Abstract
Even though light is the driving force in photosynthesis, it also can be harmful to plants. The water-splitting photosystem II is the main target for this light stress, leading to inactivation of photosynthetic electron transport and photooxidative damage to its reaction center. The plant survives through an intricate repair mechanism involving proteolytic degradation and replacement of the photodamaged reaction center D1 protein. Based on experiments with isolated chloroplast thylakoid membranes and photosystem II core complexes, we report several aspects concerning the rapid turnover of the D1 protein. (i) The primary cleavage step is a GTP-dependent process, leading to accumulation of a 23-kDa N-terminal fragment. (ii) Proteolysis of the D1 protein is inhibited below basal levels by nonhydrolyzable GTP analogues and apyrase treatment, indicating the existence of endogenous GTP tightly bound to the thylakoid membrane. This possibility was corroborated by binding studies. (iii) The proteolysis of the 23-kDa primary degradation fragment (but not of the D1 protein) is an ATP- and zinc-dependent process. (iv) D1 protein degradation is a multienzyme event involving a strategic (primary) protease and a cleaning-up (secondary) protease. (v) The chloroplast FtsH protease is likely to be involved in the secondary degradation steps. Apart from its significance for understanding the repair of photoinhibition, the discovery of tightly bound GTP should have general implications for other regulatory reactions and signal transduction pathways associated with the photosynthetic membrane.
Chloroplasts of higher plants perform the conversion of light energy into chemically stored energy. However, light also can be harmful to plants. Light-induced inactivation of the photosynthetic process (photoinhibition) is an essential factor that limits plant growth with important physiological and agricultural consequences, particularly when combined with other abiotic stress factors (1, 2).
The primary target for photoinhibition is the oxygen-evolving photosystem II (PSII) (1–3), which is a membrane-bound protein complex consisting of at least 25 different subunits (4). Despite the large number of proteins, only two subunits, namely, the reaction center II D1 and D2 proteins, each possessing five trans-membrane domains, harbor all the redox ligands necessary for the PSII electron transport. Light stress to PSII results in the inactivation of electron transport and photooxidative damage to the reaction center (5–7). The plants survive this light stress by a repair mechanism in which damaged D1 protein is replaced by a new protein copy. Thus, the turnover rate of the D1 protein is several orders of magnitude higher than any other chloroplast protein (8, 9). A key step in the repair process is the degradation of the damaged D1 protein (2, 10, 11). Relatively little is known about this proteolytic process, which can be followed both in vivo and in vitro. A 23.5-kDa N-terminal fragment of the D1 protein was detected early in vivo by Greenberg et al. (12), and, later, the corresponding 10-kDa C-terminal fragment was identified by Canovas and Barber (13). This breakdown pattern indicates that the primary cleavage of the D1 protein occurs in the stromal loop between the transmembrane D and E helices exposed at the outer thylakoid surface. The two primary degradation products of the D1 protein also have been identified in isolated systems such as thylakoid membranes and oxygen-evolving PSII core and reaction center complexes (14–16).
There are sufficient experimental data to support the concept of a light-induced, structural modification of the D1 protein initiating the proteolytic process once oxidative damage has occurred (15, 17, 18). The degradation of the damaged protein is thought to occur via proteolysis (10, 14–16, 19) although the protease(s) involved have not been identified. Chemical cleavage of the D1 protein by active oxygen species (20, 21) also has been proposed.
In support of a degradation mechanism involving enzymatic cleavage, the photoinactivation of PSII electron transport can proceed readily at low temperatures, whereas the degradation of the D1 protein is a temperature-sensitive and light-independent subsequent event (10). Furthermore, the activity of the primary proteolytic cleavage has been demonstrated to be of serine type, membrane-bound, and most likely closely associated with the PSII complex (11, 15, 16). The involvement of nuclear- as well as chloroplast-encoded proteolytic systems has been suggested (22, 23). The proteolytic process has been considered not to require ATP (10, 14, 15).
Several proteolytic activities present in the thylakoid membrane, as in other biological membranes, are dependent on ATP (24–26). The precise roles of nucleotides and their hydrolysis in proteolysis are poorly understood. The most relevant suggestion, at least in the case of ATP, is the requirement of unfolding of the protein to allow access of a protease to a certain peptide bond of a membrane protein. Therefore, it is surprising that a hydrophobic membrane protein as the D1 protein appears to be degraded independently of ATP.
In this work, we have investigated the requirements for nucleotides and other cofactors during the light-induced proteolysis of the D1 protein. Our results reveal that the primary cleavage of the D1 protein is a GTP-dependent process whereas the secondary cleavage steps require ATP and zinc ions. Furthermore, these results give unexpected evidence for the existence of tightly bound GTP to the thylakoid membrane, potentially implying a more general role for this nucleotide in the regulation of photosynthetic activities.
MATERIALS AND METHODS
Plant Material.
Thylakoid membranes were isolated from 5-week-old spinach leaves as described in ref. 27. The thylakoids were washed in 50 mM Hepes⋅NaOH, pH 7.4, containing 0.4 M sucrose, 15 mM NaCl, and 2 mM EDTA (unless otherwise stated). The final resuspension was made in 50 mM Hepes⋅NaOH, pH 7.4, containing 0.4 M sucrose, 15 mM NaCl, and 5 mM MgCl2 (incubation buffer). PSII-enriched membranes, prepared according to ref. 28, were used for the isolation of oxygen-evolving PSII core complexes by perfusion chromatography (29). The final resuspension was made in 50 mM Hepes⋅NaOH, pH 7.4, containing 0.4 M sucrose, 15 mM NaCl, and 5 mM MgCl2 and supplemented with 5 mM CaCl2 and 0.01% dodecyl maltoside. Chlorophyll (Chl) was determined according to Arnon (30).
Photoinhibition Assays.
Photoinhibition experiments were performed essentially according to ref. 10, with minor modifications. The isolated thylakoid membranes or PSII core preparations (0.2 mg of Chl per ml) were incubated in buffer supplemented with 20% (wt/vol) glycerol. The suspensions were illuminated at −1°C in a thermostated beaker, with heat-filtered visible light (5,000 μmol of photons m−2⋅s−1) supplied with a 250-W projector lamp. When 50% photoinactivation of PSII electron transport was reached, aliquots were withdrawn and mixed with an equal volume of the incubation buffer supplemented with indicated additions. The temperature of the samples was raised to 22°C followed by incubation in complete darkness for 30 and 90 min. The following nucleotides were added at the beginning of the dark incubation period: ATP, adenosine 5′-O-(γ-thiotriphosphate) (ATP[γS]), β,γ-imidoadenosine 5′-triphosphate (AppNHp); GTP, GDP, GMP, guanosine 5′-O-(γ-thiotriphosphate) (GTP[γS]), β,γ-imidoguanosine 5′-triphosphate (GppNHp), guanosine 5′-O-(3-methyl-triphosphate) (GTP[γCH3]), UTP, and CTP. In certain experiments, nucleotides were hydrolyzed by addition of apyrase (EC 3.6.1.5, from potato).
Binding of externally added GTP to apyrase-treated thylakoids (0.2 mg of Chl per ml) was estimated by the addition of [α-32P]GTP (Amersham) to a final concentration of 0.4 mM (50 μCi per μmol; 1 μCi = 37 kBq) to dark- and photoinactivated samples. After 30 min of incubation on ice in darkness, the thylakoid suspensions were centrifuged and the pellets were washed three times with cold incubation buffer. The radioactivity in the supernatants and pellets was determined by scintillation counting in distilled water.
In addition to the nucleotides, salts of several divalent cations (zinc acetate, MnCl2, CaCl2, and FeSO4) or EDTA were added to the photoinactivated thylakoids at the beginning of the dark incubation period.
Protein Analysis.
SDS/PAGE was performed in 14% polyacrylamide gels containing 6 M urea (31). After electrotransfer of the resolved proteins to poly(vinylidene difluoride) membranes (PVDF-Plus 0.45 μm; Micron Separations, Westboro, MA) (32), the D1 protein and its 23-kDa N-terminal fragment were identified by immunoblotting. The D1 protein was detected by using an antibody raised against the entire spinach protein, whereas the 23-kDa fragment was detected with an antibody raised against amino acids 234–242 of the protein. The immunoreaction was visualized by enhanced chemiluminescence (ECL, Bio-Rad). For quantification, thylakoids and PS II core complexes containing 0.40 and 0.25 μg of Chl, respectively, were applied per lane, allowing the immunostaining response to be in the linear range. Higher amounts of Chl (0.8 μg) were loaded per lane to detect breakdown products of the D1 protein. The data presented are the average of three independent experiments, with typical SDs of 7% and 15% for the D1 protein and its 23-kDa N-terminal fragment, respectively.
Measurements of Photosynthetic Activity.
PSII electron transport was measured as steady-state oxygen evolution with a Clark-type electrode (Hansatech Instruments, Pentney King’s Lynn, U.K.) under saturating visible light at 22°C by using 0.5 mM phenyl-p-benzoquinone as electron acceptor.
RESULTS
Slow Proteolysis of Photodamaged D1 Protein in Vitro.
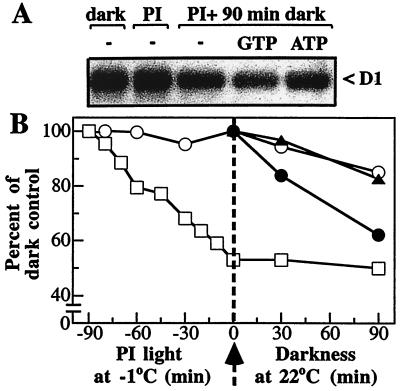
To investigate the requirements for the light-induced D1 protein degradation, isolated thylakoid membranes and PSII core complexes were photoinactivated at −1°C and subsequently incubated in the dark at 22°C after the addition of various effectors. The advantage of this experimental two-step strategy is that the process of D1 proteolysis can be studied separately from the light-dependent inactivation of PSII electron transport. As illustrated in Fig. 1, illumination of isolated thylakoid membranes with 5,000 μmol of photons m−2⋅s−1 at −1°C for 90 min resulted in a 50% inactivation of oxygen evolution. Degradation of the D1 protein did not occur at this low temperature but could be observed after the transfer of the photoinactivated thylakoid samples to 22°C (Fig. 1A). After 90 min of incubation in darkness at 22°C, approximately 15% of the initial amount of the D1 protein was degraded (Fig. 1B). The initial rate constant of 1.6 × 10−3 min−1 indicates a rather slow proteolytic reaction.
Figure 1.
Effect of ATP and GTP on the proteolysis of the D1 protein in photoinactivated thylakoids. (A) Autoradiogram showing the level of the D1 protein in EDTA-washed thylakoids before and after photoinactivation (PI) for 90 min followed by 90 min of darkness. (B) Time course for the photoinactivation of oxygen evolution (□) and degradation of the D1 protein in the absence (○) or presence of either 1 mM ATP (▴) or 0.2 mM GTP (●). The amount of the remaining D1 protein was normalized to that in the nonirradiated thylakoids and plotted as a function of time. The maximal rate of oxygen evolution was 350 μmol of O2 per mg of Chl per h in dark-control thylakoids (100%). The arrow indicates the transfer of samples photoinactivated at −1°C to darkness at 22°C.
GTP Stimulates the Primary Cleavage of the D1 Protein.
It is currently assumed that light-induced D1 protein degradation does not require ATP (10, 14). Our data presented in Fig. 1B corroborate this observation because added ATP has an effect neither on the rate nor on the extent of the D1 protein loss. However, surprisingly, added GTP significantly accelerated the degradation process. The calculated initial rate constant of 5.6 × 10−3 min−1 reveals a 3.5-times-faster proteolytic reaction in the presence of added GTP. Furthermore, as much as 40% of the D1 protein in photoinactivated thylakoids was degraded in the samples subsequently incubated for 90 min in darkness (Fig. 1B). Other nucleotides, such as CTP and UTP, did not influence the D1 protein degradation (not shown).
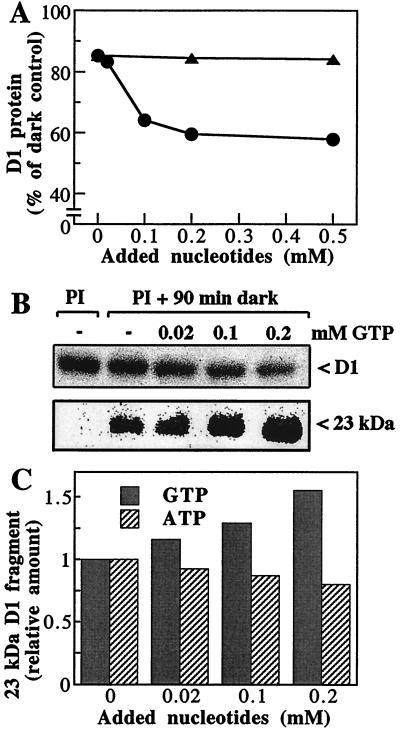
The results presented in Fig. 2 A and B revealed that the degradation of the damaged D1 protein depended on the GTP concentration up to 0.2 mM. At higher concentrations, no significant additional loss of the protein was detected, indicating that saturation of the degradation process was reached. In control experiments, it was shown that increasing concentrations of ATP had no stimulatory effect on the extent of D1 protein degradation (Fig. 2A). As will be discussed below, the D1 protein lost in the absence of added nucleotides depends on residual GTP bound to the thylakoid membrane.
Figure 2.
D1 protein degradation in photoinactivated thylakoids—dependence on nucleotide concentration. (A) Effect on the extent of D1 protein loss. The level of remaining D1 protein was determined by Western blotting in photoinactivated samples that were incubated in darkness for 90 min in the presence of the indicated concentrations of ATP (▴) or GTP (●). (B) Autoradiograms showing the level of D1 protein and its 23-kDa fragment in samples photoinactivated (PI) and subsequently dark-incubated with concentrations of up to 0.2 mM GTP. (C) Effect on the accumulation of the 23-kDa fragment of the D1 protein. The relative amounts of the proteolytic fragment were determined in samples treated as in A.
Degradation of the D1 protein in the thylakoid membranes generates a 23-kDa N-terminal primary fragment (Fig. 2B), and the correlation between the loss of the D1 protein and the accumulation of this degradation fragment was studied at different concentrations of added nucleotides. Concentrations of GTP up to 0.2 mM increased significantly the amount of the 23-kDa fragment (Fig. 2 B and C), providing additional evidence for the GTP requirement of the primary proteolytic step during the D1 protein degradation.
Even though the addition of ATP did not affect the degradation of the D1 protein (Fig. 2A), it led to a pronounced decrease in the amount of the 23-kDa fragment (Fig. 2C). These data suggest that, in contrast to the primary cleavage step, the secondary proteolysis yielding smaller fragments is ATP-dependent (see further below).
Degradation of the D1 Protein Requires Hydrolysis of GTP Tightly Bound to the Thylakoid Membrane.
In an attempt to show whether the degradation of the D1 protein involves binding and/or hydrolysis of GTP, we studied the time course for the loss of the D1 protein in photoinactivated thylakoids transferred to the dark in the presence of GTP[γS], a nonhydrolyzable analogue of GTP (33). As illustrated in Fig. 3, GTP[γS] did not lead to any stimulation of the degradation. In contrast, the proteolysis was inhibited to a level below that seen in the absence of any additions, virtually leaving all D1 protein undegraded. Similar results were obtained in the case of other nonhydrolyzable GTP analogues such as GppNHp, GTP[γCH3], and GMP (not shown). GDP, however, did not affect the degradation kinetics as compared with the samples without additions. The pronounced inhibitory effects of the nonhydrolyzable GTP analogues demonstrate that D1 protein degradation depends on GTP hydrolysis. Furthermore, these inhibitory effects reveal the unanticipated existence of membrane-bound GTP that should be responsible for the observed slow degradation of the D1 protein in absence of any additions (Fig. 1).
Figure 3.
Time course of D1 protein degradation determined by Western blotting in thylakoids photoinactivated at −1°C and then incubated in darkness for 0, 30, and 90 min at 22°C in the absence (○) and presence of 0.2 mM GTP (●) or 0.2 mM GTP[γS] (■), or 5 units of apyrase per ml (⋄). The arrow indicates the transfer of samples photoinactivated at −1°C to darkness at 22°C.
To analyze this possibility further, the thylakoid membranes were treated with apyrase, an enzyme that hydrolyzes β- and γ-phosphate bonds, yielding nucleoside monophosphates (34). Apyrase was added to the photoinactivated samples in the absence or presence of GTP at the beginning of the dark period after the photoinhibitory treatment in the cold. As shown in Fig. 3, apyrase completely blocked the D1 protein degradation, giving independent support for the existence of membrane-bound GTP and its functional role in the D1 protein turnover.
To investigate whether this GTP binding is tight or simply a result of unspecific membrane attachment, we performed initial binding studies by adding radioactive GTP to apyrase-treated thylakoids. On the basis of the measured radioactivity retained in membranes washed several times, 0.28 nmol of GTP per mg of Chl was determined. In the samples that had been photoinactivated 25%, 0.46 nmol of GTP per mg of Chl was obtained. These binding results suggest that GTP is tightly bound to the thylakoid membranes, light-dependent, and approximately correlated with the extent of PSII inactivation.
ATP and Zinc Ions Stimulate Secondary Cleavage of the D1 Protein.
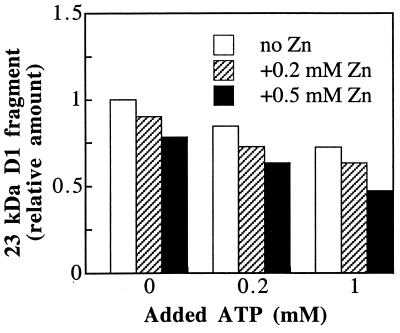
Addition of ATP, in contrast to GTP, at the onset of the dark incubation after strong illumination has no effect on the primary cleavage of the D1 protein (Fig. 1). However, as shown in Fig. 2C, increasing concentrations of ATP promoted a gradual disappearance of the 23-kDa D1 fragment, whose accumulation was stimulated by added GTP. Notably, this secondary proteolysis was stimulated further by the addition of zinc ions (Fig. 4). Thus, more than 50% of the fragment was degraded in the presence of 1 mM ATP and 0.5 mM zinc acetate.
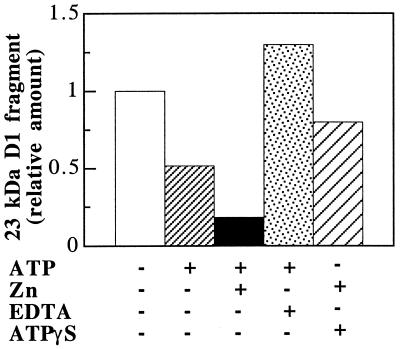
Figure 4.
Effect of ATP and zinc ions on the degradation of the 23-kDa D1 protein fragment. Thylakoids were photoinactivated and subsequently incubated in darkness for 90 min in the presence of the indicated concentrations of ATP and zinc acetate. The level of the proteolytic fragment was expressed relative to the amount detected in the absence of any additions.
To test the specific requirement for zinc as a cofactor of the secondary proteolytic activity, the effect of other divalent cations like calcium, manganese, and iron at a final concentration of 0.5 mM also has been investigated. It was found that, indeed, zinc ions stimulated the degradation to the highest extent. Manganese and iron showed only moderate stimulation whereas calcium had no effect (not shown).
These observations led us to suggest the involvement of an ATP- and zinc-dependent protease in the secondary cleavage of the D1 protein. Interestingly, 20% of the fragment was degraded in the presence of ATP or zinc alone (Fig. 4), indicating the functional significance of residual ATP and zinc ions bound to the thylakoid membrane. EDTA is a commonly used inhibitor of metalloproteases, chelating preferentially zinc and iron ions. In thylakoids not washed with buffer containing 2 mM EDTA, 50% loss of the 23-kDa fragment is observed in the presence of ATP alone during the dark incubation (Fig. 5). Addition of 0.5 mM zinc acetate to such a sample promotes almost a complete disappearance of the fragment. The addition of 2 mM EDTA at the onset of the dark incubation leads to an increased accumulation of the 23-kDa fragment (Fig. 5). Addition of ATP[γS], a nonhydrolyzable analogue of ATP (35), in the absence or presence of zinc ions, was found to inhibit the degradation of the 23-kDa fragment (Fig. 5). Similar effects were observed when AppNHp was used (not shown). Taken together, the results shown in Fig. 4 and 5 suggest that the secondary cleavage of the D1 protein requires hydrolysis of ATP bound to the thylakoid membrane.
Figure 5.
Comparative levels of the 23-kDa D1 protein fragment in thylakoids not EDTA-washed before photoinactivation and subsequently incubated in darkness for 90 min in the absence (−) or presence (+) of 0.5 mM ATP, 0.5 mM zinc acetate, 2 mM EDTA, and 0.5 mM ATP[γS].
Even though GTP induces an enhancement in the amount of the 23-kDa fragment, the addition of ATP and zinc ions promoted degradation of the fragment to the same extent as in the photoinactivated samples that did not contain externally added GTP (not shown). We conclude that D1 protein degradation proceeds in at least two distinct proteolytic steps with specific requirements: GTP for the primary cleavage and ATP and zinc ions for the secondary cleavages. Moreover, the degradation of the D1 protein is much slower than that of its 23-kDa fragment, suggesting that the primary cleavage is the rate-limiting step in the multienzyme event of D1 protein degradation.
The PSII Core Complexes Can Perform the GTP-Dependent Primary Cleavage, but Lack the Enzyme(s) Involved in the Secondary Cleavages.
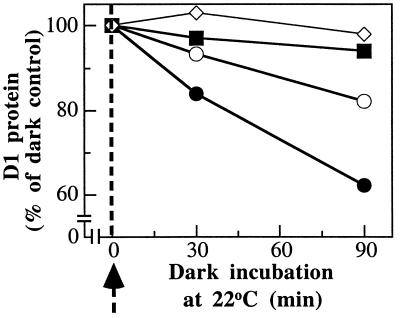
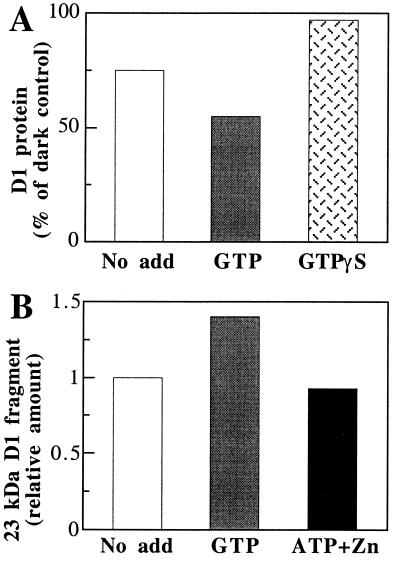
It has been reported that D1 protein degradation can proceed in isolated PSII complexes (15, 16). It was therefore of interest to study the effect of the various nucleotides in PSII core complexes by using the experimental system of photoinactivation at a low temperature followed by subsequent incubation at room temperature in the dark. To reach 50% loss of the oxygen-evolution activity, isolated PSII core complexes were irradiated with 5,000 μmol of photons m−2⋅s−1 at −1°C for 45 min followed by a dark incubation period of 90 min at 22°C. No loss of the D1 protein was observed during the irradiation at −1°C whereas degradation started after transfer of the photoinactivated sample to darkness at 22°C (not shown). As in the case of thylakoids, GTP stimulated the degradation of the D1 protein, i.e., 45% of the initial D1 protein was lost as compared with 25% in the absence of any additions (Fig. 6A). GTP[γS] induced a complete inhibition of D1 protein degradation. These data suggest that the GTP-dependent proteolytic activity involved in D1 protein degradation is associated with the PSII core complex, which appears to contain a site for tight binding of GTP. In contrast, ATP and zinc ions did not enhance degradation of the accumulated 23-kDa D1 fragment (Fig. 6B). This suggests that the secondary proteolytic activity involved in the degradation of the D1 protein is not associated with the PSII core complex.
Figure 6.
Effect of GTP, ATP, and zinc ions on D1 protein degradation in photoinactivated PSII core complexes. (A) Extent of D1 protein degradation determined by Western blotting. Isolated PSII core complexes were photoinactivated for 45 min at −1°C and subsequently incubated in darkness for 90 min in the absence and presence of 0.2 mM GTP or GTP[γS]. (B) Levels of the 23-kDa D1 fragment determined in samples of PSII core complexes, treated as in A, incubated in the absence and in the presence of 0.2 mM GTP or 1 mM ATP and 0.5 mM zinc acetate.
DISCUSSION
The turnover rates of different proteins show a considerable variation and also may be influenced by the developmental stage or the physiological condition of a particular organism. Today we have a wealth of information concerning the biosynthetic steps during protein turnover, and the number of different components required for the assembly of a protein complex is impressive (36). In contrast, the disassembly phase of protein turnover is considerably less well characterized, although evidence is accumulating to suggest that protein degradation is a complex and highly specific process and that it shows a high degree of regulation (24, 25).
In chloroplasts, the light-dependent, rapid turnover of the D1 protein of the PSII reaction center has been studied from both biochemical and physiological points of view (11). This process offers a unique model system to follow the degradation and replacement of a single subunit in a multisubunit and membrane-bound protein complex carrying an array of different redox-active ligands. Despite the considerable experimental efforts, the enzymatic system catalyzing the D1 protein degradation and its regulation have not been established (19). One of the unsolved questions concerns the nucleotide requirement for the proteolytic process in light of the size as well as complicated folding and assembly of the D1 protein substrate. As reviewed by Gottesman and Maurizi (37), an efficient degradation of large proteins requires continuous nucleotide hydrolysis, whereas the cleavage of smaller peptides readily proceeds in the absence of nucleotides.
By taking advantage of an in vitro experimental system that separates the two stages of the photoinhibition process, the inactivation of electron transport (illumination at low temperature) and proteolysis (subsequent incubation at room temperature in the dark), we have been able to demonstrate an unexpected and complicated nucleotide requirement for the D1-degradation process. Our results demonstrate that in photoinactivated thylakoids the degradation of the D1 protein involves at least two principal enzymatic steps that can be resolved in terms of their different nucleotide requirement (Fig. 7). The primary cleavage of the D1 protein was shown to possess an unexpected GTP dependence whereas further secondary proteolysis of the degradation fragments depended on ATP and zinc ions. The primary GTP-dependent cleavage was found to be associated with isolated PSII complexes in contrast to the secondary ATP-dependent cleavages, which could be detected only in unfractionated thylakoid membranes. Notably, the maximal stimulation of the D1 protein degradation was obtained at 0.2 mM, known to correspond to the physiological concentration of GTP in the chloroplast stroma (38).
Figure 7.
Model for the proteolytic events occurring during D1 protein degradation after photoinactivation and photodamage to the PSII reaction center.
The results also demonstrate the existence of a light-dependent tight binding of GTP to thylakoid membranes and PSII. Based on the binding data and assuming an equal distribution of Chl between the two photosystems, 0.14 and 0.23 nmol of GTP per nmol of PSII could be calculated for the dark- and photoinactivated thylakoids, respectively. The existence of tightly bound GTP should be of general significance for regulatory reactions and signal transduction pathways during photosynthesis in addition to the presently described repair of photodamaged PSII reaction centers. The elucidation of such potential GTP-dependent processes and the identification of putative thylakoid G proteins should be a challenging task for future biochemical and genomic research in the area. Notably, despite the crucial role of GTP in most biological systems, the involvement of this nucleotide in processes associated with the photosynthetic light reaction and the thylakoid membrane largely has been ignored. Early observations by Millner (39), who suggested the involvement of GTP in the regulation of thylakoid kinase activity, was never substantiated (40). More recently, Hoffman and Franklin (41) demonstrated the requirement for stromal GTP in the integration of chlorophyll-binding proteins in the thylakoid membrane by a chloroplast homologue of the mammalian signal-recognition particle (SRP 54).
The molecular mechanism behind the nucleotide requirement during D1 protein degradation remains to be elucidated. Most likely, the nucleotide hydrolysis provides energy necessary for unfolding of the D1 protein and its N-terminal, 23-kDa fragments because both of these protein substrates contain large loops connecting trans-membrane helices that may hide proteolytic sites. Structural changes associated with the so-called DE loop of the D1 protein and its plastoquinone/herbicide-binding domain has been suggested previously to influence initiation or triggering of D1 protein degradation (17, 18). In addition, the nucleotides may be required for the partial disassembly of the PSII complex occurring during replacement of the D1 protein (2), in particular, the reversible dissociation of the chlorophyll a-binding protein CP43 (42).
The involvement of GTP rather than ATP during the primary protein cleavage probably represents a need for a very strict regulation of this first, committed or strategic step in the turnover of the D1 protein. The subsequent secondary degradation steps may, in contrast, be less specific or regulated, representing a more conventional cleaning-up, ATP-requiring proteolysis (25). That the D1 protein degradation proceeds in a stepwise manner and the initial cleavage is the rate-limiting step have been suggested previously from in vivo studies (12).
Several proteases in chloroplasts have been shown to be homologous to bacterial enzymes (25) such as the ClpAP (43) and FtsH (44) proteases. In light of these findings and the prokaryotic nature of chloroplast, it is of interest that a GTP-dependent proteolytic system has been reported for Escherichia coli (45). The presence of the bacterial homologue FtsH protease in thylakoid membranes is of further relevance to our present work. FtsH is the only known ATP- and zinc-dependent protease in thylakoid membranes (44) and was shown recently to perform clean-up proteolysis of unassembled Rieske Fe-S protein (46). This protease is not associated with PSII complexes and is located exclusively in the stroma-exposed thylakoid regions (44), where the final steps during D1 protein degradation are known to occur (19, 42). These previous experimental data and our present observation on an ATP- and zinc-dependent proteolysis of the N-terminal, 23-kDa fragment strongly suggest that FtsH is participating in the secondary cleaning-up proteolysis during D1 protein degradation (Fig. 7).
Acknowledgments
We thank Professors F. Ekstein (University of Göttingen) and R. Barbato (University of Alessandria) for generous gifts of GTP[γCH3] and the D1 protein antibody, respectively. We are also grateful to Professor I. Ohad (The Hebrew University of Jerusalem) for valuable discussions and suggestions. This work was supported by grants from the Swedish Natural Science Research Council. C.S. was a recipient of fellowships from the Swedish Foundation for International Cooperation in Research and Higher Education and the Wenner–Gren Foundation.
ABBREVIATIONS
- ATP[γS]
adenosine 5′-O-(γ-thiotriphosphate)
- Chl
chlorophyll
- D1
32-kDa protein of the photosystem II reaction center
- GTP[γS]
guanosine 5′-O-(γ-thiotriphosphate)
- PS
photosystem
References
- 1.Barber J, Andersson B. Trends Biochem Sci. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- 2.Prasil O, Adir N, Ohad I. In: Topics in Photosynthesis. Barber J, editor. Vol. 11. Amsterdam: Elsevier; 1992. pp. 295–348. [Google Scholar]
- 3.Andersson B, Salter A H, Virgin I, Vass I, Styring S. J Photochem Photobiol B. 1992;15:15–31. [Google Scholar]
- 4.Hankamer B, Barber J, Boekema E J. Annu Rev Plant Physiol Mol Biol. 1997;48:641–671. doi: 10.1146/annurev.arplant.48.1.641. [DOI] [PubMed] [Google Scholar]
- 5.Vass I, Styring S, Hundal T, Koivuniemi A, Aro E-M, Andersson B. Proc Natl Acad Sci USA. 1992;89:1408–1412. doi: 10.1073/pnas.89.4.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telfer A, Bishop S M, Phillips D, Barber J. J Biol Chem. 1994;269:13244–13253. [PubMed] [Google Scholar]
- 7.Hideg É, Kálai T, Hideg K, Vass I. Biochemistry. 1998;37:11405–11411. doi: 10.1021/bi972890+. [DOI] [PubMed] [Google Scholar]
- 8.Kyle D J, Ohad I, Arntzen C J. Proc Natl Acad Sci USA. 1984;81:4070–4074. doi: 10.1073/pnas.81.13.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattoo A K, Hoffman-Falk H, Marder J B, Edelman M. Proc Natl Acad Sci USA. 1984;81:1380–1384. doi: 10.1073/pnas.81.5.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aro E-M, Hundal T, Carlberg I, Andersson B. Biochim Biophys Acta. 1990;1019:269–275. [Google Scholar]
- 11.Andersson B, Barber J. In: Advances in Photosynthesis. Baker N R, editor. Vol. 5. Dordrecht, The Netherlands: Kluwer; 1996. pp. 101–121. [Google Scholar]
- 12.Greenberg B M, Gaba V, Mattoo A K, Edelman M. EMBO J. 1987;6:2865–2869. doi: 10.1002/j.1460-2075.1987.tb02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canovas P M, Barber J. FEBS Lett. 1993;324:341–344. doi: 10.1016/0014-5793(93)80147-m. [DOI] [PubMed] [Google Scholar]
- 14.Ohad I, Kyle D J, Hirschberg J. EMBO J. 1985;4:1655–1659. doi: 10.1002/j.1460-2075.1985.tb03833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salter A H, Virgin I, Hagman Å, Andersson B. Biochemistry. 1992;31:3990–3998. doi: 10.1021/bi00131a014. [DOI] [PubMed] [Google Scholar]
- 16.De Las Rivas J, Shipton C A, Ponticos M, Barber J. Biochemistry. 1993;32:6944–6950. doi: 10.1021/bi00078a019. [DOI] [PubMed] [Google Scholar]
- 17.Jansen A K, Depka B, Trebst A, Edelman M. J Biol Chem. 1993;268:21246–21252. [PubMed] [Google Scholar]
- 18.Zer H, Prasil O, Ohad I. J Biol Chem. 1994;26:17670–17676. [PubMed] [Google Scholar]
- 19.Andersson B, Aro E-M. Physiol Plant. 1997;100:780–793. [Google Scholar]
- 20.Mishra N P, Frank C, van Gorkom H J, Ghanotakis D F. Biochim Biophys Acta. 1994;1186:81–90. [Google Scholar]
- 21.Miyao M. Biochemistry. 1994;33:9722–9730. doi: 10.1021/bi00198a043. [DOI] [PubMed] [Google Scholar]
- 22.Bracht E, Trebst A. Z Naturforsch. 1994;49c:439–446. [Google Scholar]
- 23.Gong H. Biochim Biophys Acta. 1994;1188:422–426. [Google Scholar]
- 24.Goldberg A L. Eur J Biochem. 1992;203:9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- 25.Adam Z. Plant Mol Biol. 1996;32:773–783.25. doi: 10.1007/BF00020476. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl M. Ph.D. thesis. Kista Snabbtryck, Stockholm: Stockholm University; 1997. [Google Scholar]
- 27.Andersson B, Åkerlund H E, Albertsson P-Å. Biochim Biophys Acta. 1976;423:122–132. doi: 10.1016/0005-2728(76)90106-7. [DOI] [PubMed] [Google Scholar]
- 28.Ghanotakis D F, Babcock G T, Yocum C F. Biochim Biophys Acta. 1984;765:388–398. doi: 10.1016/s0005-2728(89)80164-1. [DOI] [PubMed] [Google Scholar]
- 29.Roobol-Bóza M, Andersson B. Anal Biochem. 1996;235:127–133. doi: 10.1006/abio.1996.0104. [DOI] [PubMed] [Google Scholar]
- 30.Arnon D I. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4353. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilman A G. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 34.Sakahibara T, Murakami S, Hattori N, Nakajima M, Imai K. Anal Biochem. 1997;250:157–161. doi: 10.1006/abio.1997.2217. [DOI] [PubMed] [Google Scholar]
- 35.Ekstein F. Annu Rev Biochem. 1985;54:367–375. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 36.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 37.Gottesman S, Maurizi M R. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause G H, Heber U. In: Topics in Photosynthesis. Barber J, editor. Vol. 1. Amsterdam: Elsevier; 1976. pp. 171–214. [Google Scholar]
- 39.Millner P A. FEBS Lett. 1987;226:155–160. [Google Scholar]
- 40.Gal A, Zer H, Ohad I. Physiol Plant. 1997;100:869–885. [Google Scholar]
- 41.Hoffman N E, Franklin A E. Plant Physiol. 1994;105:295–304. doi: 10.1104/pp.105.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbato R, Friso G, Rigoni F, Dalla Vecchia F, Giacometti G M. J Cell Biol. 1992;119:325–335. doi: 10.1083/jcb.119.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vierstra R D. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- 44.Lindahl M, Tabak S, Cseke L, Pichersky E, Andersson B, Adam Z. J Biol Chem. 1996;271:29329–29334. doi: 10.1074/jbc.271.46.29329. [DOI] [PubMed] [Google Scholar]
- 45.Gottesman S. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 46.Osterzetzer O, Adam Z. Plant Cell. 1997;9:957–965. doi: 10.1105/tpc.9.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]