation experiments on S-phase arrested HeLa cells. These cells possess a stringent block to centriole reduplication (Balczon et al., 1995; Loncarek et al., 2008). However, if a daughter centriole is removed from the diplosome of an l by laser micro-irradiation, the mother centriole initiates a new round of centriole duplication (Loncarek et al., 2008). This clearly demonstrates that the capacity of the mother centriole to duplicate is based solely on whether or not it is engaged. Even artificial removal of the daughter centriole from the diplosome is sufficient to induce formation of a new daughter, as long as cytoplasmic conditions remain permissive.
Molecular mechanisms responsible for centriole disengagement are poorly understood. It has been suggested that natural centriole disengagement results from the proteolysis of a proteinaceous link between mother and daughter centrioles at the time of the metaphase-anaphase transition (Tsou and Stearns, 2006b). This cleavage may be mediated by separase, the enzyme responsible for sister chromatid separation. Separase activity is inhibited during S, G2, and the first part of mitosis. At the metaphase-anaphase transition, separase becomes activated via by the anaphase-promoting complex/cyclosome (APC/C). The activated separase cleaves cohesin, the protein that glues two sister chromatids together; the cleavage then allows the chromatids to separate. This separase hypothesis, although attractive, is at odds with the observation that centrioles can disengage under conditions when separase should not be globally active. For instance, centrioles have been seen to disengage during prolonged mitotic arrest in several systems (Mazia, 1960; Keryer et al., 1984; Sluder and Begg, 1985). During ciliogenesis, numerous daughter centrioles disengage from their mothers within the interphase during which they were formed and they then move to the cell surface (Dawe et al., 2007; Dirksen, 1991). In addition, in cell line CHO, where centrioles reduplicate during prolonged S-phase-arrest induced by hydroxyurea (Balczon et al., 1995; Loncarek et al., 2008), initially duplicated centrioles disengage before a new duplication cycle is initiated within the same interphase (Loncarek et al., 2008). Moreover, in S-phase-arrested cells, individual diplosomes disengage asynchronously, indicating that the engagement/disengagement state is intrinsic to the individual centrosome. A possibility that separase can exert its activity locally (rather than cell-wide) cannot be ruled out in the examples listed above; nevertheless we need to account for these findings. It is also noteworthy that natural disengagement can be mimicked via physical removal of the daughter centriole from the diplosome. These data imply that engagement is not based on specific proteinaceous link between the centrioles. Instead, the centrosome somehow “senses” the presence of an immature centriole within the PCM cloud. It is a challenge for the future, for us to understand how diplosome configuration translates into the regulatory signal, and which components of the centrosome play active roles in the process.
While one centriolar cycle per DNA cycle is the general rule for centriole propagation, there are examples of specialized cell types in various organisms, where the two cycles are not strictly coordinated. For instance, several consecutive rounds of DNA replication can occur in Drosophila endoreduplicating follicle cells that have their centriolar cycles attenuated, or even their centrioles lost, despite high Cdk2/cyclin E activity (Mahowald et al., 1979). Further, in most animal species, unfertilized eggs lack centrioles, and either one or two centrioles are delivered into the egg via fertilization by the sperm. There in the egg, centrioles recruit pericentriolar components abundantly present in the cytoplasm, and they reconstitute the first centrosome(s) (Delattre and Gonczy, 2004; Manandhar et al., 2005). As a consequence of asymmetrical centriole inheritance, centrioles often have to be duplicated independently of chromosome duplication, during first mitotic divisions of the zygote, in order to compensate for the improper centrosome/chromosome ratio inherited via fertilization (Manandhar et al., 2005). The mechanisms controlling the centriolar and nuclear cycles during these special cell cycle programs are as yet unknown. Repetitive centriolar cycles within the single cell cycle can also be experimentally induced in embryos and somatic brain cells of Drosophila, upon overexpression of Sak or SAS-6 (Peel et al., 2007c; Rodrigues-Martins et al., 2007b). It is not clear how overexpression of either of these proteins decouple the centriolar cycle from the nuclear cycle. Multiple rounds of centriole duplication have been also observed in human tumor cells after expression of the viral E7 oncogene (Duensing and Munger, 2003).
One daughter centriole per mother centriole
Given the perfectly nine-fold symmetry of the centriole, it seems counterintuitive that only one daughter centrioles forms near the wall of the mother. One explanation for this symmetry-breaking mechanism is that centrioles carry an undetected intrinsic asymmetry, or a site at/or close to the wall of the mother centriole that determines the birth place of the future daughter (Jones and Winey, 2006; Tsou and Stearns, 2006a). This hypothesis is compatible with the idea of an intrinsic block to centriole duplication; as long as the site is occupied, the new daughter centriole cannot form on the same mother centriole. However, the hypothesis is not consistent with several observations outlined below.
The one-mother-one-daughter rule does not apply to certain modes of basal body formation. For example, during ciliogenesis in tracheal cells, up to 9 daughter centrioles can simultaneously form around one mother (Dawe et al., 2007; Dirksen, 1991). An analysis of centrioles in reduplication during S-phase arrest of CHO cells has also shown that one mother centriole can form more than one daughter. Further, the number of daughters forming on the same mother varies in each round of reduplication (Loncarek et al., 2008). Thus, daughter centrioles can be formed at more than one specific site near the wall of the mother.
It is possible that numerical control of procentriole formation is based on a finely balanced stoichiometry among the centrosomal proteins. Indeed, formation of multiple daughter centrioles on a single mother can be induced by overexpression of Plk4, in cycling or S- phase-arrested U2OS cells (Kleylein-Sohn et al., 2007). Under these conditions daughter centrioles are organized in a flower-petal configuration forming a ‘rosette’. Formation of centriolar rosettes also occurs in HeLa or U2-OS cells upon overexpression of SAS-6 (Duensing et al., 2007; Strnad et al., 2007). These observation seem to create a paradox because overexpression of either Plk4 (considered to be regulatory protein) and SAS-6 (a structural protein) forces cells to similarly loose the numerical control of centriole duplication. It is noteworthy, that expression level of SAS-6 should not be directly affected by overexpression of Plk4 and vice versa. Thus, endogenous levels of either of these two proteins are sufficient to support simultaneous formation of multiple daughter centrioles. Yet, under normal circumstances one and only one daughter forms. Interestingly, overexpression the PCM protein, pericentrin (Young et al., 2000), in S-phase-arrested CHO cells increases the size of the PCM that surrounds the mother centriole. This condition also results in the simultaneous formation of numerous daughter centrioles Loncarek et al., 2008). Pericentrin is not directly involved in the formation of daughter centrioles, and depletion of pericentrin does not block centriole duplication. Thus, formation of multiple daughters in cells overexpressing pericentrin is likely to be an indirect consequence of the PCM enlargement. If so, the size of the PCM cloud surrounding the mother centriole would appear to control the number of centrioles formed within the centrosome (Loncarek et al., 2008). Intriguingly, daughter centrioles formed inside the enlarged PCM cloud are not oriented orthogonal to their mother centriole, nor is their distance from the mother centriole constant. Perhaps, then, centriole orientation within the diplosome is due to PCM constrains instead of direct connections between centrioles (Loncarek et al., 2008).
De novo - centriole assembly in the absence of maternal organelles
While duplication of mother centrioles represents the most common pathway for centriole propagation, it is not the only one. Centrioles can also form in the absence of any preexisting centrioles. This de novo mode of centriole assembly is widely utilized in various organisms.
An intriguing feature of organism development in higher animals is that oocytes in most species lack centrioles. Upon fertilization the sperm introduces paternal centrioles that subsequently duplicates in the zygote (Manandhar et al., 2005). Alternatively, in a number of species both maternal and paternal centrioles are lost. For instance, the mouse zygote initially does not contain centrioles, and fist embryonic divisions are acentrosomal. Then, during the blastomere stage of development (16 to 32 cells), each cell mysteriously assembles centrioles in the proper number; these centrioles continue to segregate and duplicate by the canonical pathway thereafter (Szollosy et al., 1972). Almost nothing is known about molecular mechanisms responsible for this type of centriole de novo assembly. However, it is clear that this pathway is ubiquitous as centrioles can assemble de novo during parthenogenesis even in those species where centrioles are normally contributed by the sperm (Manandhar et al., 2005; Riparbelli and Callaini, 2003; Szillosi and Ozil, 1991). Further, hundreds of centrioles can form in a differentiated ciliated epithelial cells around amorphous EM-dense granules composed of various centrosomal proteins (Dawe et al., 2007; Vladar and Stearns, 2007). These granules eventually fuse into the larger structures called deuterosomes (Sorokin, 1968).
The de novo centriole assembly can be activated in a cycling vertebrate somatic cell if all resident centrioles are removed by microsurgery or ablated by a laser microbeam (Khodjakov et al., 2002; La Terra et al., 2005; Uetake et al., 2007). Interestingly, after activation of the de novo pathway in a somatic cell, the numerical control over centriole formation is lost, resulting in a variable number of centrioles.
EM analysis of de-novo formed centrioles in somatic cells revealed various intermediate stages, ranging from electron-dense amorphous clouds associated with sporadic microtubule blades, to apparently morphologically complete centrioles (Khodjakov et al., 2002; La Terra et al., 2005). De novo centriole assembly in somatic cells occurs throughout the cytoplasm, and the forming centrioles do not appear to be associated with any structures resembling dense granules or deuterosomes. Whether this reflects a difference in the de novo pathway between ciliated epithelia and cultured somatic cells, is not yet clear.
In general, the de novo centriole assembly pathway is far less well documented than is the centriole duplication pathway, especially when the de novo one has been induced in somatic cycling cells. However, the induced de novo pathway is an excellent model for use in centriole biogenesis studies, since the events and components important specifically for centriole formation are not obscured by other centrosomal functions present during centriole duplication.
How does the cell decide: ab ovo or de novo?
Lack of numerical control during de novo centriole assembly would have deleterious consequences for the cell, given that supernumerary centrioles often form multipolar mitotic spindles. Fortunately, the de novo mode of centriole assembly is suppressed in cycling somatic cells as long as a single mature centriole is present in the cytoplasm (La Terra et al., 2005; Uetake et al., 2007). This raises the interesting question of how an individual cell “decides” which pathway to employ, and how a cells switches between the two modes of centriole assembly. As different as they seem, centriole duplication and the de novo pathway display common molecular requirements. If entry into S-phase is blocked, the de novo assembly is prevented (La Terra et al., 2005a; Uetake et al., 2007b), suggesting that the two pathways require the same cytoplasmic conditions. The same proteins that are essential for canonical centriole duplication, such as Sak/Plk4, SAS-6 or SAS-4/CPAP, are also essential for de novo centriole assembly, in Drosophila and in mammals (Bettencourt-Dias et al., 2005; Peel et al., 2007; Rodrigues-Martins et al., 2007; Vladar and Stearns, 2007). We do not yet know which component of the centrosome - the centriole itself, or its surrounding PCM is responsible for the suppression of the de novo pathway. It is noteworthy that even strong overexpression of centriolar proteins such as Sak or SAS-6 do not activate the de novo pathway in centriole-containing somatic cells or embryos (Peel et al., 2007; Rodrigues-Martins et al., 2007). Instead, overabundance of these proteins results in the formation of multiple daughter centrioles on the same mother or repetitive re-duplication of mother centrioles in the same cell cycle (Peel et al., 2007; Rodrigues-Martins et al., 2007, Duensing et al., 2007). At the same time, physical removal of centrioles from the cell induces de novo assembly of numerous centrioles in spite the low amounts of Plk4 and SAS-6. Currently we can only speculate on how the presence of even a single centrosome within the cytoplasm suffices to prevent activation of the de novo pathway.
Centrosomes may actively participate in the attenuation of the de novo pathway by maintaining a well-defined cloud of PCM, where critical protein-protein interactions or protein activations needed for initiation of procentriole formation are promoted. In this scenario, an procentriole formation would be restricted to the vicinity of the existing centrioles. It has been shown that small aggregates of centrosomal proteins that can self-assemble the cytoplasm are actively transported towards and incorporated into the centrosome via dynein-mediated transport on microtubules (Young et al., 2000). This mechanism would prevent spontaneous de novo assembly of centrioles as long as a resident centrosome is present in the cell. In contrast, removing the centrosome would affect microtubule organization and allow irregular self-assembled PCM clouds to develop into centers for centriole formation. However, in lieu of experimental support this scenario remains a rampant speculation.
Mechanistic similarities between the two modes of centriole formation render the distinction between them fuzzy and it is plausible that the two modes are alternative manifestations of the same process. Is canonical centriole duplication a spatially-restricted and more stringently controlled version of centriole de novo assembly? If centriole duplication provides far more precise control of centriole numbers, then why do so many organisms employ centriole de novo formation, in such critical events as early embryogenesis? It is fascinating that, whenever centriole de novo formation occurs naturally during development or differentiation, the process is always fine tuned, and it results in a precisely correct number of centrioles. In contrast, in all examples of experimentally induced centriole de novo assembly numerical control over the centrioles appears to be lost.
Numerous studies conducted during the last century have brought us to a better understanding of centriole biology. But much remains to be discovered in the decipherment of this complex and mysterious organelle.
References
- 1.Alvey PL. An investigation of the centriole cycle using 3T3 and CHO cells. J Cell Sci. 1985;78:147–162. doi: 10.1242/jcs.78.1.147. [DOI] [PubMed] [Google Scholar]
- 2.Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 Knockout mice are viable. Cur Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 4.Bettencourt-Dias M, Carvalho-Santos Z. Double life of centrioles: CP110 in the spotlight. Trends in Cell Biol. 2008;18:8–11. doi: 10.1016/j.tcb.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. Sak/Plk4 is required for centriole duplication and flagella development. Cur Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 6.Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- 7.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent Cdk substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 10.Chretien D, Buendia B, Fuller SD, Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. J Struct Biol. 1997;120:117–133. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- 11.Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 14.Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Cur Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 15.Delattre M, Gonczy P. The arithmetic of centrosome biogenesis. J Cell Sci. 2004;117:1619–1630. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- 16.Dippell R. The development of basal bodies in Paramecium. PNAS. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirksen ER. Centriole and basal body formation during ciliogenesis revisited. Biol Cell. 1991;72:31–38. doi: 10.1016/0248-4900(91)90075-x. [DOI] [PubMed] [Google Scholar]
- 18.Dix CI, Raff JW. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Cur Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duensing S, Munger K. Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J Virol. 2003;77:12331–12335. doi: 10.1128/JVI.77.22.12331-12335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duensing A, Liu Y, Perdreau SA, Kleylein-Sohn J, Nigg EA, Duensing S. Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene. 2007;26:6280–6288. doi: 10.1038/sj.onc.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutcher SK. Finding treasures in frozen cells: new centriole intermediates. Bioessays. 2007;29:630–634. doi: 10.1002/bies.20594. [DOI] [PubMed] [Google Scholar]
- 22.Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. PNAS. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 24.Fuller SD, Gowen BE, Reinsch S, Sawyer A, Buendia B, Wepf R, Karsenti E. The core of the mammalian centriole contains γ-tubulin. Cur Biol. 1995;5:1384–1393. doi: 10.1016/s0960-9822(95)00276-4. [DOI] [PubMed] [Google Scholar]
- 25.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 26.Hinchcliffe EH, Sluder G. It takes two to tango: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001a;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- 27.Hinchcliffe EH, Sluder G. Centrosome duplication: Three kinases come up a winner! Cur Biol. 2001b;11:R698–R701. doi: 10.1016/s0960-9822(01)00412-2. [DOI] [PubMed] [Google Scholar]
- 28.Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Cur Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 29.Hook SS, Lin JJ, Dutta A. Mechanisms to control re-replication and implications for cancer. Cur Op in Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones MH, Winey M. Centrosome duplication: is asymmetry the clue? Cur Biol. 2006;16:R808–810. doi: 10.1016/j.cub.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 31.Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of Mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 33.Keryer G, Ris H, Borisy GG. Centriole distribution during tripolar mitosis in Chinese hamster ovary cells. J Cell Biol. 1984;98:2222–2229. doi: 10.1083/jcb.98.6.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkham M, Müller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 Is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 36.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuriyama R, Borisy GG. Cytasters induced within unfertilized sea-urchin eggs. J Cell Sci. 1983;61:175–189. doi: 10.1242/jcs.61.1.175. [DOI] [PubMed] [Google Scholar]
- 39.La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 41.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahowald AP, Caulton JH, Edwards MK, Floyd AD. Loss of centrioles and polyploidization in follicle cells of Drosophila melanogaster. Exp Cell Res. 1979;118:404–410. doi: 10.1016/0014-4827(79)90167-8. [DOI] [PubMed] [Google Scholar]
- 43.Manandhar G, Schatten H, Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol Reprod. 2005;72:2–13. doi: 10.1095/biolreprod.104.031245. [DOI] [PubMed] [Google Scholar]
- 44.Marshall WF. Centriole Assembly: The origin of nine-ness. Cur Biol. 2007;17:R1057–R1059. doi: 10.1016/j.cub.2007.10.038. [DOI] [PubMed] [Google Scholar]
- 45.Marshall WF. The cell biological basis of ciliary disease. J Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Cur Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto Y, Maller JL. A centrosomal localization signal in cyclin E required for cdk2-independent S phase entry. Science. 2004;306:885–888. doi: 10.1126/science.1103544. [DOI] [PubMed] [Google Scholar]
- 48.Mazia D. The multiplicity of the mitotic centers and the time-course of their duplication and separation. Biophys Biochem Cytol. 1987;7:1–20. doi: 10.1083/jcb.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 50.Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- 51.Moudjou M, Bordes N, Paintrand M, Bornens M. γ-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109:875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- 52.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Cur Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 53.Nigg EA. Centriole duplication: of rules and licenses. Trends in Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 54.O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans Zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- 55.O’Toole ET, Giddings TH, McIntosh JR, Dutcher SK. Three-dimensional organization of basal bodies from wild-type and δ-tubulin deletion strains of Chlamydomonas reinhardtii. Mol Biol Cell. 2003;14:2999–3012. doi: 10.1091/mbc.E02-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, et al. Nucleophosmin/B23 is a target of CDK2/Cyclin E in centrosome duplication. Cell. 2000b;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 57.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Cur Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pelletier L, ’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 59.Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Cur Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 60.Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 61.Riparbelli MG, Callaini G. Drosophila parthenogenesis: a model for de novo centrosome assembly. Dev Biol. 2003;260:298–313. doi: 10.1016/s0012-1606(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 62.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 63.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 Organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Cur Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 64.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Cur Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 65.Silflow CD, Liu B, LaVoie M, Richardson EA, Palevitz BA. γ-Tubulin in Chlamydomonas: characterization of the gene and localization of the gene product in cells. Cell Mot & Cyoskel. 1999;42:285–297. doi: 10.1002/(SICI)1097-0169(1999)42:4<285::AID-CM3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 66.Sluder G, Begg DA. Experimental analysis of the reproduction of spindle poles. J Cell Sci. 1985;76:35–51. doi: 10.1242/jcs.76.1.35. [DOI] [PubMed] [Google Scholar]
- 67.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 68.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 Suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 69.Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends in Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 70.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szollosy D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- 73.Szollosi D, Ozil JP. De novo formation of centrioles in parthenogenetically activated, diploidized rabbit embryos. Biol Cell. 1991;72:61–66. doi: 10.1016/0248-4900(91)90079-3. [DOI] [PubMed] [Google Scholar]
- 74.Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr199 by cyclin- dependent kinase 2-cyclin E and its role in centrosome duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- 75.Tsou MFB, Stearns T. Controlling centrosome number: licenses and blocks. Cur Op in Cell Biol. 2006a;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 76.Tsou MFB, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006b;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 77.Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vorobjev IA, Chentsov Y. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winey M, Goetsch L, Baum P, Byers B. Mps1 and Mps2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winkles JA, Alberts GF. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene. 2005;24:260–266. doi: 10.1038/sj.onc.1208219. [DOI] [PubMed] [Google Scholar]
- 82.Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- 83.Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ. Cytoplasmic dynein-mediated assembly of pericentrin and γ-tubulin onto centrosomes. Mol Biol Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, Pelletier L. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Cur Biol. 2008;18:136–141. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]


