Summary
Cell fate in the Arabidopsis root is determined by positional information mediated by plant hormones and interpreted by transcriptional networks. In this review, we summarize recent advances in our understanding of the regulatory networks that control cell fate within the root meristem, and in the interplay of these networks with phytohormones. Recent work describing the importance of chromatin organization in tissue patterning is also highlighted. A new, high resolution root expression map detailing the transciptome of nearly all cell types in the Arabidopsis root across developmental timepoints will provide a framework for understanding these networks.
Introduction
Transcriptional networks form the basis of cellular processes throughout the biological world, from unicellular algae to mammals. In multicellular organisms, these regulatory networks act in concert with positional signals to specify cell fate and tissue organization. An understanding of how these networks act to establish cell fate is emerging in the reference plant Arabidopsis.
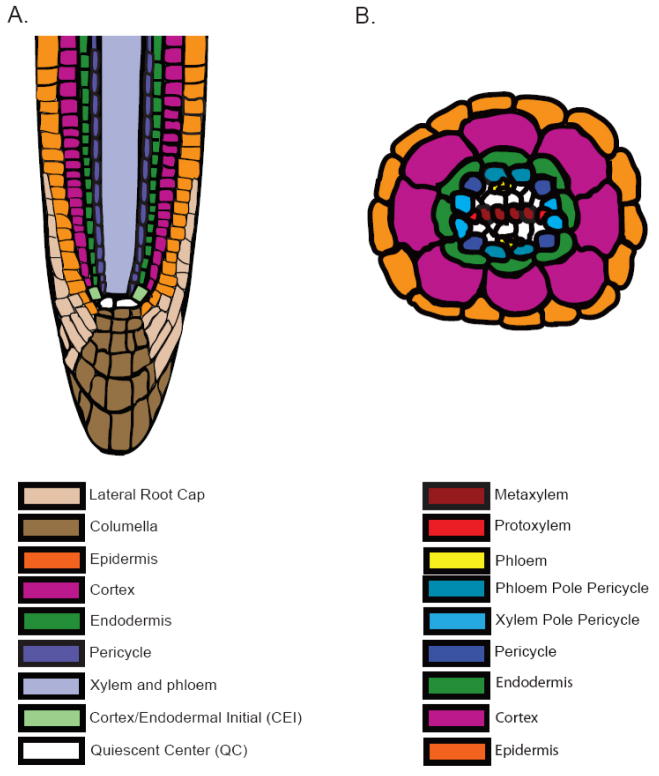
The simple structure and stereotyped development of the Arabidopsis root make it a tractable system to study cell fate specification. The root is composed of approximately 15 cell types that can be simplified to a set of concentric tissues arranged around a radial axis (Figure 1). As in animal cells, the plant stem cell niche defines the region that maintains the stem cells, undifferentiated cells with the capacity for self-renewal [1, 2]. In the root apex, four sets of stem cells, or initials, surround the quiescent center (QC), less mitotically active cells required for stem cell maintenance. Laser ablation of the QC results in the differentiation of the surrounding initials [3]. Stem cells can be grouped into two sets. Asymmetric division of the provascular, cortex/endodermal, and epidermal/lateral root initials of the meristem produce daughter cells that will divide again to form the vasculature, ground tissue, epidermis, and lateral root cap, respectively. Divisions of the columella initials result in daughter cells that will differentiate into columella cells without a subsequent division [2]. Position is key, as stem cell daughters can adopt different fates depending on their location [4]. Since plant cells do not move, new cells arising from the stem cell niche form files of cells up the root. As new cells form, they add onto their older counterparts pushing downwards into the soil. Further along the longitudinal axis they elongate and differentiate. Cell lineages can thus be easily followed along this developmental timeline, where position indicates age.
Figure 1.
Longitudinal (A) and transverse (B) sections of the Arabidopsis root
Over the past 15 years, mutant analysis has led to the identification of various transcription factors (TFs) responsible for radial patterning of the root and positioning of the stem cell niche. However, recent advances in transcriptional profiling, microarray analyses, and systems biology have allowed the elucidation of new interactions, and complex networks that define cell fate in the root are emerging [5, 6]. In this review, we focus on regulatory networks in post-embryonic cell fate specification in the stem cell niche, ground tissue, stele and epidermis. The interaction with phytohormones and intersection with epigenetic modifications involved in cell fate specifications of each tissue are also discussed.
1.1 Stem cell niche specification requires polar auxin transport and the parallel action of two pathways
1.1.1 Auxin and root meristem patterning
The phytohormone auxin is responsible for a wide range of developmental processes in the plant, including cell division, expansion, tropisms, vascular differentiation, and root meristem (RM) maintenance. This is accomplished through its ability to change transcriptional programs of specific cell types and transmit spatial information. Although auxin can diffuse throughout the plant, most processes it influences are dependent upon its polar transport. Auxin is thought to be synthesized in the aerial organs [7] and transported acropetally in the stele to the RM, resulting in an auxin maximum in the root tip. This asymmetric auxin distribution affects root patterning, and is needed for the specification of distal cell types [8–12].
Polar auxin transport (PAT) is made possible largely through the action of PIN proteins, auxin efflux carriers that are polarly localized to plant membranes. The localization depends on the cell type, and is in line with the direction of auxin flow. Of the eight member PIN family, five have been extensively characterized, and contribute to embryonic development, root patterning, and tropic growth [13]. PAT results in a distal auxin maximum in the root tip; changing the maximum induces a new QC where the new maximum occurs [8]. Examination of PIN expression and localization in single and multiple pin mutants showed cell fate and morphological changes in response to alterations in auxin distribution, and suggested that a graded distribution of auxin may be important for patterning [9, 11, 14].
The distal maxima is achieved via an auxin reflux loop in the RM. The distribution of the PINs in the columella and epidermis redirects the auxin at the tip laterally and basipetally to the proximal meristerm, and then back down again through PINs in the stele [14, 15]. In line with this, decapitated roots are able to maintain the QC; laser ablation of the QC in such roots revealed new QC formation and auxin maxima within three hours [15]. Thus repatterning can occur without any additional shoot derived auxin. Using PIN localization data and a simplified version of the root, Grieneisen et al. (2007) developed a model to simulate auxin flow within the root. They postulated that auxin biosynthesis and decay are less important than its flux through the PIN network. In this way the root is like a capacitor that efficiently stores auxin. The root maintains the maxima, and in turn the meristem, through the PIN-directed auxin flux. By including terms for known auxin effects on cell elongation and division in their model, the authors provided evidence for gradients within the root. In model simulations the auxin maxima is maintained and stable in the QC but dynamic around the elongation zone/meristematic zone border (the transition zone ‘TZ’), thus allowing for different levels of growth and expansion in the root. Capacitance is dependent on the PIN layout within the root, and only drastic changes in the layout can alter the auxin distribution.
1.2 PLETHORA1 and 2: Master regulators of root development
A promoter trap screen identified two AP2/EREBP TF family members, PLETHORA 1 and 2, that are redundantly required for QC identity and stem cell maintenance [16]. The PLT1/2 family contains two other AP2 TFs, PLT3 (AININTEGUMENTA 6) and BABYBOOM (BM) [17]. Double plt1 plt2 mutants have shorter roots with more lateral roots than wild type (wt), and their small root meristems differentiate within 6–8 days. PLT1/2 are induced by auxin, and embryonic transcription is dependent on the auxin response factors MONOPTEROS (MP) and NON-PHOTOTROPIC HYPOCOTYL4 (NPH4). Expression of multiple PINs is redundantly controlled by PLT1, 2 and 3 in the embryonic and post-embryonic stage [17]. True to their description as ‘master regulators’ of root development, PLT genes can induce root meristems, QC and initial formation when ectopically expressed in the stem [16, 17].
The four PLT genes have different but overlapping expression domains; protein patterns reflect the expression domain for each [17]. These expression patterns exhibited a gradient in the RM, with maxima of each localized to the stem cell niche. Altering the amount and expression pattern of different PLT genes in wt and mutant plants demonstrated that each has to be expressed at the right place and level for the correct output to occur, as different levels were instructive for different outcomes (cell expansion vs. cell proliferation, for example). These experiments showed that different levels of PLT genes define the stem cell domain, the cell proliferation zone in the meristem, and the transition zone [17].
The finding that the auxin-induced PLT genes have a gradient of expression in the RM, analogous to that predicted for auxin [15], suggested that PLT genes may be outputs of an underlying auxin gradient [17]. Since PLT genes can control the expression of PINs, they may work as master regulators through a feedback loop in which PLTs are induced by auxin and in turn contribute to the regulation of the PIN network for auxin flux.
1.3 GRAS family transcription factors regulate stem cell niche specification and ground tissue patterning
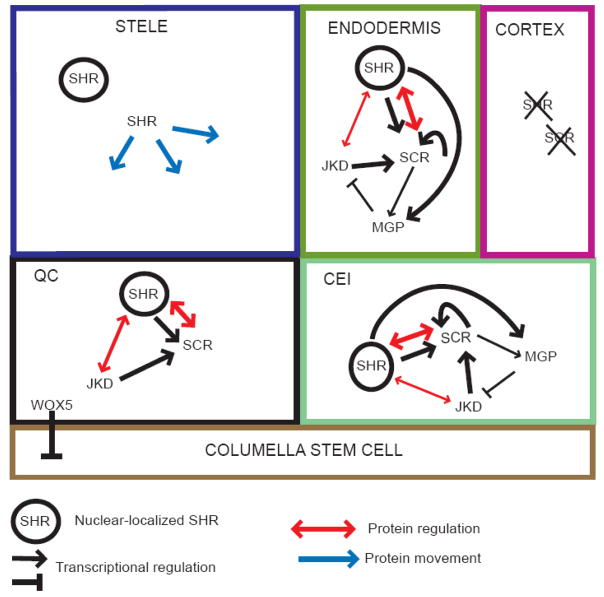
Early genetic screens for altered root development identified two GRAS family TFs important for radial patterning in the root, SHORTROOT (SHR) and SCARECROW (SCR) [18, 19]. SHR transcripts are found only in the stele [20], but the protein moves into the endodermis, QC, and cortex/endodermal initial (CEI) [21–23], where it activates expression of the downstream TF SCR [20, 23, 24] (Figure 2). In wt plants the CEI divides anticlinally, producing a daughter cell, and regenerating itself. The daughter cell then divides periclinally to produce the first cells of the cortex and endodermis. In shr mutants, this periclinal division does not occur, resulting in a mutant layer with only cortex characteristics [18, 20]. Thus SHR is necessary both for the periclinal division of the CEI daughter cell and endodermal specification. Ectopic expression of SHR results in supernumary cell layers and altered cell specification. SHR also has a role in QC specification, as evidenced by the disappearance of several QC markers in the shr mutant, and in 5 day old shr seedlings the RM begins to differentiate.
Figure 2.
Model for the SHR-SCR pathway. SHR is expressed in the stele and the protein moves into the endodermis, QC, and CEI, where it activates expression of SCR. SCR induces an asymmetric periclinal division in the CEI daughter that results in one layer of endodermis and one of cortex. In the endodermis and CEI, SCR autoinduces and interacts with SHR to sequester it in the nucleus, preventing SHR movement and the production of supernumary layers. SCR and SHR must be quickly degraded in the cortex by unknown factors. JKD may act directly with SHR or indirectly through SCR to restrict SHR movement. MGP is epistatic to JKD, and though the mechanism has yet to be determined, it may counteract the effect of JKD by interfering with a JKD-SCR-SHR complex. WOX5 acts non-autonomously to suppress columella stem cell differentiation and redundantly with other factors to suppress differentiation of the other initials. See text for additional details.
Like SHR, SCR is a member of the GRAS family of TFs [18, 25] but is expressed in the QC, CEI and the endodermis (Figure 2). SCR expression is consistently lower (although still present) in the shr mutant background, suggesting that SHR regulates SCR expression. In scr mutants, division of the CEI daughter cell does not occur, resulting in one mutant layer with mixed cortex-endodermal identity. Thus, SCR is required for the periclinal asymmetric division of the CEI daughter [25]. scr mutant plants have a partial loss of QC identity and initials eventually differentiate. scr mutants lack SCR expression in the QC, but not in the single ground tissue layer, indicating that SCR is necessary for its own transcription in the QC [25] (Figure 2). SCR is required cell-autonomously for QC identity and non- cell autonomously for stem cell specification [26, 27]. In line with this, expression of SCR in the ground tissue of scr mutants but not in the QC rescued the radial patterning defect, but not QC identity or stem cell specification [26]. The PLT genes appear to act in a parallel pathway to that of SCR and SHR, as SHR and SCR are expressed correctly in the plt1plt2 double mutant. PLT1 is expressed in the right place in scr and shr mutants, though at a lower intensity [16].
The observation that cell types differ in their ability to create supernumary layers in response to SHR [28] suggested that tissue-specific factors were important to restrict SHR movement. Several lines of evidence suggested that one of the factors might be SCR. First, ectopic expression and clonal activation or deletion of SCR showed that SCR plays a role in restricting SHR movement [27, 28]. In scr mutants, SHR is largely cytoplasmic in the mutant cell layer, and it can move beyond the mutant layer into the epidermal layer [28]. Further, co-IP and protein interaction experiments showed that SCR and SHR interact [23, 29].
Cui et al. [23] hypothesized that SCR might limit SHR movement by sequestering SHR in the nucleus. Although a large amount of SCR could be necessary to sequester SHR and prevent its movement, SHR induction of SCR [24] combined with a feedback activation loop could provide the required amount of protein [27, 28] (Figure 2). If SCR were diminished below a threshold level, SHR would no longer be sequestered, and could move into the next cell layer, activate SCR, and keep doing this until the supply of SHR were diminished as it moved along the radial zones of the root. To test this hypothesis, the authors examined RNAi lines with different levels of SCR. Using these lines, they elegantly showed that the amount of SHR movement and number of supernumary cell layers was inversely correlated with the level of SCR. The current model is that 1) SHR expression is induced in the stele, then the protein moves into the endodermis where it activates SCR expression; 2) Autoinduction of SCR provides enough protein to sequester SHR in the nucleus (Figure 2), preventing further movement, as SHR with the addition of a strong NLS cannot move from the stele [30]; 3) SCR preferentially interacts with SHR, reinforcing the feed-forward loop. This model also accounts for the fact that cortex and endodermal cell fates are quickly established after the CEI division [27]. Since SHR cannot move into the cortex cell, less SCR is induced and the SHR/SCR complex in the cortex is quickly turned over. Notably, rice orthologs of SHR and SCR interact in yeast, suggesting that this mechanism is conserved across the monocot-dicot divide and may be responsible for limiting the number of endodermal layers in plant cells [23].
SCR is not the only protein that restricts SHR movement. A screen of 15,000 promoter-GUS lines identified one with expression in the SCR domain. JACKDAW (JKD) is a C2H2 TF that interacts with both SCR and SHR [29] (Figure 2). JKD induces SCR in the QC and alters its expression in the ground tissue. Mutants in JKD have an altered QC and display ectopic periclinal divisions in the cortex, sporadically creating three layers of ground tissue. SHR is expressed in the middle cortex layer, indicating that SHR has moved past its usual boundaries into these cells. In addition, in jkd mutants, SHR is not localized to the nucleus in the QC. Whether these events are indirect effects of the decrease in SCR in the jkd mutant or a direct effect of JKD in limiting SHR movement remains to be determined.
MAGPIE (MGP), a homolog of JKD, is a target of SHR and SCR [discussed below; 23, 24, 29]. Single mgp mutants have no phenotype [29]. MGP interacts with SHR, SCR and JKD, and mgp mutants partially complement the jkd phenotype, suggesting that MGP and JKD counteract each other in the SHR/SCR network [29] (Figure 2). MGP is expressed in the stele, CEI, and CEI initial, but not in the QC. Thus, in line with the spatially separate functions for SHR/SCR in regulating QC identity, CEI asymmetric cell division and ground tissue specification, there may be different networks to sequester SHR in the QC, CEI daughter and endodermis [29].
1.3.1 Downstream targets of SHR and SCR
Meta analysis of three different microarray expression data sets identified eight putative direct targets of SHR: SCR and three additional TFs: SCARECROW-LIKE 3 (SCL-3), MAGPIE (MGP), NUTCRACKER (NUC), SNEEZY/SLEEPY2, Br6xo2, a receptor like kinase, and tropinone reductase [24]. All targets were expressed in a subset of the SHR expression domain, and a direct interaction with SHR was confirmed for four of the eight using ChIP-PCR [24]. Interestingly, SCR also binds to SCL-3, MAGPIE, and NUTCRACKER promoters at similar sites as SHR [23]. SHR binding to these sites requires SCR, as SHR binding was abolished in the scr-4 mutant background. Microarray analysis of scr and shr mutants demonstrated the functional interdependence of SCR and SHR. Nearly all of the identified SHR direct targets, as well as a number of indirect targets, had reduced expression in the scr background [23].
1.4 Additional inputs for stem cell maintenance
A short-range signal must pass from the QC to the surrounding stem cells to prevent them from differentiating. This signal may be WOX5 or its direct target. WOX5 is a homeobox TF homologous to WUSCHEL (WUS), which is involved in stem cell maintenance in the shoot [31]. WOX5 is expressed in the QC, and mutants have an abnormally shaped QC [32]. Expression of QC-specific markers in wox5 mutants is unpertubed, suggesting that WOX5 does not play a major role in QC specification. However, columella stem cells differentiate in the mutant and when introgressed into scr shr or plt1 plt2 double mutants the wox5 mutation causes the proximal meristem to differentiate. Thus WOX5 is required non-cell autonomously to keep the distal stem cells undifferentiated, and is either redundantly required to keep the proximal meristem undifferentiated or has a stem cell independent function in the proximal meristem [32] (Figure 2). Notably, WOX5 and WUS are interchangeable in stem cell maintenance, as expression of WUS cDNA from the WOX5 promoter completely rescues stem cell maintenance in the root meristem [32].
Recent work has demonstrated similarities between animal and plant stem cell maintenance. In animals, RETINOBLASTOMA (RB) regulates the decision to enter the cell cycle at the G1 restriction point; RB family members are major players in stem cell biology [33]. Using tissue specific RNAi, Wildwater et al. (2005) showed that modulation of Arabidopsis RETINOBLASTOMA-RELATED (RBR) activity is crucial for stem cell maintenance, as reduction in RBR transcript levels results in an increase in the number of stem cells, and overexpression leads to stem cell differentiation. RBR expression is reduced in the scr mutant background; genetic analysis indicated that SCR functions upstream of RBR. Since SCR is required for QC identity, the authors suggest that SCR functions in the QC to downregulate RBR, which suppresses additional layers of stem cells. Analysis of RBR interactions with known cell cycle modulators suggests that RBR operates through a pathway analogous to animal RB [34].
Additional factors involved in cell division and cell cycle regulation play a role in RM regulation, but their link with stem cell maintenance is less clear [35–39].
1.4.1 Chromatin modifications in root meristem maintenance
The FASCIATA (FAS) genes, FAS1 and FAS2, encode members of the two largest subunits of the chromatin assembly complex CAF-1, and are required for RM maintenance, function and stable organization. fas mutants are defective in both the shoot and root meristems and display ectopic, stochastic SCR expression [40]. FAS may be important for stable gene expression in the RM and for maintaining epigenetic states, as genes typically silenced in wt plants are transcriptionally active at low and random levels in fas mutants [41]. Evidence for its importance in maintaining chromatin structure comes from its role in root epidermal pattening, discussed below.
1.5 Hormone interactions in root meristem maintenance: More than auxin, auxin, auxin
In addition to auxin, the phytohormones cytokinin, ethylene, gibberellic acid, and CLAVATA/ESR (CLE) peptides have been implicated in RM maintenance. Exogenous cytokinin application reduced meristem size [42], and overexpressing cytokinin oxidases resulted in a larger meristem [42, 43]. The effects of cytokinin are specific to the vascular tissue in the TZ, where cell differentiation starts. The cytokinin signaling protein AHK3, and downstream regulators ARR1 and ARR12 may regulate cell differentiation in the vasculature [42]. No changes were seen in meristem size when cytokinin oxidases were overexpressed in pin2pin3pin7, indicating that the effects of cytokinin depend on the correct distribution of auxin in the RM. Thus meristem size may be fixed by the combined inputs of auxin and cytokinin.
Cytokinins do not appear to affect QC identity or stem cell maintenance [42]. In contrast, ethylene promotes cell division in the QC. Mutants that produce excess ethylene have an increased number of cell divisions in the QC [44]. These divisions could be suppressed with an ethylene inhibitor. The extra cells maintained QC identity, and stem cells did not differentiate, indicating that low mitotic activity is not required for QC function. Ethylene plays a role in a number of biotic and abiotic stresses [45, 46] and its regulation could be one way the root modifies development under different environmental regimes [44].
Several lines of evidence suggest that gibberellins may play a role in the SHR-SCR pathway. Paquette and Benfey (2005) found that the initiation of middle cortex formation – an additional layer of cortex arising from endodermal cells later in postembryonic development – was negatively regulated by SCR and GA. SCR and GA are thought to act independently and additively in the process. scr mutants produced earlier and ectopic divisions. Middle cortex formation was delayed by high doses of GA in wt and scr, although the scr mutant was more sensitive than wt. Surprisingly, the shr mutant does not have middle cortex formation, and is insensitive to GA, suggesting that SHR plays a SCR-independent role in middle cortex formation. This suggests that SHR may be involved in the GA pathway. GA may act to promote middle cortex formation by repressing SHR action (or a function that requires SHR), independent of SCR [47]. Additional evidence for the role of GA in the SHR-SCR pathway comes from observations that the SHR target gene, SNEEZY, plays a role in the GA pathway [48].
CLE peptides are small proteins (<15kD) with a 14AA CLE motif. A member of the family, CLV3, regulates the size of the stem cell pool in the shoot meristem [49]. Several groups have shown that overexpression or root-specific expression of CLE peptides affects RM maintenance [50–53]. Since the SAM and RM may have similar stem cell regulatory mechanisms [2, 32], this suggests that there may be a receptor for the CLE peptides in the RM. Some CLE peptides also play a role in secondary xylem differentiation [54].
2.1 Vascular patterning: xylem or phloem?
A plant’s vascular system connects its aerial organs to each other and to their underground counterparts, and transports water (through the xylem) and nutrients (through the phloem). The stele is the central core of vascular tissue in the Arabidopsis root, and consists of multiple cell types that arise from the provascular initials proximal to the QC [55] (Figure 1).
In contrast to the initial cells of the ground tissue and epidermis, little is known about the transcriptional networks that determine cell fate in the stele. Forward and reverse genetic screens have identified a large number of mutants with defects in vascular development, but most of these have been in the stem or leaves, or have not been well characterized in the root. These analyses have shown that hormones such as auxin and brassinosteroids, and three classes of TFs (class III HD-Zip, KANADI, and YABBY) play roles in vascular patterning [56, 57]. Here we discuss transcriptional networks and hormone signaling in the early events of postembryonic vascular differentiation in the root. For a comprehensive review discussing secondary vascular development see Fukuda et al. (2004).
Unlike the outer layers of the root, the stele has bilateral symmetry with two diametrically opposed xylem poles (Figure 1). Surrounding the central xylem and phloem is the pericycle, a heterogenous tissue consisting of two cell types, xylem pole pericycle (XPP) and phloem pole pericyle (PPP). Post-embyronic divisions in XPP cells result in lateral root outgrowth [58], while cells of the PPP are mainly quiescent [59].
One well-characterized mutant with deficiencies in the root vasculature is Altered Phloem Development (APL) (Table 1). APL encodes a coiled-coil MYB TF that is required both for phloem related cell divisions and phloem differentiation [60]. The apl phenotype displays treachery element-like (TE; a xylem cell type) cells in the phloem position, and is seedling lethal. Expression of APL throughout the stele does not result in ectopic phloem, suggesting that APL is required but not sufficient for phloem formation. Moreover, ectopic expression of APL under the WOODEN LEG (WOL) promoter represses TE differentiation, suggesting that APL plays a role in repressing the early stages of xylem differentiation [60]. Recently, screening of enhancer trap lines for protophloem marker expression identified four genes involved in protophloem development [61]. One of these genes encodes BREVIS RADIX, a novel transcriptional activator that quantitatively contributes to root length [62].
TABLE 1.
Major Vascular Development Genes in the Arabidopsis Root
| Gene | Mutant Phenotype | Ectopic or overexpression phenotype | Identity | Functions | References |
|---|---|---|---|---|---|
| ALTERED PHLOEM DEVELOPMENT (APL) | Defective phloem differentiation; xylem TE- like cells in phloem pole position; seedling lethal | Expression throughout the stele does not result in ectopic phloem, but represses TE differentiation | MYB transcription factor (TF) | Promotes phloem differentiation; represses stages of xylem differentiation | 60, 61 |
| WOODEN LEG (WOL)/CRE1/AHK4 (ARABIDOPSIS HISTIDINE KINASE 4) | Short, determinate root with few vascular initials. Vascular tissue differentiates only as protoxylem. | Cytokinin receptor and histidine kinase, CRE1/AHK4. The wol allele of CRE1 has no cytokinin binding activity and cannot phosphorylate downstream partners in the presence of cytokinin. | Controls asymmetric cell division of root vascular initials. Phosphorylates AHP proteins in the presence of cytokinin; acts as a phosphatase in the absence of cytokinin | 65–69 | |
| LONESOME HIGHWAY (LH) | Diarch symmetry lost, only one strand of provascular tissue (protoxylem and protophloem); lateral roots produced only on one side of the root; only half the wt number of cells in the stele; columella stem cells differentiate at 13 days | Plant specific protein with some similarity to bHLH Tfs. Can activate transcription in the Y2H system. | Required to establish and maintain normal vascular cell numbers and for meristem maintenance | 75 | |
| VASCULAR RELATED NAC DOMAIN 6 (VND6) | No phenotype (T-DNA insertion lines) | Induces metaxylem formation in vascular and nonvascular cells of root | NAC domain TF | Promotes metaxylem cell fate; master regulator of metaxylem development? | 76 |
| VASCULAR RELATED NAC DOMAIN 7 (VND7) | No phenotype (T-DNA insertion lines) | Induces protoxylem formation in vascular and nonvascular cells of root | NAC domain TF | Promotes protoxylem cell fate; master regulator of protoxylem cell fate? | 76 |
| ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6) | Sporadic protoxylem differentiation, disrupted cytokinin signaling, identified in a screen for wol suppressor mutations | 1 of 6 AHPs in Arabidopsis. In response to cytokinin, AHPs are phosphorylated by AHKs, and in turn, phosphorylate B-type ARRs. AHP6 is likely not a true AHP as it inhibits phosphotransfer from AHPs to ARRs | Promotes protoxylem development by restricting cytokinin signaling to specific domains within the stele. | 64 |
Several lines of evidence suggest a role for SHR in vascular patterning. Three of the SHR direct targets are expressed in the stele, and one, Br6ox2 is involved in biosynthesis of brassinosteroids [24]. Additionally, shr mutants have altered vascular initials and subtle alterations in the stele [24].
2.2. Cytokinin signaling and the root vasculature
Cytokinin signaling is mediated in several steps through a two-component histidine kinase (HK) pathway [63]. The ARABIDOPSIS HISTIDINE KINASE (AHK) cytokinin receptors are activated by cytokinin and phosphorylate ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEINs (AHPs) [64], which translocate to the nucleus and phosphorylate B-type ARABIDOPSIS RESPONSE REGULATORS (ARRs). B-type ARRs in turn activate transcription of downstream signaling genes. Some of these, such as the A-type ARRs, act as negative feedback signals on the cytokinin pathway [63]. In the stele, cytokinin negatively regulates protoxylem formation, and is necessary for the formation and maintenance of cell types in the stele other than protoxylem [65] (Figure 1).
One of the first hints of the importance of cytokinin signaling in vascular differentiation came with the identification of the wol mutant [19, 65]. WOL controls the asymmetric cell division of root vascular initials [65] (Table 1). Mutants have a short, determinate root, with fewer vascular initials. Vascular tissue differentiates only as protoxylem, as the periclinal divisions required for procambium differentiation are absent [65–69]. WOL encodes a cytokinin receptor, known as CRE1 [68, 69]. CRE1 is a histidine kinase with a putative extracellular cytokinin binding domain and intracellular kinase signaling and receiver domains [68]. The WOL allele of CRE1 encodes a kinase with a single amino acid change [CRE1(T278I)] that eliminates the cytokinin binding activity [65, 69]. In response to cytokinin, CRE1 phosphorylates downstream AHP proteins. In the absence of cytokinins, CRE1 acts as a phosphatase [66] (Table 1).
There are two additional cytokinin binding receptors in Arabidopsis, AHK2 and AHK3, and single or double mutants in either of these have normal root lengths and vascular cell numbers [66]. Roots of plants with triple knockouts of all three receptors resemble wol, suggesting that these genes act in a redundant manner [64, 66, 70, 71]. Surprisingly, null alleles at the CRE1 locus can complement the wol phenotype, and a screen for suppressor mutants of wol identified several with stop codons in CRE1[66, 67]. The cytokinin induced gene ARR15 [72] is expressed in the cre1–2 null allele (although lower than wt), but is further reduced in the wol mutant [66]. These results suggest that although wol is genetically recessive, it exerts negative effects on cytokinin signaling [66]. In line with this, regions of the root where CRE1 is expressed more strongly than either AHK2 or AHK3 are those in which the wol phenotype is evident [66].
In a recent study, Mähönen et al. (2006b) showed how this negative signaling occurs. In vitro kinase assays demonstrated that CRE1 has cytokinin-dependent kinase activity and cytokinin-independent phosphatase activity. Although CRE1 complemented a yeast histidine kinase mutant in the presence of cytokinin, wild-type yeast expressing CRE1 were not viable in the absence of the hormone, presumably because the phosphatase activity of CRE1 interfered with a signaling pathway. Thus in the presence of cytokinin, the kinase activity of CRE1can overcome its phosphatase activity. The WOL protein, CRE1(T278I), has weak kinase activity, and could not complement the yeast histidine kinase mutant in the presence of cytokinins. Moreover, wild-type yeast expressing the mutant protein were not viable in the presence or absence of cytokinins, demonstrating that the negative activity of CRE1l(T278I) is constitutively active. This indicates that protein encoded by the wol allele is in a “locked” position, which cannot phosphorylate downstream partners nor properly mediate cytokinin signaling [66].
Similar assays showed that the two other AHKs in Arabidopsis, AHK2 and AHK3, have kinase activity but lack observable phosphatase activity. The in planta and yeast assays suggested that the members of the AHK family have different kinase and phosphatase activities in response to diverse levels of cytokinin [66].
Identification of the ARRs that play a role in cell fate in the stele is still at an early stage, but recent work suggests that the B-type ARRs ARR10 and ARR12 might act downstream of AHK3 and AHK4 [42, 73], and the A-type ARRs ARR15 and ARR16 may be downstream of AHK4 [72, 74]. The differential activities of AHKs in response to distinct cytokinin levels and the differential interaction of AHKs with different ARRs may account for the wide range of cytokinin responses in the plant [66].
The screen for wol suppressor mutations identified another downstream component of the cytokinin signaling pathway. In contrast to wol, ahp6 mutants display sporadic protoxylem differentiation, while metaxylem differentiates normally [64] (Table 1). AHP6 transcription is repressed by cytokinin. As described above, AHPs are components of the cytokinin signaling pathway that are phosphorylated by AHKs and in turn phosphorylate ARRs. However, unlike the other AHPs in Arabidopsis, AHP6 is likely not a true phosphotransfer protein, but instead functions to impede cytokinin signaling by inhibiting phosphotransfer from AHPs to ARRs. In accordance with this hypothesis, cytokinin signaling is disrupted in ahp6 mutants. In wt roots AHP6 expression is restricted to protoxylem and associated pericycle, but in roots of wol and cre1 ahk2 ahk3 triple mutants, the AHP6 expression domain extends throughout the vascular bundle. Moreover, plants expressing cytokinin oxidase under the aph6 promoter rescue the APH6 phenotype. Thus, APH6 promotes protoxylem development by restricting cytokinin signaling to specific domains within the stele [64]. In turn, cytokinin regulates the spatial domain of AHP6. The regulatory circuit in which cytokinin restricts the domain of its target, AHP6, which in turn restricts the domain of cytokinin signaling is reminiscent of the interaction between auxin and the PLT and PIN genes.
2.3 bHLH and NAC domain TFs contribute to vascular cell fate
A screen of 48,000 M2 lines for mutations in the enhancer trap J0121:GFP, which marks the xylem pole pericycle, identified five alleles of LONESOME HIGHWAY (LHW) [75]. lhw mutants lose the diarch symmetry of wt Arabidopsis roots and have just one strip of provascular tissue (protoxylem and protophloem) in both primary and lateral roots (Table 1). Consistent with this, mutants produce lateral roots only on one side of the root. Closer examination of the stele revealed that mature lhw mutants consistently have half the number of cells in the stele as wt, and thus LHW is required to establish and maintain normal vascular cell number. Surprisingly, the SCR promoter (pSCR::GFP) was not expressed in the QC of seven day old lhw mutants, but was present in torpedo-stage embryos. Further analysis showed that QC markers were lost over time and that columella initials in lhw mutants differentiated by 13 days, suggesting that LHW is not necessary for formation of the root meristem but for meristem maintenance. LHW encodes a novel, plant-specific protein with minimal similarity to bHLH domain proteins. The protein can activate transcription in the Y2H system, indicating that LHW might act as a transcription factor. Double mutant analysis with wol revealed a possible role for cytokinins in the lhw phenotype. The role of cytokinin in the LHW pathway may be more complex, as cytokinins repress protoxylem differentiation [64], but LHW does not [75].
Using ATH1 gene chips and an in vitro xylem element vessel differentiation system with Arabidopsis suspension cells, Kubo et al. [76] identified two genes involved in protoxylem and metaxylem specification. VASCULAR RELATED NAC-DOMAIN 6 and VASCULAR RELATED NAC-DOMAIN 7, are, as their names suggest, members of the plant specific NAC domain transcription factor family (Table 1). Though T-DNA insertion lines in either VND6 or VND7 had no obvious phenotype, overexpression of VND6 and VND7 induced metaxylem (VND6) and protoxylem (VND7) differentiation in vascular and nonvascular cells of both the Arabidopsis root and poplar leaves. This suggests that these genes may be master regulators of xylem cell fate, and may be evolutionary conserved. VND6 and VND7 were similar to an EST identified in a similar xylem induction system in Zinnia [77]. Several other reports have used reverse genetics to identify genes involved in secondary growth in Arabidopsis, Zinnia, or poplar [78–83], and it will be interesting to learn whether any of these genes are also involved in the early events of vascular development.
3.1 A complex network mediates epidermal cell fate
The epidermis is the outermost layer of the root and functions in nutrient and water acquisition as well as protecting the root from water loss and pathogen invasion. In Arabidopsis, epidermal cells are arranged in long files along the longitudinal axis of the root, and their hair or non-hair fate depends on the arrangement of cortex cells beneath them [84]. Those that overlie two cortical cells are destined to become hair cells (H; trichoblasts), while those lying directly above one cortical cell will differentiate into non-hair cells (N; atrichoblasts). This arrangement results in approximately 16 cell files (8 hair cells and 8 non-hair cells) [85]. The correlation between cell fate and position suggests that a positional signaling mechanism operates in epidermal cells [86]. This position-dependent patterning occurs in Arabidopsis embryos, and is continued post-embryonically [87]. Consistent with this hypothesis, differences between trichoblasts and atrichoblasts are visible prior to the onset of hair formation. Compared to atrichoblasts, trichoblasts have increased cytoplasmic density [22, 88], delayed vacuolation [22], faster cell-division rate [89], smaller cell length [88, 90], and unique surface epitopes [88]. Despite the differences in cells before root hairs develop, a change in position can lead to a change in cell fate [91]. Here we focus on the network of TFs involved in epidermal cell fate specification in the root as well as recent work highlighting the importance of chromatin modification in root epidermal cell fate specification.
3. 2 Lateral inhibition and feedback loops control epidermal cell fate specification
Work over the last 20 years has identified a network of TFs involved in regulating the cell fate of epidermal cells. Genetic analysis has identified eight transcription factors required for either N or H cell fate specification [85, 86, 92]. Five genes encoding the TFs GLABRA2 (GL2), GLABRA 3 (GL3), ENHANCER of GLABRA3 (EGL3), TRANSPARENT TESTA GLABRA (TTG), and WEREWOLF (WER) are necessary for N cell fate. Mutations in these genes result in an excess of root hairs [22, 90, 93, 94]. Three TF – encoding genes, CAPRICE (CPC), TRIPTYCHON (TRY), and ENHANCER of TRIPTYCHON AND CAPRICE (ETC) are required for the specification of H cell fate [95–97]. Mutations in any of these genes result in too few root hairs compared to wild type. Current models (described below; Figure 3) suggest that epidermal patterning operates through a combination of lateral inhibition and positive and negative feedback loops among the proteins encoded by these genes.
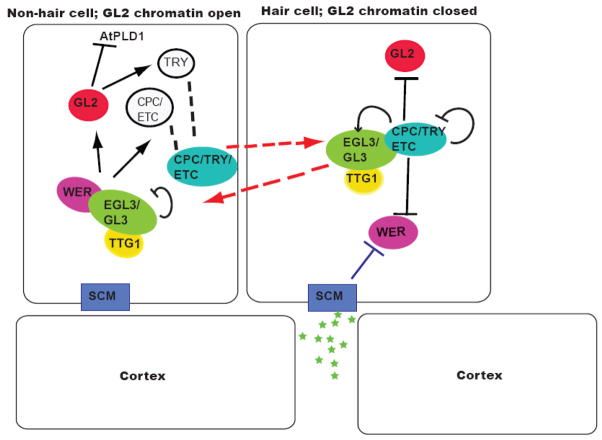
Figure 3.
A model for epidermal cell fate specification. In N cells WER promotes expression of GL2 and CPC/ETC1; GL2 in turn positively regulates TRY. CPC (and probably TRY and ETC1) moves to H cells where it (probably with TRY) inhibits WER, GL2, TRY ETC1, and its own expression. CPC, TRY, and ETC1 are partially redundant MYBs that promote H cell fate by inhibiting formation of the WER complex, which prevents the induction of GL2 and results in the specification of H cell fate. In H cells, CPC promotes expression of GL3 and EGL3, which move to N cells where they form part of the WER complex that induces CPC/ETC1 and GL2. WER-GL3-EGL3 negatively regulate GL3/EGL3 in N cells. In this way, cells with higher concentrations of the WER complex activate GL2 and form N cells, while cells with higher concentrations of the CPC complex inhibit GL2 and form H cells. The SCM kinase perceives an unknown signal and further inhibits WER in the H position. Broken arrows indicate protein movement; broken lines indicate proteins that will form a complex Stars are (unknown) positional cues. See text for additional details.
Although WER, an R2R3 MYB protein, is preferentially expressed in N cells [94], several observations have suggested that it is really the ‘master regulator’ of root epidermal cell fate. First, WER positively regulates CPC [98–100], a gene that promotes H cell formation (see below). Furthermore, overexpression of WER in the wer-1 background can uncouple cell fate from position, resulting in a variable pattern of H and N cells and suggesting that WER responds to a positional signal to pattern epidermal cells [98, 99].
Yeast two-hybrid analysis demonstrated that WER interacts with the partially redundant bHLH TFs GL3 and EGL3 [94] and that GL3 can interact with the WD40 repeat containing protein TTG1 [101, 111] (Figure 3). The WER- EGL3-GL3-TTG complex (WER complex) promotes activation of GL2 [93, 98], which encodes an HD-ZIP homeodomain TF [90, 102, 103]. WER binds to two sites in the GL2 promoter [100]. Activation of GL2 represses H cell fate in part by repressing the phospholipase AtPLDζ1, and associated signaling events [104].
In addition to the activation of GL2, the WER complex also activates CPC transcription [98–100]. WER binds to a 69 base pair region within the CPC promoter [99, 100]. CPC is a small, 94 amino acid, truncated MYB protein with no activation domain [97, 99, 105]. CPC promotes H cell fate in conjunction with two additional small MYBs, TRY and ETC1 [95, 96, 106] (Figure 3). GL2 promotes expression of TRY in cells in the N position [106]. CPC, GL2, and ETC1 transcripts are positively regulated by WER and TTG in the N cell position [106].
Although CPC expression is activated in N cells by the WER complex, the protein (and probably TRY and ETC1) moves laterally into neighboring H cells [107] where it (and probably TRY) negatively regulates expression of WER, GL2, TRY, ETC1, and itself [95–98, 105–108] (Figure 3). This movement probably requires tissue-specific factors, as the protein could not move when expressed in the stele under the SHR promoter [107]. Yeast two-hybrid analysis revealed that CPC interacts with GL3 and EGL3 [93]. This binding most likely forms a complex of CPC-GL3-EGL3-TTG that cannot activate GL2 [93, 98, 105]. Both CPC and TRY are needed to repress GL2 in H cells [106]. Without GL2, the H fate cannot be repressed, and it becomes the default pathway [109].
As well as forming a complex with GL3 and EGL3, CPC positively regulates their expression in H cells [110]. Translational fusions with GL3 showed that the protein moved into neighboring H cells [110]. Thus, GL3 and EGL3 promote transcription of CPC in the N position, and their transcription is in turn promoted by CPC in the H position [110], while WER-GL3-EGL3 negatively regulate transcription of GL3 and EGL3 in the N position [110]. Thus, the relative levels of WER and CPC, determined by lateral inhibition and feedback loops, are important for epidermal cell fate. Competition between CPC and WER for the GL3-EGL3-TTG complex likely results in the H or N cell fate outcome [93, 98, 111] (Figure 3).
3.3 Positional signaling is mediated by the SCRAMBLED kinase
To identify genes with roles in positional signaling, Kwak et al. [112] performed a screen with an EMS mutagenized pGL2::GUS line. They identified one line with random patterns of GUS expression in the epidermis, suggesting a mutation in the signal that mediates H and N cell fates. The mutant gene encoded a leucine-rich-repeat receptor kinase, SCRAMBLED (SCM) that is expressed throughout the developing root. Consistent with studies suggesting WER is the master regulator of epidermal cell fate, and possibly the target of positional cues [94, 98], SCM was shown to negatively regulate WER, and thus mediate positional signaling in part through this gene [113]. Although this brings us closer to an understanding of how epidermal cells recognize their position, we still do not know the signal to which the SCM kinase responds.
Interestingly, in an independent report, Chevalier et al. [114] identified a mutant, with a mutation in the same gene as SCM. The strubbelig (sub) mutant was characterized primarily in aerial organs, and was found to have defects in ovule development, with misoriented cell division planes and altered morphology. Additionally sub mutants are also shorter, and have stems that twist randomly. Though a similar role in positional signaling was not suggested, the fact that sub mutants display altered ovule development, normally a very stereotypical process, suggests that there may be commonalities in its pleiotropic effects [115]. Biochemical and genetic data suggest that SCM/SUB is catalytically inactive [114]. Understanding how SCM/SUB signals is an important question for the future.
3.4 Chromatin involvement in root hair patterning
Three recent reports have demonstrated the importance of epigenetics in root epidermal patterning. Costa and Shaw [116] showed that GL2 expression depended on proper chromatin confirmation. Using a GL2BAC probe in FISH experiments, the authors found that the GL2 chromatin state is open in N cells, and closed in H cells. In cpc and wer mutants, the GL2 chromatin state is open in all epidermal cells. This indicates that CPC is necessary for the closed chromatin state in H cells, and that cell fate specification is not required to maintain the open state. Presumably, the open confirmation in N cells allows WER binding, GL2 expression and subsequent repression of H fate. GL2 is ectopically expressed in the fas2 mutant (discussed above). In fas2 mutants, the wt hybridization pattern of the GL2BAC probe is lost, indicating that FAS2 is necessary for organizing the proper chromatin state around GL2. Additional assays revealed that the chromatin state around GL2 is remodeled immediately after each cell cycle, depending on underlying positional cues. Root hair formation is known to be environmentally responsive [117], and the ability to change fate might be an important factor in adapting to changing environments such as low iron or phosphorous [117–119]. Thus an epidermal cell may be able to quickly change its fate through modification of its chromatin structure.
In addition to the chromatin state, histone modifications are also involved in epidermal patterning. Treatment of Arabidopsis seedlings with trichostatin A (TSA), an inhibitor of histone deactylases (HDACs) promoted H cell fate at the N position, resulting in a remarkable increase in root hairs [120]. This effect was concentration-dependent and reversible, as plants switched to media without TSA lost the ectopic root hairs. Of eight genes involved in epidermal patterning, microarray analysis revealed that the expression of three was affected by TSA. The extra acetylation increased expression of CPC and GL2, while decreasing that of WER. Mutant analysis suggested that HDA18 is involved in the process [120].
Another report identified a role for histone modifications in epidermal cell patterning. Caro et al. (2007) [121] identified the GL2 EXPRESSION MODULATOR (GEM), a protein that interacts in the yeast two-hybrid assay with CDT1. CDT1 is a component of the pre-replication complex necessary for the initiation of DNA replication. Mutants in GEM had increased cell division levels in the epidermis and cortex and decreased numbers of root hairs. GEM levels were inversely correlated with CPC and GL2, and ChIP experiments showed that GEM bound the GL2 and CPC promoters, suggesting that GEM plays a role both in cell division and epidermal hair patterning. Further, GEM interacted with TTG1 in a pull-down assay. Genetic analysis suggested that GEM repression of CPC and GL2 occurs in part through TTG1. Examination of the methylation status of the CPC and GL2 promoters showed that in gem-1 mutants CPC and GL2 had increased levels of H3K9me3, and decreased levels of H3K9me2, marks of active and silent chromatin, respectively. Further, these changes in histone modifications were located upstream of the GL2 and CPC open reading frames. The authors propose a model in which GEM represses CPC and GL2 by altering histone modifications in their promoters. It will be interesting to understand the relationship between GEM and HDA18. It is also interesting to speculate as to what other chromatin modifications might exist, not only for epidermal patterning genes but other genes involved in cell fate specification in the Arabidopsis root.
4.1 The Root Map: High-resolution global gene expression in the root
An understanding of the transcriptional pathways that underlie developmental processes is vital if we are to fully understand the mechanisms that determine cell fate and tissue organization. The vast transcriptional complexity identified by the recent publication of a high resolution transcriptional map of nearly every cell type in the Arabidopsis root and 13 developmental time points along the longitudinal axis will greatly increase our understanding of these processes [6]. Using fluorescence activated cell sorting of cell-type specific markers [122], Brady et al. [6] profiled 14 of the 15 cell types in the root. In addition, they microdissected individual roots along the longitudinal axis into 13 developmental stages consisting of 3 to 5 cells each in length. Additionally, in line with previous work that profiled 5 root cell types [5], transcription output from individual cell types was more informative than that from the whole root. Surprisingly, analysis of gene ontology (GO) category enrichment showed that most GO terms were associated with one cell type, while relatively few generalized GO terms were associated with more than one cell type. Fifty-one radial and 40 longitudinal dominant expression patterns were identified; 17 of the 51 radial patterns were cell-type specific, and others were found among cell types with known relationships. Novel relationships among cell types were discovered, as some patterns were found to be shared between disparate cell types such as the columella and xylem. Surprisingly, 17 of the 40 longitudinal patterns showed fluctuation in expression along this axis. By identifying sets of coregulated genes within the patterns, the authors were able to verify biological processes known to correlate with specific cell types and predict novel processes and targets for cell types that share a transcriptional program [6].
5.1 Conclusions
The positional information that determines cell fate in the Arabidopsis root is established through the combinatorial action of different positive and negative signals. Common themes determining diverse cell types have emerged, including regulatory loops of TFs and hormones that serve as repressors for one cell type but activators for another. The high resolution root map will allow us to form new, testable hypotheses about the interaction of TFs with downstream developmental processes and will reveal novel networks in cell-type specific processes, leading to a heightened understanding of cell fate specification. Cell-type specific transcription profiling under different environmental conditions will identify environment-invariant TFs that may contribute to the determination of cell fate. Finally, the integration of transcriptional networks with protein and metabolic data will enrich our understanding of the overall network design necessary for the specification of each cell type in the root.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jiang K, Feldman LJ. Regulation of root apical meristem development. Annual Review of Cell and Developmental Biology. 2005;21:485–509. doi: 10.1146/annurev.cellbio.21.122303.114753. [DOI] [PubMed] [Google Scholar]
- 2.Scheres B. Stem-cell niches: nursery rhymes across kingdoms. Nature Reviews Molecular Cell Biology. 2007;8:345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- 3.vandenBerg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 4.Benfey PN, Scheres B. Primer - Root development. Current Biology. 2000;10:R813–R815. doi: 10.1016/s0960-9822(00)00814-9. [DOI] [PubMed] [Google Scholar]
- 5.Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 6.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 7.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Annals of Botany. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- 9.Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, Palme K. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108:661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 10.Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg G, Hooykaas PJJ, Palme K, Offringa R. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 11.Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. A molecular framework for plant regeneration. Science. 2006;311:385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- 13.Vieten A, Sauer M, Brewer PB, Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends in Plant Science. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 15.Grieneisen VA, Xu J, Maree AFM, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- 16.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 18.Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root Development in Arabidopsis - 4 Mutants with Dramatically Altered Root Morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- 19.Scheres B, Dilaurenzio L, Willemsen V, Hauser MT, Janmaat K, Weisbeek P, Benfey PN. Mutations Affecting the Radial Organization of the Arabidopsis Root Display Specific Defects Throughout the Embryonic Axis. Development. 1995;121:53–62. [Google Scholar]
- 20.Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 22.Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG Gene Is Required to Specify Epidermal-Cell Fate and Cell Patterning in the Arabidopsis Root. Developmental Biology. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- 23.Cui HC, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 24.Levesque MP, Vernoux T, Busch W, Cui HC, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, Scheres B, Benfey PN. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. Plos Biology. 2006;4:739–752. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiLaurenzio L, WysockaDiller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 26.Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes & Development. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes & Development. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sena G, Jung JW, Benfey PN. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131:2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- 29.Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes & Development. 2007;21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Current Biology. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 31.Carles CC, Fletcher JC. Shoot apical meristem maintenance: the art of a dynamic balance. Trends in Plant Science. 2003;8:394–401. doi: 10.1016/S1360-1385(03)00164-X. [DOI] [PubMed] [Google Scholar]
- 32.Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 33.Galderisi U, Cipollaro M, Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25:5250–5256. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- 34.Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 35.Willemsen V, Wolkenfelt H, de Vrieze G, Weisbeek P, Scheres B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development. 1998;125:521–531. doi: 10.1242/dev.125.3.521. [DOI] [PubMed] [Google Scholar]
- 36.Blilou I, Frugier F, Folmer S, Serralbo O, Willemsen V, Wolkenfelt H, Eloy NB, Ferreira PCG, Weisbeek P, Scheres B. The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes & Development. 2002;16:2566–2575. doi: 10.1101/gad.237302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serralbo O, Perez-Perez JM, Heidstra R, Scheres B. Non-cell-autonomous rescue of anaphase-promoting complex function revealed by mosaic analysis of HOBBIT, an Arabidopsis CDC27 homolog. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13250–13255. doi: 10.1073/pnas.0602410103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inagaki S, Suzuki T, Ohto M, Urawa H, Horiuchi T, Nakamura K, Morikami A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18:879–892. doi: 10.1105/tpc.105.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki T, Inagaki S, Nakajima S, Akashi T, Ohto M, Kobayashi M, Seki M, Shinozaki K, Kato T, Tabata S, Nakamura K, Morikami A. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant Journal. 2004;38:673–684. doi: 10.1111/j.1365-313X.2004.02074.x. [DOI] [PubMed] [Google Scholar]
- 40.Kaya H, Shibahara K, Taoka K, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/s0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 41.Ono T, Kaya H, Takeda S, Abe M, Ogawa Y, Kato M, Kakutani T, Scheid OM, Araki T, Shibahara K. Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes to Cells. 2006;11:153–162. doi: 10.1111/j.1365-2443.2006.00928.x. [DOI] [PubMed] [Google Scholar]
- 42.Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 43.Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ortega-Martinez O, Pernas M, Carol RJ, Dolan L. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science. 2007;317:507–510. doi: 10.1126/science.1143409. [DOI] [PubMed] [Google Scholar]
- 45.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 46.van Loon LC, Geraats BPJ, Linthorst HJM. Ethylene as a modulator of disease resistance in plants. Trends in Plant Science. 2006;11:184–191. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Paquette AJ, Benfey PN. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiology. 2005;138:636–640. doi: 10.1104/pp.104.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12771–12776. doi: 10.1073/pnas.0404287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laux T. The stem cell concept in plants: A matter of debate. Cell. 2003;113:281–283. doi: 10.1016/s0092-8674(03)00312-x. [DOI] [PubMed] [Google Scholar]
- 50.Hobe M, Muller R, Grunewald M, Brand U, Simon R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Development Genes and Evolution. 2003;213:371–381. doi: 10.1007/s00427-003-0329-5. [DOI] [PubMed] [Google Scholar]
- 51.Casamitjana-Martinez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Current Biology. 2003;13:1435–1441. doi: 10.1016/s0960-9822(03)00533-5. [DOI] [PubMed] [Google Scholar]
- 52.Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 2005;17:2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fiers M, Ku KL, Liu CM. CLE peptide ligands and their roles in establishing meristems. Current Opinion in Plant Biology. 2007;10:39–43. doi: 10.1016/j.pbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 54.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 55.Fukuda H. Signals that control plant vascular cell differentiation. Nature Reviews Molecular Cell Biology. 2004;5:379–391. doi: 10.1038/nrm1364. [DOI] [PubMed] [Google Scholar]
- 56.Ye ZH. Vascular tissue differentiation and pattern formation in plants. Annual Review of Plant Biology. 2002;53:183–202. doi: 10.1146/annurev.arplant.53.100301.135245. [DOI] [PubMed] [Google Scholar]
- 57.Carlsbecker A, Helariutta Y. Phloem and xylem specification: pieces of the puzzle emerge. Current Opinion in Plant Biology. 2005;8:512–517. doi: 10.1016/j.pbi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, Bennett M. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig S, Helariutta Y, Haseloff J, Chriqui D, Beeckman T, Nussaume L. Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiology. 2007;146:140–148. doi: 10.1104/pp.107.107870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonke M, Thitamadee S, Mahonen AP, Hauser MT, Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426:181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- 61.Bauby H, Divol F, Truernit E, Grandjean O, Palauqui JC. Protophloem differentiation in early Arabidopsis thaliana development. Plant and Cell Physiology. 2007;48:97–109. doi: 10.1093/pcp/pcl045. [DOI] [PubMed] [Google Scholar]
- 62.Mouchel CF, Briggs GC, Hardtke CS. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes & Development. 2004;18:700–714. doi: 10.1101/gad.1187704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller B, Sheen J. Advances in cytokinin signaling. Science. 2007;318:68–69. doi: 10.1126/science.1145461. [DOI] [PubMed] [Google Scholar]
- 64.Mahonen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Tormakangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 65.Mahonen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes & Development. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mahonen AP, Higuchi M, Tormakangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Current Biology. 2006;16:1116–1122. doi: 10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 67.de Leon BGP, Zorrilla JMF, Rubio V, Dahiya P, Paz-Ares J, Leyva A. Interallelic complementation at the Arabidopsis CRE1 locus uncovers independent pathways for the proliferation of vascular initials and canonical cytokinin signalling. Plant Journal. 2004;38:70–79. doi: 10.1111/j.1365-313X.2004.02023.x. [DOI] [PubMed] [Google Scholar]
- 68.Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 69.Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant and Cell Physiology. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- 70.Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuroha T, Ueguchi C, Sakakibara H, Satoh S. Cytokinin receptors are required for normal development of auxin-transporting vascular tissues in the hypocotyl but not in adventitious roots. Plant and Cell Physiology. 2006;47:234–243. doi: 10.1093/pcp/pci240. [DOI] [PubMed] [Google Scholar]
- 72.Kiba T, Yamada H, Mizuno T. Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant and Cell Physiology. 2002;43:1059–1066. doi: 10.1093/pcp/pcf121. [DOI] [PubMed] [Google Scholar]
- 73.Yokoyama A, Yamashino T, Amano YI, Tajima Y, Imamura A, Sakakibara H, Mizuno T. Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant and Cell Physiology. 2007;48:84–96. doi: 10.1093/pcp/pcl040. [DOI] [PubMed] [Google Scholar]
- 74.Kiba T, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T. The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant and Cell Physiology. 2003;44:868–874. doi: 10.1093/pcp/pcg108. [DOI] [PubMed] [Google Scholar]
- 75.Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development. 2007;134:2959–2968. doi: 10.1242/dev.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kubo M, Udagawa M, Nishibubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes & Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Demura T, Tashiro G, Horiguchi G, Kishimoto N, Kubo M, Matsuoka N, Minami A, Nagata-Hiwatashi M, Nakamura K, Okamura Y, Sassa N, Suzuki S, Yazaki J, Kikuchi S, Fukuda H. Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15794–15799. doi: 10.1073/pnas.232590499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, Van Montagu M, Sandberg G, Olsson O, Teeri TT, Boerjan W, Gustafsson P, Uhlen M, Sundberg B, Lundeberg J. Gene discovery in the wood-forming tissues of poplar: Analysis of 5,692 expressed sequence tags. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Milioni D, Sado PE, Stacey NJ, Domingo C, Roberts K, McCann MC. Differential expression of cell-wall-related genes during the formation of tracheary elements in the Zinnia mesophyll cell system. Plant Molecular Biology. 2001;47:221–238. [PubMed] [Google Scholar]
- 80.Oh S, Park S, Han KH. Transcriptional regulation of secondary growth in Arabidopsis thaliana. Journal of Experimental Botany. 2003;54:2709–2722. doi: 10.1093/jxb/erg304. [DOI] [PubMed] [Google Scholar]
- 81.Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell. 2004;16:2278–2292. doi: 10.1105/tpc.104.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao CS, Craig JC, Petzold HE, Dickerman AW, Beers EP. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiology. 2005;138:803–818. doi: 10.1104/pp.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ko JH, Beers EP, Han KH. Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Molecular Genetics and Genomics. 2006;276:517–531. doi: 10.1007/s00438-006-0157-1. [DOI] [PubMed] [Google Scholar]
- 84.Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annual Review of Plant Biology. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- 85.Serna L. Epidermal cell patterning and differentiation throughout the apical-basal axis of the seedling. Journal of Experimental Botany. 2005;56:1983–1989. doi: 10.1093/jxb/eri213. [DOI] [PubMed] [Google Scholar]
- 86.Dolan L. Positional information and mobile transcriptional regulators determine cell pattern in the Arabidopsis root epidermis. Journal of Experimental Botany. 2006;57:51–54. doi: 10.1093/jxb/erj037. [DOI] [PubMed] [Google Scholar]
- 87.Lin Y, Schiefelbein J. Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development. 2001;128:3697–3705. doi: 10.1242/dev.128.19.3697. [DOI] [PubMed] [Google Scholar]
- 88.Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. Clonal Relationships and Cell Patterning in the Root Epidermis of Arabidopsis. Development. 1994;120:2465–2474. [Google Scholar]
- 89.Berger F, Hung CY, Dolan L, Schiefelbein J. Control of cell division in the root epidermis of Arabidopsis thaliana. Developmental Biology. 1998;194:235–245. doi: 10.1006/dbio.1997.8813. [DOI] [PubMed] [Google Scholar]
- 90.Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW. The homeobox gene GLABRA 2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 91.Berger F, Haseloff J, Schiefelbein J, Dolan L. Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Current Biology. 1998;8:421–430. doi: 10.1016/s0960-9822(98)70176-9. [DOI] [PubMed] [Google Scholar]
- 92.Schiefelbein J. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Current Opinion in Plant Biology. 2003;6:74–78. doi: 10.1016/s136952660200002x. [DOI] [PubMed] [Google Scholar]
- 93.Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- 94.Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- 95.Kirik V, Simon M, Huelskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPCl gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Developmental Biology. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 96.Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO Journal. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 98.Lee MM, Schiefelbein J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell. 2002;14:611–618. doi: 10.1105/tpc.010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ryu KH, Kang YH, Park YH, Hwang D, Schiefelbein J, Lee MM. The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development. 2005;132:4765–4775. doi: 10.1242/dev.02055. [DOI] [PubMed] [Google Scholar]
- 100.Koshino-Kimura Y, Wada T, Tachibana T, Tsugeki R, Ishiguro S, Okada K. Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant and Cell Physiology. 2005;46:817–826. doi: 10.1093/pcp/pci096. [DOI] [PubMed] [Google Scholar]
- 101.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rerie WG, Feldmann KA, Marks MD. The GLABRA2 Gene Encodes a Homeo Domain Protein Required for Normal Trichome Development in Arabidopsis. Genes & Development. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- 103.DiCristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant Journal. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- 104.Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T. Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science. 2003;300:1427–1430. doi: 10.1126/science.1083695. [DOI] [PubMed] [Google Scholar]
- 105.Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development. 2002;129:5409–5419. doi: 10.1242/dev.00111. [DOI] [PubMed] [Google Scholar]
- 106.Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidennal patterning. Developmental Biology. 2007;311:566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 107.Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, Sato S, Tabata S, Okada K, Wada T. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development. 2005;132:5387–5398. doi: 10.1242/dev.02139. [DOI] [PubMed] [Google Scholar]
- 108.Esch JJ, Chen MA, Hillestad M, Marks MD. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant Journal. 2004;40:860–869. doi: 10.1111/j.1365-313X.2004.02259.x. [DOI] [PubMed] [Google Scholar]
- 109.Schiefelbein JW. Constructing a plant cell. The genetic control of root hair development. Plant Physiology. 2000;124:1525–1531. doi: 10.1104/pp.124.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bernhardt C, Zhao MZ, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- 111.Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 112.Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- 113.Kwak SH, Schiefelbein J. The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Developmental Biology. 2007;302:118–131. doi: 10.1016/j.ydbio.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 114.Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K. STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9074–9079. doi: 10.1073/pnas.0503526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guimil S, Dunand C. Patterning of Arabidopsis epidermal cells: epigenetic factors regulate the complex epidermal cell fate pathway. Trends in Plant Science. 2006;11:601–609. doi: 10.1016/j.tplants.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 116.Costa S, Shaw P. Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature. 2006;439:493–496. doi: 10.1038/nature04269. [DOI] [PubMed] [Google Scholar]
- 117.Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- 118.Schikora A, Schmidt W. Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiology. 2001;125:1679–1687. doi: 10.1104/pp.125.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Muller M, Schmidt W. Environmentally induced plasticity of root hair development in arabidopsis. Plant Physiology. 2004;134:409–419. doi: 10.1104/pp.103.029066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu CR, Liu C, Wang YL, Li LC, Chen WQ, Xu ZH, Bai SN. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14469–14474. doi: 10.1073/pnas.0503143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Caro E, Castellano MM, Gutierrez C. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature. 2007;447:213–U6. doi: 10.1038/nature05763. [DOI] [PubMed] [Google Scholar]
- 122.Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN. Cell type-specific expression profiting in plants via cell sorting of protoplasts from fluorescent reporter lines. Nature Methods. 2005;2:615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]