Abstract
Histone-methyl transferases (HMTs) are key enzymes that post-translationally modify histones, and serve key role in gene expression, epigenetic regulation, and as determinants of survival in malignant cells. Recent studies have shed light on the role of hSET1 which is a key element of highly conserved multi-protein HMT complex that catalyze methylation of histone H3 lysine 4 (H3K4) regulating expression of specific proteins important for the malignant phenotype. To understand the importance of differential expression of H3K4 HMTs in cancer, we specifically down-regulated hSET1 the only H3K4 specific histone methyl transferase present in yeast as well as in human that is directly involved in gene expression. hSET1 has been shown to be differentially over-expressed in the malignant cells as compared to the normal cells at the RNA as well as protein level. In a wide array of normal and malignant cells it has been demonstrated that phosphorothioate antisense against hSET1 (DN5) caused selective and differential apoptosis in malignant cells only while the normal cells remains unaffected. Downregulation of hSET1 leads to rapid and complete regression of SW480 colon xenograft in mice model. These findings demonstrate that hSET1 over-expression promotes cell proliferation and cancer cell survival, and may be a novel target for cancer therapy.
Keywords: cancer, xenografts, histone methyl transferase, hSET1, epigenetic regulation
1. Introduction
In the recent years, cancer is recognized as a genetic and epigenetic disease and great effort has been directed towards understanding the role of establishment and relevance of aberrant epigenetic patterns in human tumors [1–5]. Post-translational modifications of histones are recognized as having a primary role in the control of gene expression and chromatin structure. Among the modifications, histone lysine methylation is considered to be critical for transcriptional regulation and methylation of histone 3 at position lysine 4 (H3K4) is known to be associated with the epigenetic control of heterochromatin assembly and with DNA methylation-induced gene silencing in cancer cells [6–10]. In particular, H3K4 trimethylation is associated with promoters and early transcribed regions of active genes [6,7]. There are several known H3K4 specific histone methyl transferases such as MLL1 (mixed lineage leukemia 1), MLL2, MLL3 and hSET1 (human SET1) in mammalian cells which are shown to be present as distinct multi-protein complexes with several highly conserved protein complexes and methylate H3K4. Amongst them SET1 is the only H3K4 specific histone methyl-transferase which is present in yeast as well as human and is directly involved in gene expression [11]. Yeast SET1 is a multi-protein complex (also known as COMPASS) containing subunits SET1, Bre2, Spp1, Swd1, Swd2, Swd3, and Sdc1 and its HMT activity is fully active only in the context of a multi-subunit complex [11–14]. Recently, it has also been demonstrated that SET1, in yeast, is required for methylation of conserved lysines in a kinetochore protein, Dam1, suggesting its important role in mitosis [15]. SET1 mediated histone methylation is functionally coupled with histone H2B monoubiquitination, Paf1 complex and other factors involved in transcriptional regulation [16,17].
Though the importance of above histone methyl-transferases in determining the malignant phenotype has been recognized as a phenomenon, the nature and significance of differential patterns of histone methylation, their biochemical roles in regulating gene expression and the mechanism through which these HMTs determine differential gene expression in cancer cells is largely unknown [18,19]. Recently, MLL1, MLL2 and hSET1 enzyme complexes has been purified from human cells and demonstrated that human CGBP (CpG binding protein, also known as CFP1) which is a well characterized protein containing a DNA binding “CXXC” finger domain which binds with unmethylated CpG dinucleotides and plays a critical role in gene expression and mammalian development, is a component of all of the above three HMT complexes [20]. These HMTs (MLL1, MLL2 and hSET1) are also shown to be physically interact with RNA polymerase II (RNAP II) suggesting their direct roles in transcription and possibly in mRNA processing, although, their exact roles in transcription are yet to be understood [20,21–23].
To understand the importance of differential expression of H3K4 HMTs in cancer, we specifically down-regulated hSET1 which is a key element of highly conserved multi-protein HMT complex that catalyze methylation of histone H3 lysine 4 (H3K4) using a phosphorothioate antisense (DN5) in cell culture models of malignant and non-malignant cells and extended these studies in-vivo using a SW480 colon cancer xenograft model. Suppression of hSET1 was found to cause tumor-selective apoptosis in cultured cells and complete sustained remission of subcutaneously implanted colon cancer xenografts by hSET1 depletion. hSET1 has been shown to be differentially over-expressed in a series of cultured malignant cells as compared to the normal cells at the RNA as well as protein level.
2. Materials and methods
2.1. Materials
RPMI-1640, Ham’s F12 K and DMEM medium, McCoy’s 5a medium, PBS, trypsin-EDTA, penicillin/streptomycin solution (P/S), fetal bovine serum (FBS), and trypan blue were purchased from Life Technologies, Inc.(Grand Island, NY). Medium EGM-2 Bullet Kit and MEGM Bullet kit was purchased from Cambrex BioScience (Walkersville, MD). Reagents for SDS-PAGE were purchased from Bio-Rad Laboratories (Hercules, CA). Polyclonal rabbit anti-hSET1 IgG was purchased from Bethyl Laboratories (Montgomery, TX). Rhodamine-x and FITC-conjugated goat anti-rabbit antibodies were purchased from Vector Laboratories Inc. (Burlingame, CA). RNeasy kit for RNA purification and one step RT-PCR kits were purchased from Qiagen (Valencia, CA). Fluorescein terminal deoxynucleotidyl transferase (TdT)-mediated nick end labeling (TUNEL) apoptosis assay kit was procured from Promega (Madison, WI). Maxfect transfection reagent was from MoleculA (Columbia, MD). Dimethylsulphoxide (DMSO) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetra-zolium bromide (MTT) were obtained from sigma (St. Louis, MO). Gene specific primers, phosphorothioate oligonucleotides against hSET1 and scrambled antisense were purchased from Biosynthesis Inc. (Lewisville, TX) and the sources of other chemicals used in this study were same as described previously [24].
2.2. Cell lines and cultures
Human colon cancer cell line (SW480 and HT29), small cell lung cancer (SCLC) lines H1618, and non-small cell lung cancer (NSCLC) lines H358 (bronchio-alveolar), H226 (squamous cell carcinoma), H2347 (adenocarcinoma), PC-3 (human prostate), MCF10a (breast epithelial), MDA-MB231 (breast adenocarcinoma) and K562 (human erythroleukemia) cell line were purchased from American Type Culture Collection (Manassas, VA). Human aortic vascular smooth muscle cells (HAVSMC) and human breast cancer (MCF7 and MCF7-VP) cells were kindly donated by Dr. Paul Boor, University of Texas Medical Branch at Galveston, Galveston, TX. Human umbilical vascular endothelial cells (HUVEC) were from Dr. Fiemu Nwariaku (UTSWMC, Dallas, TX). Human ovary carcinoma OVCAR-3 and SKOV-3 cells were kindly provided by Dr. Andras Lacko, University of North Texas Health Science Center, Fort Worth, TX. All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2 in the appropriate medium: RPMI-1640 (SCLC, NSCLC, K562, SW480, HT29, MDA-MB231 and MCF7), DMEM (HAVSMC, MCF7-VP), RPMI-1640 supplemented with 0.01 mg/ml bovine insulin (OVCAR-3), EGM-2 bullet kit (HUVEC), MEGM bullet kit (MCF10a), McCoy’s 5a (SKOV-3), Ham’s F12 K (PC-3) medium supplemented with 10% (v/v) heat-inactivated FBS and 1% (v/v) P/S solution.
2.3 Expression of hSET1 in cultured cells
The expression of hSET1 was determined in crude homogenates of malignant (H358, OVCAR-3, MCF7, PC-3, K562 and SW480) and non-malignant cells (HUVEC and HAVSMC) by Western blot analysis as described previously [24]. Briefly, cells were pelleted and washed with balanced salt solution (138 mM NaCl, 5 mM KCl, 0.3 mM KH2PO4, 0.3 mM Na2HPO4, 4 mM NaHCO3, and 5.6 mM glucose, pH 7.4) three times and were lysed in 10 mM Tris-HCl, pH 7.4, containing 1.4 mM β-mercaptoethanol (BME), 0.1mM phenylmethanesulphonylfluoride (PMSF), 0.05 mM butylated hydroxytoluene (BHT), 0.1% polidocanol and 0.1mM EDTA (lysis buffer). Lysates were sonicated three times for 30 sec at 50W and the resultant preparations were centrifuged at 100,000-x g for one hour. Total protein was quantified by the method of Minamide and Bamburg [25]. 200 μg total crude proteins were used for SDS-PAGE and Western blotting, using rabbit anti-human SET1 polyclonal antibodies. Peroxidase conjugated goat-anti-rabbit IgG was used as a secondary antibody. The blots were developed using 4-chloro-1-naphthol as chromogenic substrate. β-actin was used as an internal control.
Expression of hSET1 was evaluated at the RNA level by RT-PCR analysis. The RNA was prepared by RNeasy kit (Qiagen Inc.), quantified and purity was determined by measuring optical density at 260 and 280 nm. hSET1 gene-specific primers [1422–1439 bp (upstream) 5′: GACACAAGCTTCTCCAGCA and 1602–1621 (downstream) 5′: TGAAGATGCAGAGAAGT-GGC, spanning 1422 to 1621 in open reading frame of hSET1 (KIAA0339) gene were used. RT-PCR was performed using the one step RT-PCR kit according to the manufacturer’s instructions (Qiagen Inc.). Bands were quantified by scanning densitometry using Innotech Alpha Imager HP.
2.4. hSET1 phosphorothioate antisense (DN5) preparation
The region spanning nucleotides 1420–1640 starting from 1 AUG codon in the open reading frame) in the N-terminal region of hSET1 was chosen as the target region for synthesis of phosphorothioate antisense. The oxygen in the backbone of the DNA molecules was replaced by sulfur in each phosphate group, which makes the DNA backbone resistant to nucleases. The selected DNA sequence was subjected to BLAST search (National Center for Biotechnology Information database) against expressed sequence tag libraries to ensure that only the selected gene was targeted. Chemically synthesized phosphorothioate DNA in desalted form was purchased from Biosynthesis, Inc. (Lewisville, TX). A 21-nucleotide long scrambled phosphorothioate DNA was used as a control. The scrambled DNA sequence was not homologous with hSet1 cDNA in a BLAST search against hSET1. The targeted cDNA sequence (ATCCCCCAAGGAACCCT) corresponds to nt 1488–1504. The corresponding phosphorothioated DNA sequence was AAGGGGGTTCCTTGGGA. The sequence of the scrambled DNA was CATCGAAATCGTTGCAGTTAC.
2.5. Transfection of hSET1 phosphorothioate DNA in cultured cells
Cells were transfected with hSET1 phosphorothioate DNA (DN5) using Maxfect transfection reagent (MoleculA) following manufactures instructions. Depletion of hSET1 expression in cells by hSET1 antisense was measured as follows: cells were incubated for 6 h with 0–10 μg/ml antisense in Maxfect transfection reagent (MoleculA), according to the manufacturer provided protocol. Levels of hSET1 protein in control and transfected clones were measured in normal (HAVSMC) and malignant (SW480) cells by immunoassay using anti-hSET1 IgG after 24 hours. The time dependent effect of hSET1 antisense (fixed 10 μg/ml final conc.) was also evaluated using three malignant (H358, PC-3 and MCF7) and two non-malignant cells (HUVEC and MCF10a) at 0, 12, 24 and 48 h after treatment with antisense for hSET1 by Western blot analysis. 200 μg crude proteins were used for SDS-PAGE and Western blotting, using rabbit-anti-human SET1 polyclonal antibodies. Peroxidase conjugated goat-anti-rabbit IgG was used as a secondary antibody. The blots were developed using 4-chloro-1-naphthol as chromogenic substrate. β-actin was used as an internal control.
2.6. Cell survival (MTT) assay
Cell number/ml in an aliquot of cells growing in log phase was determined by counting trypan blue excluding cells in a hemocytometer and 20,000 cells were plated into each well of 96-well flat-bottomed micro-titer plates. After 24 h incubation at 37 °C, cells were transfected with the scrambled antisense or antisense against hSET1 (0–10 μg/ml final conc.) using Maxfect transfection reagent (MoleculA) and after 6 hours, medium was changed and incubated for 24 hour. 20 μl of 5 mg/ml MTT were then introduced to each well and incubated for 2 h. The plates were centrifuged and medium was decanted. Cells were subsequently dissolved in 100 μl DMSO with gentle shaking for 2 h at room temperature, followed by measurement of OD570 [24, 26, 27]. Eight replicate wells were used in each point in each of three separate measurements. Measured absorbance values were directly linked with a spread sheet for calculation of cell survival.
2.7. Immuno-cytological localization of hSET1 in colon cancer cells
Immuno-cytological localization of hSET1 was performed to assess whether cellular levels of hSET1 could be depleted by antisense against hSET1 in cultured colon cancer cells by the method described previously with slight modifications [28]. Cells were grown on cover slips and were treated with scrambled as well as hSET1 antisense (10 μg/ml final conc.). After 24 h incubation, the cells were washed with PBS and were fixed by using methanol: acetic acid in the ratio of 3:1. Non-specific antibody interactions were minimized by pre-treating cells with 10% goat serum in PBS for 60 min at room temperature. The cells were incubated with primary antibodies, anti-hSET1 IgG, for overnight at 4 °C in a humidified chamber. After washing off the primary antibody with PBS (10 times, 3 min each), rhodamine red-x-conjugated goat anti-rabbit IgG (1:50 dilution in PBS) as secondary antibodies were added and incubated for 1 h at room temperature in a humidified chamber. The unbound secondary antibodies were removed by washing with PBS (10 times, 3 min each) and nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). Cover slips were air-dried and mounted on the slides with Vectashield mounting medium for fluorescence (Vector Laboratories, CA). Slides were photographed at 400 × magnifications using a LEICA DMLB fluorescence microscope (Germany). Photographs were taken with 1-second integration.
2.8. Effect of hSET1 antisense on apoptosis by TUNEL assay
To check whether suppression of hSET1 is correlated with apoptosis was assessed by the TUNEL assay. The SW480 colon cancer cells (1 × 106 cells) were grown on the cover slips. The cells were treated with hSET1 antisense (10 μg/ml final conc.). After 24 h incubation, the medium was removed, and cells were washed with PBS. TdT-mediated dUTP nick end labeling (TUNEL) assay was performed using Promega fluorescence detection kit according to the protocol provided by the manufacturer and described by us previously [24,27]. Briefly, cells were fixed by immersing slides in freshly prepared 4 % paraformaldehyde solution for 30 min at 4 °C followed by washing with PBS. The cells were permeabilized by immersing the slides in 0.2% Triton X-100 solution in PBS for 5 min followed by washing with PBS. Cells were equilibrated with equilibration buffer (provided by Promega) for 10 min. The equilibrated areas were blotted with tissue paper and TdT incubation buffer was added to the cells, and placed in humidified chamber. The chamber was covered with aluminum foil to protect from direct light. Slides were incubated at 37 °C for 60 min and the reaction was terminated by immersing the slides in 2 × SSC buffer for 15 min followed by washing with PBS. The slides were stained in propidium iodide solution for 10 min in the dark and washed with distilled water several times. Slides were analyzed under a fluorescence microscope using a standard fluorescein filter set to view the green fluorescence at 520 nm, and red fluorescence of propidium iodide at > 620 nm. Fluorescence micrographs were taken using Zeiss LSM 510 META (Germany) laser scanning fluorescence microscope at 400 × magnification.
2.9. Animal model
Hsd: Athymic nude nu/nu mice were obtained from Harlan (Indianapolis, IN). All animal experiments were carried out in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC). Twenty-four 12 week old mice were divided into three groups of 8 animals treated with PBS, scrambled antisense, and phosphorothioate antisense DNA against hSET1. All 24 animals were injected with 2 × 106 SW480 colon cancer cell suspensions in 100 μl of PBS, subcutaneously. Animals were examined daily for signs of tumor growth. Treatment was administered when the tumor surface area exceeded 42 mm2 (day 26). Treatment consisted of 200 μg phosphorothioate antisense against hSET1 in 100 μl PBS. Control groups were treated with 200 μg/100 μl scrambled antisense or diluent (PBS) alone. Tumors were measured in two dimensions using calipers.
2.10. Statistical methods
All data were evaluated with a two-tailed unpaired student’s t test or compared by one-way ANOVA and are expressed as the mean ± SD. A value of P < 0.05 was considered statistically significant.
3. Results and discussion
3.1. Expression of hSET1 in malignant and normal cells
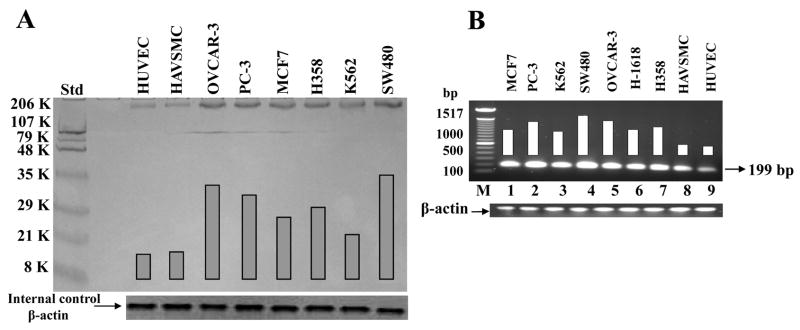
Histone methyl transferases are known to be differentially expressed in cancer cells and promote cell proliferation, colony formation and increased invasion of benign cells in-vitro. We therefore, quantitated hSET1 in a wide array of cultured cell lines of different origin. Western blot analyses of the total crude protein extracts from a panel of cultured human malignant cell lines including human lung (H358), ovarian (OVCAR-3) breast (MCF7), prostate (PC-3), colon (SW480), erythroleukemia (K562) and non-malignant human cell lines of endothelial (HUVEC) and aortic vascular smooth muscle (HAVSMC) were performed. Each lane was loaded with equal amounts of crude protein (200 μg) for SDS-PAGE and Western blot analyses. Western blot performed against anti-hSET1 IgG, detected a single band of ~ 186 kDa characteristic of hSET1. These results showed the presence of hSET1 in all cell lines and indicated relatively larger expression of hSET1 in malignant vs normal cells (Fig. 1A). The expression of hSET1 was more in colon cancer (SW480) and ovarian cancer (OVCAR-3) as compared to breast cancer (MCF7). It is noteworthy that hSET1 over-expressed in solid tumor as well as in human erythroleukemia (K562) cells.
Figure 1. Comparison of hSET1 levels in cultured malignant vs non-malignant cells.
Aliquots of total crude protein of malignant cells (human prostate, PC-3; non-small cell lung, H358; colon, SW480; breast, MCF7; ovarian,OVCAR-3; leukemia, K562) and non-malignant cells (human aortic vascular-smooth-muscle cells, HAVSMC, and human umbilical vascular-endothelial cells, HUVEC) containing 200 μg protein were used for SDS-PAGE and Western blot was performed against anti-hSET1 IgG as primary antibody and horseradish peroxidase-conjugated goat-anti-rabbit-IgG as secondary antibody and developed with 4-chloro-1-naphthol as chromogenic substrate. Intensity of the full-length hSET1 protein (~186 kDa) band was quantified by scanning densitometry using Innotech Alpha Imager HP. β-actin was used as an internal control (panel A). The level of hSET1 in various human cultured cells (PC-3, H358, H1618, SW480, MCF7, OVCAR-3, K562, HAVSMC and HUVEC) was also evaluated by RT-PCR analysis. RNA was prepared by RNeasy kit (Qiagen) and RT-PCR was performed using hSET1 gene specific primers [1422–1439 bp (upstream) and 1602–1621 (downstream)]. The bands were quantified by densitometry using Innotech Alpha Imager HP. β-actin was used as a loading control (panel B).
The expression of hSET1 was further confirmed at the transcription level by RT-PCR analysis using the gene specific primers for hSET1 (Fig. 1B) in a panel of malignant (H358, H1618, OVCAR-3, MCF7, PC-3, K562 and SW480) and non-malignant (HUVEC, HAVSMC) cell lines. The RNA was prepared by RNeasy kit (Qiagen Inc.) and RT-PCR was performed by using hSET1 gene-specific primers [1422–1439 bp (upstream) and 1602–1621 (downstream) spanning 1422 to 1621 in open reading frame of hSET1 gene], using the single-step RT-PCR kit (Qiagen Inc.). The bands quantified by scanning densitometry indicated that hSET1 over-expressed in malignant cells as compared to the normal cells (Fig. 1B). Our results clearly indicated that hSET1 differentially expressed in normal and malignant cells both at the transcription as well as translation level.
3.2. hSET1 suppression caused preferential cytotoxicity in malignant cells
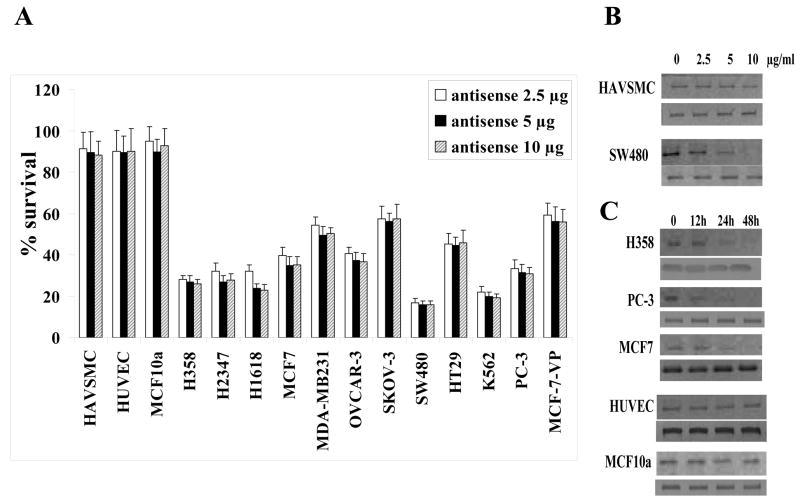
As histone methyl transferases over-expressed in the malignant cells and promotes cell proliferation, knock-down of hSET1 should result in the growth arrest. The cytotoxic effects of hSET1 suppression by using the phosphorothioate antisense against it, was assessed by an established MTT cell survival assay [24,26,27]. In cell survival assay, suppression of hSET1 was preferentially directed to the malignant cells as compared with non-malignant cells. All non-malignant cells of different origin (HUVEC, HAVSMC, as well as breast epithelial cells, MCF10a) were resistant to the suppression of hSET1. The suppression of hSET1 causes more cytotoxicity in human colon (SW480), erythroleukemia cells (K562), and lung (H358, H2347, H1618) cancer cells with ~ 70–80% cell death in concentration dependent manner (Fig. 2A) as compared to ovarian (OVCAR-3) where about 50% cell death was observed. Breast cancer cells (MCF7 and MDA-MB231) and drug-resistant cell lines (MCF7-VP and SKOV-3) were more resistant to the suppression and behave more like the non-malignant cells. Studies of concentration dependent effects of hSET1 antisense (DN5) revealed almost complete depletion of hSET1 protein in SW480 after 24 hours at a concentration of 10 μg/ml (Fig. 2B). The non-malignant cells (HAVSMC) were somewhat less sensitive to hSET1 depletion as compared with SW480 cells. The relative resistance of non-malignant cells to hSET1 depletion was also observed in the time-dependent studies (Fig. 2C) in which non-malignant cells (HUVEC and MCF10a) were relatively less affected as compared to malignant cells (H358, PC-3 and MCF7).
Figure 2. Selective toxicity of hSET1 antisense towards cancer cells.
Effect of phosphorothioate antisense against hSET1 (0–10 μg/ml final conc.) on cell survival was determined by MTT assay. A panel of cells were transfected with hSET1 antisense (0–10 μg/ml final conc.) using Maxfect Transfection-Reagent (Molecula, Inc.), according to the manufacturer’s instructions. Cell survival was measured by MTT cytotoxicity assay 24 h after treatment [26, 27]. The values are presented as mean ± SD from two separate determinations with eight replicates each (n =16). White bars, 2.5 μg/ml; black bars, 5 μg/ml; and hatched bars, 10 μg/ml final concentration. Concentration dependent effects of hSET1 antisense was performed by incubating normal (HAVSMC) and malignant (SW480) cells with phosphorothioate antisense against hSET1 (ranging from 0–10 μg/ml final conc.) in Maxfect Transfection Reagent followed by 24 hours of incubation at 37 °C in medium before Western blotting with anti-hSET1 IgG as primary antibody (panel B). The time dependent effect of hSET1 antisense (10 μg/ml final conc.) were also evaluated using three malignant and two non-malignant cells by determining hSET1 protein levels by Western blot analyses at 0, 12, 24, and 48 h, after treatment with hSET1 antisense (panel C). In panels B and C, internal control (β-actin) is shown below the respective Western blots from each cell line.
3.3. Immuno-cytological localization of hSET1
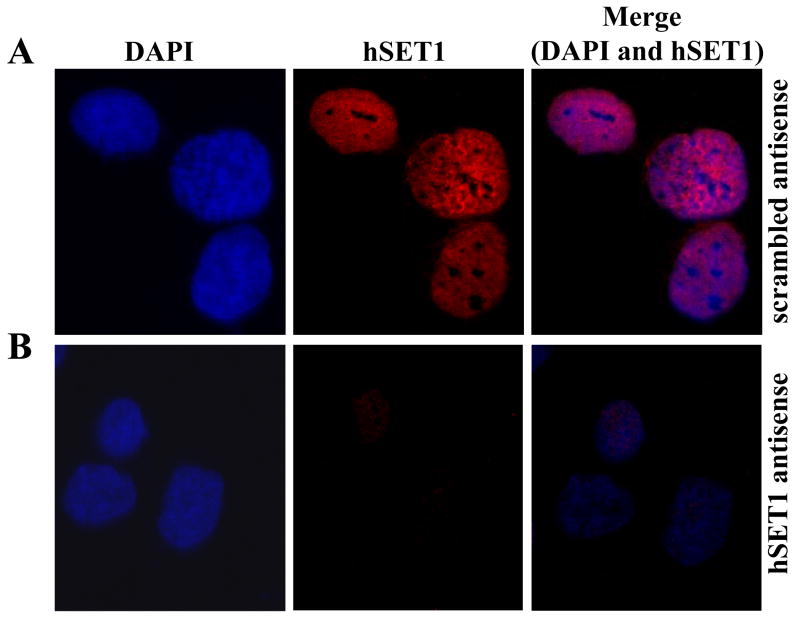
To check the effect of DN5 on suppression and localization of hSET1, SW480 cells were transfected with 10 μg/ml scrambled or antisense against hSET1 and after 24 h, immuno-cytological localization of hSET1 was performed in fixed cells using anti-hSET1antibody and were analyzed by confocal fluorescence microscopy. These results demonstrated that the majority of hSET1 was found inside the nucleus, associated with the chromatin. There was more than 80% suppression of hSET1 in the cells treated with the hSET1 antisense clearly shows that the antisense suppresses hSET1 in SW480 cells (Fig. 3).
Figure 3. Localization of hSET1 in SW480 colon cancer cells.
Localization of hSET1 was performed on SW480 fixed cells by method described previously with slight modifications [28]. Cells were grown on glass cover slips and were transfected with 10 μg/ml scrambled antisense or antisense against hSET1 using Maxfect Transfection Reagent (Molecula). After 24 h, cells were fixed with methanol and acetic acid (3:1). Nonspecific antibody interactions were minimized by pre-treating the cells with 10% goat serum in TBS for 60 min at room temperature. The cells were subjected to immuno-cytochemistry using anti-hSET1 IgG (raised in rabbit) as a primary antibody and goat-anti-rabbit rhodamine red-x-conjugated as secondary antibody. DAPI was used as a nuclear counter-stain. Slides were analyzed by confocal laser microscope (Leica TCS-SP5, Germany). Panel A, shows the cells treated with scrambled antisense and panel B shows cells treated with antisense against hSET1.
3.4. Apoptosis caused by depletion of hSET1
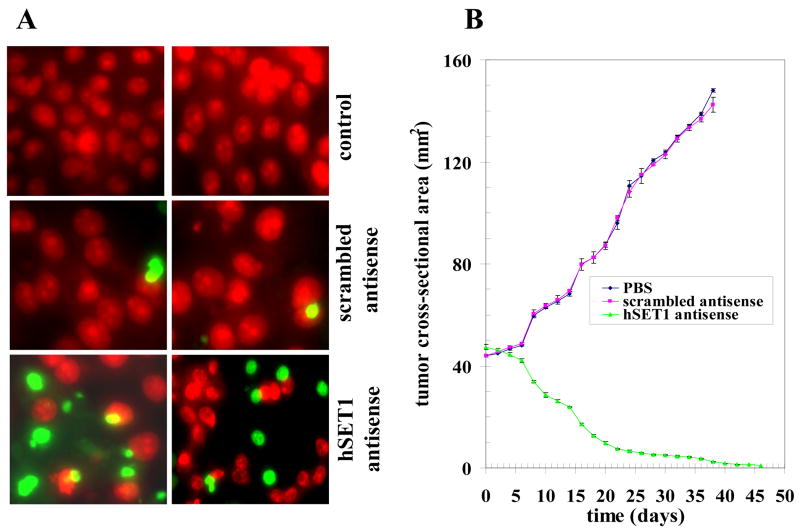
In MTT cell survival assay, suppression of hSET1 was most effective in colon and lung cancer cells (Fig. 2). The mechanism of cytotoxicity was further assessed in SW480 colon cancer cells by determining apoptosis through an immuno-histochemical TUNEL assay. The results of the TUNEL assay showed no detectable apoptosis in the SW480 cells treated with either PBS or scrambled antisense. Apoptosis was however, seen in cells treated with hSET1 antisense (Fig. 4A).
Figure 4. Effect of hSET1 antisense on apoptosis and on the regression of established SW480 colon cancer xenografts.
Effect of hSET1 antisense on apoptosis was performed by TUNEL assay. Human colon cancer cells (SW480) were grown on cover-slips. For antisense treatment, cells were transfected with 10 μg/ml scrambled antisense or antisense against hSET1 using Maxfect Transfection Reagent (Molecula, Inc.). After 24 h, TUNEL assay was performed using Promega fluorescence detection kit and examined using Zeiss LSM 510 META (Germany) laser scanning fluorescence microscope with filters 520 nm and >620 nm. Photographs taken at identical exposure at 400 x magnification are presented. Apoptotic cells showed green fluorescence (panel A). For xenografts studies, we used twenty-four 12 weeks old Hsd: Athymic nude nu/nu mice (Harlan, Indianapolis, IN) randomized 8 animals each into three groups as follow: 1) control, 2) scrambled antisense and 3) hSET1 antisense. All 24 animals were injected with 2 × 106 human colon cancer cells (SW480) suspensions in 100 μl of PBS, subcutaneously into one flank of each nu/nu nude mouse. Animals were examined daily for signs of tumor growth. When tumors reached a cross-sectional area of ~ 42 mm2 (26 days later), animals were randomized treatment groups as indicated in the figure (for animal pictures, see supplementary data). Treatment consisted of 200 μg of hSET1 antisense in 100 μl PBS. Control groups were treated with PBS or 200 μg/100 μl scrambled antisense. Tumors were measured in two dimensions using calipers. Tumor measurements are presented with all control groups (PBS or scrambled antisense) vs treated group (hSET1 antisense) (panel B).
3.5. hSET1 depletion caused regression of colon cancer xenografts in nude mice
The ultimate pre-clinical test for the potential utility of an anti-neoplastic agent is effectiveness in an animal model. The above observations of the anti-neoplastic effects of hSET1 depletion was examined in a xenografts model of colon cancer as in-vitro studies showed there is more than 80% cells death on suppression of hSET1. Tumor-bearing animals with established s.c. implanted tumors (~ 42 mm2) were treated with PBS, 200 μg of either scrambles antisense or antisense against hSET1 by i.p. injection. Treated animals had rapid and dramatic reductions in tumors, whereas uncontrolled growth was observed in the control groups. The remarkable contrast in the outcome of tumors in animals treated with hSET1 antisense vs PBS or scrambled antisense was clearly evident for colon cancer cell lines (Fig. 4B) (for animal pictures, see supplementary data). Weight gain was comparable to non-tumor bearing controls, and no overt-toxicity was evident. Present studies demonstrate nearly complete and sustained regression of xenografts of SW480 colon cancer by targeted depletion of the histone methyl transferase hSET1.
The marked effectiveness of this targeted therapy compares quite favorably with other promising targeted agents in colon cancer [29–31]. Suppression of human colon cancer tumors in nude mice by siRNA CD44 gene therapy indicated increased apoptosis of tumor cells involving caspase 3 and poly (ADP-ribose) polymerase (PARP) [29]. Fusion of enhanced carcinoembryonic antigen promoter (CEA) to a suicide gene and delivery of CEA using calcium phosphate nanoparticles together with prodrug 5-FC treatment, results in significant tumor growth delay in xenograft human colon carcinoma [30]. The in-vitro cytotoxicity and in-vivo efficacy of TRA-8, a mouse monoclonal antibody that binds to the DR5 death receptor for tumor necrosis factor-related apoptosis-inducing ligand (also called Apo2L), alone and in combination with CPT-11, has been shown to be very effective against human colon cancer cells and xenografts [31].
In the field of epigenetics, several enzymes involved in histone acetylation and DNA methylation serve as current drug targets in the treatment of cancer. The HDAC inhibitor suberoylanilidehydroxamic acid (SAHA) has recently been approved for the treatment of advanced T cell lymphoma, while other HDAC inhibitors are in clinical trials [32–35]. Compared to the field of epigenetics, the knowledge on histone methylation and the consequences of its inhibition is still behind. For lysine methyltransferases to date, only a handful of promising compounds are available. Chaetocin a fungal mycotoxin, was found to inhibit the drosophila melanogaster histone methyltransferase Su(var)3–9 in-vitro as well as in-vivo but it did not seem to inhibit the lysine methyltransferases like EZH2 or SET7/9 [36] while another inhibitor, BIX-01294 showed a reduction in histone H3 lysine 9 (H3K9) dimethylation but the mono- or trimethyl stages appeared unaffected [37].
4. Conclusions
The results of the present studies have shown that human cancers (lung, colon, breast, ovarian, leukemia, and prostate) express higher levels of hSET1 than non-cancerous cell, and that depletion of hSET1 in these cancer cell types caused marked apoptosis, whereas non-cancerous cells were spared. Because human colon cancers have a relatively high expression of hSET1, we reasoned that it may be functioning as a defense mechanism in colon cancer cells. Studies demonstrating the marked regression of tumor in colon cancer xenografts (SW480) by concomitant depletion of hSET1 have confirmed the in-vivo relevance of these observations. As the knowledge on histone methylation and the consequences of its inhibition is still behind, these outstanding findings will certainly add to the field of epigenetics, and will have obvious translational implications for the treatment of colon cancer and potentially other malignancies.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants CA 77495 and CA 104661, Cancer Research Foundation of North Texas, Institute for Cancer Research and the Joe & Jessie Crump Fund for Medical Education.
Abbreviations
- HMTs
histone-methyl transferases
- H3K4
histone H3 lysine 4
- hSET1
human SET1
- TUNEL
TdT-mediated dUTP nick end labeling assay
- MLL
mixed lineage leukemia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–35. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 3.Sims RJ, Reinberg D. From chromatin to cancer: a new histone lysine methyltransferase enters the mix. Nat Cell Biol. 2004;6:685–7. doi: 10.1038/ncb0804-685. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 6.Sims RJ, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–86. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 7.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 8.Dutnall RN. Cracking the histone code: one, two, three methyls, you’re out! Mol Cell. 2003;12:3–4. doi: 10.1016/s1097-2765(03)00282-x. [DOI] [PubMed] [Google Scholar]
- 9.Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, Issa JP. Down-regulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS ONE. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 11.Fingerman IM, Wu CL, Wilson BD, Briggs SD. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem. 2005;280:28761–65. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogenesis to covalent modifications of chromatin. J Cell Biochem. 2005;95:429–36. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- 13.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6–Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–42. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 14.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–56. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–34. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–9. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 17.Shahbazian MD, Zhang KL, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Molecular Cell. 2005;19:271–7. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol. 2005:143–5. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 20.Ansari KI, Mishra BP, Mandal SS. Human CpG binding protein interacts with MLL1, MLL2 and hSet1 and regulates Hox gene expression. Biochim Biophys Acta. 2008;1779:66–73. doi: 10.1016/j.bbagrm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Shukla N, Stanojevic Z, Shadle Duan T, Bhaumik SR. Functional analysis of H2B-Lys-123 ubiquitination in regulation of H3-Lys-4 methylation and recruitment of RNA polymerase II at the coding sequences of several active genes in vivo. J Biol Chem. 2006;281:19045–54. doi: 10.1074/jbc.M513533200. [DOI] [PubMed] [Google Scholar]
- 22.Kulaeva OI, Gaykalova DA, Studitsky VM. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat Res. 2007;618:116–29. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 24.Singhal SS, Awasthi YC, Awasthi S. Regression of Melanoma in a Murine Model by RLIP76. Depletion Cancer Res. 2006;66:2354–60. doi: 10.1158/0008-5472.CAN-05-3534. [DOI] [PubMed] [Google Scholar]
- 25.Minamide LS, Bamburg JR. A filter paper dye-binding assay for quantitative determination of protein without interference from reducing agents or detergents. Anal Biochem. 1990;190:66–70. doi: 10.1016/0003-2697(90)90134-u. [DOI] [PubMed] [Google Scholar]
- 26.Singhal SS, Yadav S, Singhal J, Zajac E, Awasthi YC, Awasthi S. Depletion of RLIP76 sensitizes lung cancer cells to doxorubicin. Biochem Pharmacol. 2005;70:481–8. doi: 10.1016/j.bcp.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Singhal SS, Yadav S, Drake K, Singhal J, Awasthi S. Hsf-1 and POB1 induce drug-sensitivity and apoptosis by inhibiting Ralbp1. J Biol Chem. 2008;283:19714–29. doi: 10.1074/jbc.M708703200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav S, Singhal SS, Singhal J, et al. Identification of Membrane Anchoring Domains of RLIP76 using Deletion Mutants Analyses. Biochemistry. 2004;43:16243–53. doi: 10.1021/bi0482811. [DOI] [PubMed] [Google Scholar]
- 29.Subramaniama V, Vincenta IR, Gilakjana M, Jothy S. Suppression of human colon cancer tumors in nude mice by siRNA CD44 gene therapy. Exp Mol Pathol. 2007;83:332–40. doi: 10.1016/j.yexmp.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Zhang G, Liu T, Chen Y, Chen Y, Xu M, Peng J, Yu S, Yuan J, Zhang X. Tissue specific cytotoxicity of colon cancer cells mediated by nanoparticle-delivered suicide gene in-vitro and in-vivo. Clin Cancer Res. 2009;15:201–7. doi: 10.1158/1078-0432.CCR-08-1094. [DOI] [PubMed] [Google Scholar]
- 31.Oliver PG, LoBuglio AF, Zinn KR, Kim H, Nan L, Zhou T, Wang W, Buchsbaum DJ. Treatment of human colon cancer xenografts with TRA-8 anti-death receptor 5 antibody alone or in combination with CPT-11. Clin Cancer Res. 2008;14:2180–9. doi: 10.1158/1078-0432.CCR-07-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakshmikanthan V, Kaddour-Djebbar I, Lewis RW, Kumar MV. SAHA-sensitized prostate cancer cells to TNFalpha-related apoptosis-inducing ligand (TRAIL): mechanisms leading to synergistic apoptosis. Int J Cancer. 2006;119:221–8. doi: 10.1002/ijc.21824. [DOI] [PubMed] [Google Scholar]
- 33.Butler LM, Liapis V, Bouralexis S, Welldon K, Hay S, Thai le M, Labrinidis A, Tilley WD, Findlay DM, Evdokiou A. The histone deacetylase inhibitor, suberoylanilide hydroxamic acid, overcomes resistance of human breast cancer cells to Apo2L/TRAIL. Int J Cancer. 2006;119:944–54. doi: 10.1002/ijc.21939. [DOI] [PubMed] [Google Scholar]
- 34.Butler LM, Zhou X, Xu W, Scher HI, Rifkind RA, Marks PA, Richon VM. Individual the histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci. 2002;99:11700–5. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 36.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. Embo J. 2002;21:1121–31. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spannhoff A, Sippl W, Jung M. Cancer treatment of the future: Inhibitors of histone methyltransferases. Int J Biochem Cell Biol. 2009;41:4–11. doi: 10.1016/j.biocel.2008.07.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.