Abstract
A key challenge of the post-genomic era is the identification of the function(s) of all the molecules in a given organism. Here, we review the status of sequence and structure-based approaches to protein function inference and ligand screening that can provide functional insights for a significant fraction of the ∼50% of ORFs of unassigned function in an average proteome. We then describe FINDSITE, a recently developed algorithm for ligand binding site prediction, ligand screening and molecular function prediction, which is based on binding site conservation across evolutionary distant proteins identified by threading. Importantly, FINDSITE gives comparable results when high-resolution experimental structures as well as predicted protein models are used.
Keywords: protein function prediction, ligand binding site prediction, virtual ligand screening, protein structure prediction, low-resolution protein structures
INTRODUCTION
Over the past decade, catalyzed by the sequencing of the genomes of hundreds of organisms [1–4], biology is undergoing a revolution comparable to what physics underwent in the early 20th century. The emphasis is shifting from the study of individual molecules to the large-scale examination of all genes and gene products in an organism and comparative genomics studies of multiple organisms [5–9]. Here, the goal is to understand the function of all molecules in a cell and how they interact on a system-wide level; this perspective has given birth to the new field of Systems Biology [10, 11]. Of course, biological function is multifaceted, ranging from biochemical to cellular to phenotypical [12,13]. By detecting evolutionary relationships between proteins of known and unknown function, sequence-based methods can provide insights into the function of about 50% of the ORFs in a given proteome [14–20], with the remainder believed to be too evolutionarily distant to infer their function [21]. Thus, the prediction of the function of the remaining 50% of unannotated ORFs remains an outstanding challenge. However, since protein structure is more conserved than protein sequence [22–24], it can play an essential role in annotating genomes [13,25–31]. In addition, protein structure should assist in lead compound identification as part of the drug discovery process [11,32–34]. A key question is whether one can use low-to-moderate resolution predicted structures which can be provided for about 70% of the protein domains in a proteome [35] or if high-resolution experimental structures are required [36,37]. This issue also has implications for the requisite scope of structural genomics that aims for high-throughput protein structure determination [38–44]. If low-to-moderate resolution models were to prove useful for functional inference, then the value of contemporary protein structure prediction approaches would be significantly enhanced [45].
One of the more disappointing aspects of protein structure-based functional inference has been the relatively minor marginal impact it has had to date relative to sequence-based methods that rely on inferring function on the basis of the evolutionary relationship between proteins of known and unknown function. Here, however, caution needs to be exercised; just because a pair of proteins are evolutionarily related does not imply that they have the identical function [20]. Proteins can add additional functions during the course of their evolution or can modify their function from that of their ancestors [46–49]. On the other hand, especially for binding site prediction and ligand screening, as proteins become evolutionary more distant, it is unclear what features are conserved and what have become modified [50]. Here, one might imagine that conservation of the protein's binding site in the structure and conservation of ligand binding features and associated ligands could prove useful. Indeed, just as protein homology modeling as extended by threading [51] (note that the most successful threading approaches have a strong evolution based component [52]) has proven to be a very powerful tool in protein function prediction, one would like to exploit these ideas for the prediction of protein function, binding site prediction and ligand screening [37]. That is to say, we wish to exploit the signal averaging provided by evolution to identify the conserved/variable functional features that can be used to infer the functional properties of proteins of unknown function and to do so by automatic approaches suitable for proteomes.
Based on the above, there is a pressing need for the development of more powerful approaches to protein function prediction that can be applied on a proteome scale. In that regard, in this Briefings in Bioinformatics article, we first summarize the status of sequence-based approaches to functional inference [14, 19, 53–55] that provide the baseline against which protein structure-based approaches are compared. Next, there is the issue of the utility of protein structure for functional inference. Is it marginal? Being bounded at higher levels of sequence identity (>20%) by purely sequence-based approaches [14,19] and at low levels of sequence identity by the inability to transfer function by interference [20] (which is the only effective means of function prediction), can one effectively exploit the insights provided by protein structure? If so, what is the quality of protein structure required for functional inference in general, and for binding site and ligand screening in particular? Can predicted, low-resolution protein models be used or are we limited to high-resolution, experimental structures [36,56,57]? Here, we focus in particular on a newly developed, powerful threading-based approach to protein function prediction, FINDSITE [37], that shows considerable promise in its ability to exploit both experimental and predicted protein structures for the inference of protein function, the prediction of protein binding sites as well as for providing guidance in small molecule ligand screening. These issues are discussed below.
SEQUENCE-BASED FUNCTIONAL INFERENCE
The biological function of a protein can be defined in physiological, developmental, cellular or biochemical contexts [58]. To characterize these facets of protein functions, a number of ontologies have been developed, including those in GO [59], KEGG [60] and MIPs [61]. However, even having an appropriate description of protein function, performing experimental assays on all the uncharacterized proteins provided by the hundreds of ongoing genome sequencing projects is impractical. Thus, computational tools are needed [62]. In fact, in newly sequenced genomes, the functional annotations of the vast majority of genes are not based on experiment but are inferred on the basis of the sequence similarity to previously characterized proteins [58,63,64].
The fundamental assumption of this strategy, termed ‘annotation transfer by homology’ [65], is that sequence similarity is equivalent to functional similarity. However, sequence similarity based function transfer is complicated by numerous factors; most critical is the functional divergence of highly similar sequences, a problem exhibited by many protein families [54,55]. Here, permissive criteria to assess the significance of the similarity between proteins can lead to wrong annotations. For example, depending on the protein family, detailed biochemical function is not completely conserved between similar proteins even when their pairwise sequence identity is 60% [20]. Despite this fact, much lower sequence similarity thresholds have been used in the functional annotation of some genomes [66]. This issue can be partly addressed by introducing family specific sequence identity thresholds [20], and especially at lower pairwise sequence identity levels (20–30%), enhanced specificity and coverage can be achieved by exploiting the conservation of functionally determining residues [14,19,67,68]. Indeed, the sequence-based method EFICAz for enzyme function inference [14,19] shows quite high levels of precision, sensitivity and specificity even at the levels of 20% sequence identity between pairs of enzyme sequences. It works because a combination of criteria designed to give a low false positive rate is used. Here, the use of functionally discriminating residues that act as a filter once a sequence is assigned as being evolutionary related to sequences of known enzymatic function is of importance.
STRUCTURE-BASED FUNCTIONAL INFERENCE
Active and binding site prediction
Within a protein family, the global fold is more strongly conserved than protein sequence [69]. Thus, the inference of protein biochemical function should benefit by the inclusion of structural information [13]. However, divergent and convergent evolution results in a non-unique relationship between protein structure and protein function; i.e. the structure of a protein in and of itself is insufficient for correct function prediction [70,71]. As in highly accurate sequence-based approaches [19], additional information is required. Three-dimensional descriptors or templates of biologically relevant sites [26,72–81] are one example of such a filter. As demonstrated for 4 enzyme systems [82], local 3D motifs frequently outperform global similarity searches using protein structure [83] or sequence [84] alone. Furthermore, the Evolutionary Trace (ET) approach shows that the accuracy of 3D templates can be further increased by selecting evolutionarily relevant residues [85,86]. In addition to these active site descriptors designed to capture the geometric features of known catalytic residues, a number of structure-based approaches have been developed to identify ligand binding sites [87]. Many focus on the recognition of particular ligand, e.g. adenylate [81], calcium [88] or DNA [89,90], with more general methods mainly tested on a few ligand types [75,91]. Of interest is the PINTS [30] approach designed to perform database searches against a collection of ligand-binding sites excised from the PDB [92] and the ProFunc server that combines a collection of sequence- and structure-based methods to identify close relationships to functionally characterized proteins [93].
Geometric methods locate putative binding residues by searching for cavities/pockets in the protein's structure [94–97]. Comprehensive benchmarks carried out for the unbound/bound protein crystal structures reveal that among the best of these pocket-detection algorithms is LIGSITECSC [96], an extension of LIGSITE [95]. LIGSITECSC calculates surface-accessibility on the Connolly surface [98] and then re-ranks the identified pockets by the degree of conservation of identified surface residues. Other methods calculate theoretical microscopic titration curves [99], analyze the spatial hydrophobicity distribution [100] or identify electrostatically destabilized residues [101]. In all these methods, the ligand itself is ignored; rather the focus is on the structural features of the protein surface.
Ligand docking algorithms
Given a protein structure, one should not only be able to identify the functional site, but also be able to predict which ligands (for enzymes, substrates) bind to that site. There are two key elements of any docking approach: First, a scoring function is required that accurately ranks the generated set of solutions. In that regard, blind docking can be used to elucidate some general features of binding ligands (or more practically, drug candidates) [102,103], even if one lacks the ability to correctly rank known binding ligands. Second, a fast and effective search algorithm is necessary to explore the conformational space of protein–ligand interactions. Efficiency is especially important in virtual screening experiments [104,105], where millions of possible ligands need to be docked into a receptor structure in an acceptable amount of time. Thus, as a practical matter, for each ligand, the docking cannot require more than a few minutes of CPU time on a state-of-the-art computer.
The past years have seen the development of a number of algorithms for docking small molecules into receptor proteins [106–109]. These approaches have been evaluated in terms of ligand binding pose accuracy and the ability to predict binding affinities [110–113]. However, it is evident that most contemporary approaches have significant problems with ligand ranking, and most require high-resolution, experimentally determined protein structures [36,111]. Thus, while considerable progress has been made, significant issues remain.
Utility of predicted structures for functional inference
A number of protein structure-based function inference methods have been reasonably successful when applied to high-resolution structures [26, 72–81, 89, 90]. Given the recent improvements in protein structure prediction algorithms [45, 114–123], it is important to establish if lower resolution predicted structures are useful. A structure-based method for protein function prediction that does not require high-resolution structures would be of significant practical value, especially since the best structure prediction approaches can produce low-resolution or better models for ∼2/3 of the proteins in a given proteome [35, 124–126].
The key issue is to establish the quality of structure required to transfer a given biochemical function at a specified level of accuracy. While there have been attempts to address this issue for enzymes using active site template matching [13,127], further investigation is required. Most often, ligand docking programs typically utilize high-resolution receptor structures determined by experiment or theoretical modeling [128–130]. Virtual screening reveals that the success rate decreases from ligand-bound to ligand-free to modeled structures [131] and is correlated with the degree of protein movement in the binding site; protein binding site rearrangements greater than 1.5 Å lead to almost complete lack of recovery of the ‘true’ binding mode [132]. Furthermore, decoy-docking experiments using deformed trypsin structures with a Cα root-mean-square deviation, RMSD from the native structure in the range of 1–3 Å for the docking of 47 ligands experimentally known to bind trypsin reveal that specific ligand–receptor contacts are rapidly lost with increasing receptor structure deformation [111].
Different docking techniques have been developed to address this problem. Most account for receptor flexibility/distortion by docking ligands against a precalculated ensemble of receptor conformations [133] or by softening the criterion for the steric fit between the ligand and receptor [134]. Other docking techniques capable of dealing with significant structural inaccuracies employ a low-resolution representation of the protein designed to accommodate structural distortions. For example, an ultra low-resolution (∼7 Å) protein representation that averages all high-resolution structural details dramatically improves the tolerance to receptor deformation [135,136]. A similar approach demonstrated that even low-quality receptor structures could be utilized [57].
Another, newly developed, low-resolution docking approach that uses a reduced ligand and protein representation is Q-dock [36]. Self-docking using crystal structures revealed ligand pose prediction accuracy comparable to all-atom docking. All-atom models reconstructed from Q-Dock's low-resolution models can be further refined by simple all-atom energy minimization. In decoy docking against distorted receptor models with a backbone RMSD from native of ∼3 Å, Q-Dock recovers on average 15–20% more specific contacts and 25–35% more binding residues than all-atom methods. Q-Dock also gives encouraging results for ligand screening against predicted protein structures whose average global backbone RMSD is 5 Å (Brylinski & Skolnick, unpublished results). Thus, the possibility of using low-resolution predicted structures for binding pose identification and ligand screening appears quite promising. In this spirit, we next turn to an automated approach that can predict ligand binding sites, binding ligands as well the molecular function of proteins, even when low-resolution protein structures are used.
FINDSITE: A threading based method for ligand binding site prediction/functional annotation
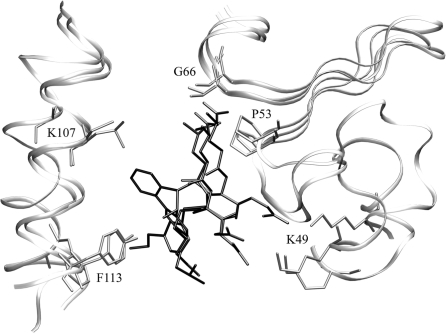
The comprehensive examination of known protein structures grouped according to SCOP [137] reveals the tendency of certain protein folds to bind substrates at a similar location, suggesting that very distantly homologous proteins often have common binding sites [138]. That is, evolution tends to conserve the functionally important region in the protein structure and conserves a subset of ligand binding features as well. For example, as shown in Figure 1, the localization of the binding pocket as well as the local geometry and the binding mode of the ligands are remarkably well conserved in glutathione S-transferase family despite the low sequence identities between family members. Hence, it should be possible to develop an approach for ligand binding site identification that is less sensitive than pocket-detection methods to structural distortions of the protein, as these distortions are present in the set of evolutionarily distant protein structures.
Figure 1:
Binding pockets in threading templates: glutathione S-transferase (GST) from R. norvegicus (PDB-ID: 1b4p), Z. mays (PDB-ID: 1bye) and H. sapiens (PDB-ID: 17gs) upon the global superposition onto the target structure, GST from E. coli (PDB-ID: 1a0f, not shown). Template-bound ligands are presented as black sticks. Selected binding residues are shown as gray sticks and labeled by the equivalent positions in the target sequence. The sequence identity to the target as well as the pairwise sequence identities between the templates is in the range of 15–25%.
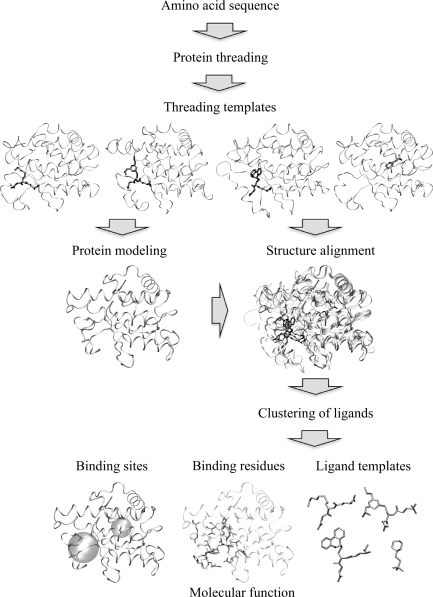
In this spirit, we developed FINDSITE [37], an algorithm for protein functional annotation that is based on binding site similarity among superimposed groups of template structures identified from threading [51]. Threading is of importance in that it acts as a filter to establish that the set of protein structures are evolutionarily related. A schematic overview of the FINDSITE methodology is shown in Figure 2. For a given target protein, the threading algorithm PROSPECTOR_3 [52] identifies protein structure templates with bound ligands. Then, these holo-templates are superimposed onto the predicted (or experimental, if available) target protein structure using the TM-align protein structure alignment algorithm [139]. Upon superimposition, the clustered centers of mass of the ligands bound to the threading templates identify putative binding sites, and the predicted sites are ranked according to the number of templates that share a common binding pocket. As suggested by Figure 1, FINDSITE also specifies the chemical properties of the ligands that likely occupy the binding site and provides a collection of ligand templates for use in fingerprint-based virtual screening.
Figure 2:
Overview of the FINDSITE approach.
To assess the general validity of FINDSITE, we employed a representative set of 901 proteins with <35% sequence identity to their templates (with a mean target-template pair-wise sequence identity of 20%) and generated models using TASSER [35, 123, 140, 141]. As demonstrated below, we find that FINDSITE operates satisfactorily in the ‘twilight zone’ of sequence similarity [142], which covers ∼2/3 of known protein sequences [143]. No experimental structure of the target protein is required; high accuracy and ability to correctly rank the identified binding sites are sustained when protein models instead of target crystal structures are used for template superimposition. Use of consensus ligands extracted from the binding sites is quite useful in ligand screening. In most cases, FINDSITE accurately assigns a molecular function to the protein model. These features should enhance the utility of low-to-moderate quality protein models in ligand screening and structure-based drug design.
Binding site prediction results
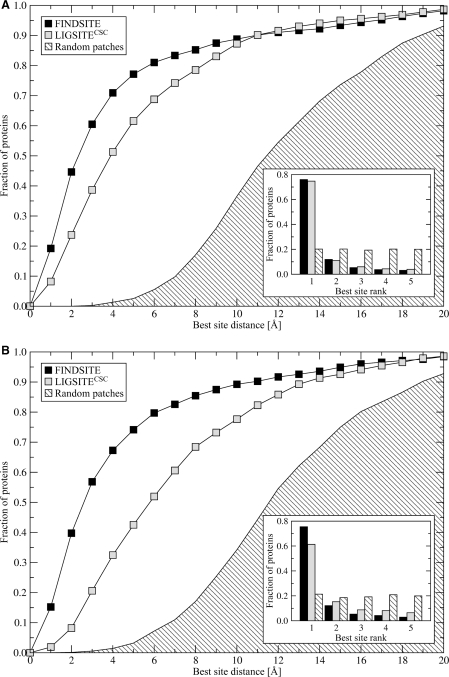
Figure 3 shows ligand binding site prediction results carried out for the 901 benchmark proteins. Here LIGSITECSC identifies possible binding pockets in the target structure (either the crystal structure or predicted model). Using FINDSITE, the set of predicted template models (where the target has a sequence identity <35% to all template structures) is superimposed onto the target structure. In Figure 3A, the target protein's crystal structure is used. In terms of both overall accuracy and pocket ranking ability, FINDSITE performs better than LIGSITECSC. Using the native structure, the success rate (where the centers of mass of the predicted and native binding sites are ≤4 Å) using the best of top five identified binding pockets is 70.9% and 51.3% for FINDSITE and LIGSITECSC, respectively. For those proteins where a binding pocket is correctly identified, the ranking of both methods is comparable; 76.0% and 74.7% of the best pockets are ranked as the top solutions by FINDSITE and LIGSITECSC.
Figure 3:
Ligand binding site prediction by FINDSITE, LIGSITECSC and randomly selected patches on the target protein's surface using (A) target crystal structures and (B) TASSER models. Results are presented as the cumulative fraction of proteins for which the distance between the ligand center of mass in the native complex and the center of the best of top five predicted binding sites is ≤ the distance on the x-axis with the rank of the best pocket of top five predictions in the inset.
As shown in Figure 3B, where modeled target structures are used, the prediction accuracy of LIGSITECSC falls off considerably, with its success rate decreasing from 51.3% for the target crystal structure to 32.5%, when protein models generated by TASSER are used. For TASSER models, only 61.4% of the best pockets are assigned rank 1. Thus, when models are used, LIGSITECSC results deteriorate. In contrast, with FINDSITE, both the high accuracy of ligand binding site prediction and correct binding site ranking are sustained when models instead of native structures are used as reference structures for holo-template superimposition. In 67.3% of the cases FINDSITE identifies a correct binding site, with corresponding ranking accuracy of 75.5%. Note that for both native structures and predicted models, the results using random patches are much worse than for LIGSITECSC and FINDSITE.
We find that for models with a global RMSD from the native structure ≤6 Å, FINDSITE typically predicts the center of mass of the binding site within 6 Å. This is because the binding sites in the models have an RMSD below 3 Å. In contrast, as is evident from Figure 3, LIGSITECSC is far more sensitive to structural distortions. The average distance between the LIGSITECSC predicted and observed binding pockets is 10–13 Å when the global RMSD of the predicted model exceeds 4 Å from the native structure.
FINDSITE's overall binding site prediction accuracy depends on the number of identified ligand-bound templates with a common binding site. We can classify proteins as Easy (>125 threading templates, including homologous proteins for each template), Medium (25–125 templates) and Hard (<25 templates) targets for threading-based binding site prediction. In the 901 protein benchmark set, 9%, 47% and 44% of the proteins are assigned by FINDSITE as Easy, Medium and Hard targets, and the average distance between the centers of predicted and observed binding pockets for top-ranked FINDSITE solutions is ∼2, 5 and 10 Å, respectively. Using a cutoff distance of 4 Å between predicted and observed binding sites, the hit rates for the top-ranked predictions are 90.0%, 71.7% and 43.7% of Easy, Medium and Hard targets, respectively.
We next explored the structural diversity of the binding site residues. We calculated the average local pairwise RMSD of binding site residues for the subset of 561 target proteins that satisfy the following criteria: The best predicted pocket must be rank 1, with the number templates identified ≥5 and there must at least 10 binding site residues. With these restrictions, we find that the average pairwise RMSD of binding site residues is 2.15 ± 0.77 Å. This gives an estimate of the allowed structural degeneracy of binding residues.
Ligand virtual screening
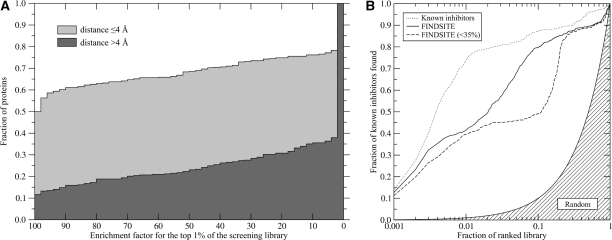
FINDSITE also extracts information about the chemical properties of the ligands bound to the consensus-binding site; we term these ‘template ligands’. Since the ‘template ligands’ are extracted form the holo templates identified by threading, only the target protein's sequence is needed for their selection. These molecules are then used to construct fingerprints that are subsequently employed in fingerprint-based similarity searching [144, 145] of the KEGG compound library, which contains 12,478 compounds [146]. For the 901 representative target proteins, all with <35% sequence identity to their closest template, Figure 4A presents the cumulative distribution of enrichment factors for the top 1% of the screening library of the ranked ligands. For accurately predicted binding sites (70.9% of the target proteins have a binding site center of mass is <4 Å of the native structure), FINDSITE performs better than random in 78% of the cases. The ideal enrichment factor (all native-like compounds in the top 1% of the ranked library) was observed for 50% of target proteins. For less accurately predicted binding pockets, the ideal enrichment factor was obtained for 12% and is better than random for 34% of the cases. Finally, in Figure 4B, a case study examined the performance of FINDSITE in virtual screening for 895 active HIV-1 protease inhibitors in a 123,331 compound library. Again, if only templates with <35% sequence identity to the target are used, the enrichment factor of the top 1% of compounds is 40.
Figure 4:
(A) Using FINDSITE selected ligand templates, cumulative distribution of enrichment factors from the ligand-based virtual screening experiment against the KEGG compound library. Depending on the whether the distance between the top-ranked pocket and the center of mass of the native ligand ≤ 4 Å and >4 Å, target proteins are divided into two subsets. (B) Enrichment behavior in virtual screening for HIV-1 protease inhibitors using ligands predicted by FINDSITE from either homologous or weakly homologous threading templates compared to that using known inhibitors and random ligand ranking.
Molecular function prediction
The relatively high accuracy of the ligand selection procedure encouraged us to investigate the transferability of specific functions from the threading templates to the target. Here, the Gene Ontology (GO) [59] description of protein molecular function is used. We selected the subset of 753 proteins from the 901 protein benchmark set for which a GO annotation is provided by Gene Ontology [59] or UniProt [125]. For each target, all GO annotations are identified for the threading templates that share the top-ranked predicted binding site. Then, the target protein is assigned a function with a probability that corresponds to the fraction of threading templates annotated with that molecular function. When at least one half of the threading holo-templates are annotated with the same GO term, the maximal Matthew's correlation coefficient of 0.64 is found. This corresponds to a precision of 0.76, and a recall of 0.54. In addition, we calculated predictive metrics with respect to individual GO identifiers. When the closest template has <35% sequence identity, FINDSITE distinguishes between enzymatic and non-enzymatic function, with a precision and sensitivity of 0.93 and 0.89, respectively. Moreover, many molecular functions that cover a broad spectrum of molecular events including both enzymatic and binding activities are accurately transferable from the templates selected by FINDSITE.
By way illustration, in Table 1, for the 753 protein benchmark set, the FINDSITE precision and sensitivity is presented for 10 most accurate predicted molecular functions as described by Gene Ontology classification and as assessed by the Matthew's correlation coefficient. Clearly, a broad spectrum of both enzymatic and non-enzymatic activities are adequately described. However, we note that since FINDSITE describes the common functional features found across an evolutionary distant but related set of proteins, it cannot describe highly specific functions such as all four EC digits of an enzyme.
Table 1:
Using the GO classification, the top 10 function predictions by FINDSITE as assessed by their Matthew's Correlation Coefficient for the 753 protein benchmark data set
| Molecular function | GO ID | Frequency in the dataset | Matthew's correlation coefficient | Precision | Sensitivity |
|---|---|---|---|---|---|
| Oxygen binding | GO:0019825 | 0.027 | 1.00 | 1.00 | 1.00 |
| Ligand-dependent nuclear receptor activity | GO:0004879 | 0.015 | 1.00 | 1.00 | 1.00 |
| Peroxidase activity | GO:0004601 | 0.005 | 1.00 | 1.00 | 1.00 |
| Dihydrofolate reductase activity | GO:0004146 | 0.004 | 1.00 | 1.00 | 1.00 |
| 3′,5′-Cyclic-nucleotide phosphodiesterase activity | GO:0004114 | 0.004 | 1.00 | 1.00 | 1.00 |
| Steroid hormone receptor activity | GO:0003707 | 0.015 | 1.00 | 1.00 | 1.00 |
| N-acyltransferase activity | GO:0016410 | 0.015 | 0.96 | 0.92 | 1.00 |
| N-acetyltransferase activity | GO:0008080 | 0.015 | 0.96 | 0.92 | 1.00 |
| Pyridoxal phosphate binding | GO:0030170 | 0.012 | 0.94 | 1.00 | 0.89 |
| Monooxygenase activity | GO:0004497 | 0.009 | 0.93 | 1.00 | 0.86 |
CONCLUSION
The most frequently used methods for protein function prediction are based on functional inference by homology [147, 148]. However, as demonstrated for enzymes [20], because of the promiscuity of protein function, care must be taken if the goal is high accuracy (an necessary condition if one wants to ascertain whether or not a specific pathway [60] is present in the proteome of interest). Moreover, current sequence-based methods become unreliable as the sequence identity between the target protein of unknown function and the template protein of known function drops below 20–30% [31]. To address this limitation, a number of structure-based approaches based on 3D geometric descriptors of enzymatic function, termed fuzzy functional forms (FFFs) were developed [26] and shown to provide high-confidence novel annotations [149]. However, they have only been successfully applied to enzymes, and typically require extensive manual intervention in their construction. In practice, their level of accuracy drops when they are applied to predicted protein models [127].
To remove these limitations, in the development of FINDSITE [37], we explored whether the conservation of binding sites among threading identified templates can be used to predict the target binding site, the ligands that bind to this site and using consensus GO molecular functions [59] of the templates, to predict the molecular function of evolutionary distant target proteins. We find that threading followed by binding site filtering to identify functionally related proteins is a very powerful approach to predict these aspects of protein function. This holds even if the sequence similarity to the target protein is well below 35% and has profound implications as to how protein molecular function has evolved. As was observed for enzymes, some functional sites in the protein structure are strongly conserved throughout evolution [150]. Not only is the protein structure conserved, but the chemical features of the ligands that bind to the protein are conserved as well. Such conservation provides a type of signal averaging that can be exploited for various applications of functional inference.
The clear advantage of FINDSITE is that predicted structures can be used. This is of importance in that state-of-the-art approaches provide predicted structures of the requisite quality for greater than 2/3 of protein domains in a given proteome [35, 143]. This work also suggests that there is a robustness to the structure and chemistry of binding sites and their associated binding ligands that needs to be more effectively exploited for both general functional inference as well as ligand screening. The fact that ‘template ligands’ from distantly related template structures conserve aspects of binding even as the binding sites become somewhat distorted (with an average local RMSD of 2.15 ± 0.77 Å), suggests that many ligand docking algorithms that require a highly accurate experimental structure [106, 151–153] are missing the essential features of binding. Nature itself tolerates binding site modifications in the range of ∼1.5–3 Å while retaining the ability to bind related ligands with strongly conserved substructures. The utility of a lower resolution description [135, 136] for docking as in Q-Dock [36] is not only of practical utility but also recapitulates aspects of the features of the ligand–receptor complex that are exhibited across evolutionary distant proteins. It is quite likely that there are other functional properties that can be detected by extensions of the FINDSITE approach. The key idea is to find a set of distantly related structures, identify common functional features and then transfer these features to the protein of interest. Thus, this is a promising avenue of investigation that holds considerable promise in extending the range and scope of structure-based approaches to protein function prediction.
Key Points.
The structural diversity of the binding site suggests binding site structural degeneracy that can be exploited in low-resolution modeling.
For a distantly related family of proteins, evolution provides signal averaging that can be employed to infer the structural and chemical features of binding sites that are strongly conserved throughout evolution.
Low-resolution predicted structures can be used for ligand docking, ranking and functional inference.
Combined evolution/structure-based approaches provide complementary information that can be exploited for quite high accuracy, proteome scale functional inference.
The FINDSITE algorithm combines these ideas into a robust evolution/structure-based approach to binding site detection, ligand virtual screening and functional inference.
FUNDING
National Institutes of Health [grant numbers GM-48835, GM-37408 to J.S.].
Biographies
Jeffrey Skolnick is Director of the Center for the Study of Systems Biology at the Georgia Institute of Technology. He has a PhD in Chemistry from Yale University. His primary interests are in the development and application of algorithms for the prediction of protein structure and function, drug discovery and cancer metabolomics.
Michal Brylinski is a Research Scientist in the Center for the Study of Systems Biology at the Georgia Institute of Technology. He has a PhD in Chemistry from Jagiellonian University. His research interests include sequence-structure-function relationship in proteins and drug discovery and design.
References
- Fraser CM, Gocayne JD, White O, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–25. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Basu MK, Carmel L, Rogozin IB, et al. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18:449–61. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Foerster H, Fulcher CA, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–31. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin V, Gelfand MS, Mironov AA, et al. A metabolic network in the evolutionary context: multiscale structure and modularity. Proc Natl Acad Sci USA. 2006;103:8774–9. doi: 10.1073/pnas.0510258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L, Heath JR, Phelps ME, et al. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–3. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- Hood L, Perlmutter RM. The impact of systems approaches on biological problems in drug discovery. Nat Biotechnol. 2004;22:1215–7. doi: 10.1038/nbt1004-1215. [DOI] [PubMed] [Google Scholar]
- Betz SF, Baxter SM, Fetrow JS. Function first: a powerful approach to post-genomic drug discovery. Drug Discov Today. 2002;7:865–71. doi: 10.1016/s1359-6446(02)02398-x. [DOI] [PubMed] [Google Scholar]
- Skolnick J, Fetrow JS. From genes to protein structure and function: novel applications of computational approaches in the genomic era. Trends Biotechnol. 2000;18:34–9. doi: 10.1016/s0167-7799(99)01398-0. [DOI] [PubMed] [Google Scholar]
- Arakaki AK, Tian W, Skolnick J. High precision multi-genome scale reannotation of enzyme function by EFICAz. BMC Genomics. 2006;7:315. doi: 10.1186/1471-2164-7-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Vandergriff J, Campbell M, et al. Assessment of genome-wide protein function classification for Drosophila melanogaster. Genome Res. 2003;13:2118–28. doi: 10.1101/gr.771603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin F, Rance B, Itoh M, et al. Using protein motif combinations to update KEGG pathway maps and orthologue tables. Genome Inform. 2004;15:266–75. [PubMed] [Google Scholar]
- Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinform. 2008;9:210–9. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- Tian W, Arakaki AK, Skolnick J. EFICAz: a comprehensive approach for accurate genome-scale enzyme function inference. Nucleic Acids Res. 2004;32:6226–39. doi: 10.1093/nar/gkh956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol. 2003;333:863–82. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J Mol Biol. 2005;346:1173–88. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Chandonia JM, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–25. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LH, Lewis TE, Addou S, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–7. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci USA. 2005;102:1029–34. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett GJ, Porter CT, Borkakoti N, et al. Analysis of catalytic residues in enzyme active sites. J Mol Biol. 2002;324:105–21. doi: 10.1016/s0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- Fetrow JS, Skolnick J. Method for prediction of protein function from sequence using the sequence-to-structure-to-function paradigm with application to glutaredoxins/thioredoxins and T1 ribonucleases. J Mol Biol. 1998;281:949–68. doi: 10.1006/jmbi.1998.1993. [DOI] [PubMed] [Google Scholar]
- Gao M, Skolnick J. DBD-Hunter: a knowledge-based method for the prediction of DNA-protein interactions. Nucleic Acids Res. 2008;36:3978–92. doi: 10.1093/nar/gkn332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit SB, Gosar D, Abhiman S, et al. SUPFAM—a database of potential protein superfamily relationships derived by comparing sequence-based and structure-based families: implications for structural genomics and function annotation in genomes. Nucleic Acids Res. 2002;30:289–93. doi: 10.1093/nar/30.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley ML, Schmidt T, Wagner C, et al. The PEDANT genome database in 2005. Nucleic Acids Res. 2005;33:D308–10. doi: 10.1093/nar/gki019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Russell RB. Annotation in three dimensions. PINTS: Patterns in Non-homologous Tertiary Structures. Nucleic Acids Res. 2003;31:3341–4. doi: 10.1093/nar/gkg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol. 2000;297:233–49. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- Schnur DM. Recent trends in library design: ‘rational design' revisited. Curr Opin Drug Discov Devel. 2008;11:375–80. [PubMed] [Google Scholar]
- Shacham S, Marantz Y, Bar-Haim S, et al. PREDICT modeling and in-silico screening for G-protein coupled receptors. Proteins. 2004;57:51–86. doi: 10.1002/prot.20195. [DOI] [PubMed] [Google Scholar]
- Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discov. 2003;2:527–41. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci USA. 2004;101:7594–99. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brylinski M, Skolnick J. Q-Dock: Low-resolution flexible ligand docking with pocket-specific threading restraints. J Comput Chem. 2008;29:1574–88. doi: 10.1002/jcc.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci USA. 2008;105:129–34. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I. Structural genomics. Acc Chem Res. 2003;36:155. doi: 10.1021/ar030023t. [DOI] [PubMed] [Google Scholar]
- Brenner SE. A tour of structural genomics. Nat Rev Genet. 2001;2:801–9. doi: 10.1038/35093574. [DOI] [PubMed] [Google Scholar]
- Burley SK, Almo SC, Bonanno JB, et al. Structural genomics: beyond the human genome project. Nat Genet. 1999;23:151–7. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- Chandonia JM, Kim SH. Structural proteomics of minimal organisms: conservation of protein fold usage and evolutionary implications. BMC Struct Biol. 2006;6:7. doi: 10.1186/1472-6807-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M, Edwards A, Arrowsmith CH, et al. Structural genomics: current progress. Science. 2003;299:1663. doi: 10.1126/science.299.5613.1663b. [DOI] [PubMed] [Google Scholar]
- Skolnick J, Fetrow JS, Kolinski A. Structural genomics and its importance for gene function analysis. Nat Biotechnol. 2000;18:283–7. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- Yee A, Gutmanas A, Arrowsmith CH. Solution NMR in structural genomics. Curr Opin Struct Biol. 2006;16:611–7. doi: 10.1016/j.sbi.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Kryshtafovych A, Fidelis K, Moult J. Progress from CASP6 to CASP7. Proteins. 2007;69(Suppl. 8):194–207. doi: 10.1002/prot.21769. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Curr Opin Chem Biol. 2006;10:492–7. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Gulick AM, Palmer DR, Babbitt PC, et al. Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida. Biochemistry. 1998;37:14358–68. doi: 10.1021/bi981123n. [DOI] [PubMed] [Google Scholar]
- Hasson MS, Schlichting I, Moulai J, et al. Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase. Proc Natl Acad Sci USA. 1998;95:10396–401. doi: 10.1073/pnas.95.18.10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang RA, Sali A, Babbitt PC. Evolutionarily conserved substrate substructures for automated annotation of enzyme superfamilies. PLoS Comput Biol. 2008;4:e1000142. doi: 10.1371/journal.pcbi.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Hadley C. Threading methods for protein structure prediction. In: Higgins D, Taylor WR, editors. Bioinformatics: Sequence, structure and databanks. Heidelberg: Springer-Verlag; 2000. pp. 1–13. [Google Scholar]
- Skolnick J, Kihara D, Zhang Y. Development and large scale benchmark testing of the PROSPECTOR_3 threading algorithm. Proteins. 2004;56:502–18. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Iliopoulos I, Kyrpides NC, et al. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Can sequence determine function? Genome Biol. 2000;1:REVIEWS0005. doi: 10.1186/gb-2000-1-5-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A, Cravatt BF. Assignment of protein function in the postgenomic era. Nat Chem Biol. 2005;1:130–42. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]
- Bindewald E, Skolnick J. A scoring function for docking ligands to low-resolution protein structures. J Comput Chem. 2005;26:374–83. doi: 10.1002/jcc.20175. [DOI] [PubMed] [Google Scholar]
- Wojciechowski M, Skolnick J. Docking of small ligands to low-resolution and theoretically predicted receptor structures. J Comput Chem. 2002;23:189–97. doi: 10.1002/jcc.1165. [DOI] [PubMed] [Google Scholar]
- Rost B, Liu J, Nair R, et al. Automatic prediction of protein function. Cell Mol Life Sci. 2003;60:2637–50. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Dietmann S, Frishman D, et al. MIPS: analysis and annotation of genome information in 2007. Nucleic Acids Res. 2008;36:D196–201. doi: 10.1093/nar/gkm980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg I, Jambon M, Godzik A. New avenues in protein function prediction. Protein Sci. 2006;15:1527–9. doi: 10.1110/ps.062158406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Koonin EV. Predicting functions from protein sequences—where are the bottlenecks? Nat Genet. 1998;18:313–8. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- Ouzounis CA, Karp PD. The past, present and future of genome-wide re-annotation. Genome Biol. 2002;3:COMMENT2001. doi: 10.1186/gb-2002-3-2-comment2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–31. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Ouzounis CA. Whole-genome sequence annotation: Going wrong with confidence. Mol Microbiol. 1999;32:886–7. doi: 10.1046/j.1365-2958.1999.01380.x. [DOI] [PubMed] [Google Scholar]
- del Sol Mesa A, Pazos F, Valencia A. Automatic methods for predicting functionally important residues. J Mol Biol. 2003;326:1289–1302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- Yao H, Kristensen DM, Mihalek I, et al. An accurate, sensitive, and scalable method to identify functional sites in protein structures. J Mol Biol. 2003;326:255–61. doi: 10.1016/s0022-2836(02)01336-0. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–6. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi H, Gerstein M. The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J Mol Biol. 1999;288:147–64. doi: 10.1006/jmbi.1999.2661. [DOI] [PubMed] [Google Scholar]
- Kihara D, Skolnick J. Microbial genomes have over 72% structure assignment by the threading algorithm PROSPECTOR_Q. Proteins. 2004;55:464–473. doi: 10.1002/prot.20044. [DOI] [PubMed] [Google Scholar]
- Fleming K, Kelley LA, Islam SA, et al. The proteome: structure, function and evolution. Philos Trans R Soc Lond B Biol Sci. 2006;361:441–51. doi: 10.1098/rstb.2005.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelryck T. Efficient identification of side-chain patterns using a multidimensional index tree. Proteins. 2003;51:96–108. doi: 10.1002/prot.10338. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ. Recognition of spatial motifs in protein structures. J Mol Biol. 1999;285:1887–97. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- Liang MP, Brutlag DL, Altman RB. Automated construction of structural motifs for predicting functional sites on protein structures. Pac Symp Biocomput. 2003;8:204–15. doi: 10.1142/9789812776303_0020. [DOI] [PubMed] [Google Scholar]
- Oldfield TJ. Data mining the protein data bank: residue interactions. Proteins. 2002;49:510–28. doi: 10.1002/prot.10221. [DOI] [PubMed] [Google Scholar]
- Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13:121–30. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Pazos F, Sternberg MJ. Automated prediction of protein function and detection of functional sites from structure. Proc Natl Acad Sci USA. 2004;101:14754–9. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RB. Detection of protein three-dimensional side-chain patterns: new examples of convergent evolution. J Mol Biol. 1998;279:1211–27. doi: 10.1006/jmbi.1998.1844. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Borkakoti N, Thornton JM. TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 1997;6:2308–23. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Morris GM, Olson AJ, et al. Recognition templates for predicting adenylate-binding sites in proteins. J Mol Biol. 2001;314:1245–55. doi: 10.1006/jmbi.2000.5201. [DOI] [PubMed] [Google Scholar]
- Polacco BJ, Babbitt PC. Automated discovery of 3D motifs for protein function annotation. Bioinformatics. 2006;22:723–30. doi: 10.1093/bioinformatics/btk038. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–47. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen DM, Ward RM, Lisewski AM, et al. Prediction of enzyme function based on 3D templates of evolutionarily important amino acids. BMC Bioinformatics. 2008;9:17. doi: 10.1186/1471-2105-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RM, Erdin S, Tran TA, et al. De-orphaning the structural proteome through reciprocal comparison of evolutionarily important structural features. PLoS ONE. 2008;3:e2136. doi: 10.1371/journal.pone.0002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AT, Jackson RM. Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening. Curr Protein Pept Sci. 2006;7:395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- Wei L, Huang ES, Altman RB. Are predicted structures good enough to preserve functional sites? Structure. 1999;7:643–50. doi: 10.1016/s0969-2126(99)80085-9. [DOI] [PubMed] [Google Scholar]
- Jones S, Shanahan HP, Berman HM, et al. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–98. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilagyi A, Skolnick J. Efficient prediction of nucleic acid binding function from low-resolution protein structures. J Mol Biol. 2006;358:922–33. doi: 10.1016/j.jmb.2006.02.053. [DOI] [PubMed] [Google Scholar]
- Peters KP, Fauck J, Frommel C. The automatic search for ligand binding sites in proteins of known three-dimensional structure using only geometric criteria. J Mol Biol. 1996;256:201–13. doi: 10.1006/jmbi.1996.0077. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser F, Morris RJ, Najmanovich RJ, et al. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62:479–88. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- Hendlich M, Rippmann F, Barnickel G. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model. 1997;15:359–63,389. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–97. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly M. Analytical molecular surface calculation. Journal of Applied Crystallography. 1983;16:548–58. [Google Scholar]
- Ondrechen MJ, Clifton JG, Ringe D. THEMATICS: a simple computational predictor of enzyme function from structure. Proc Natl Acad Sci USA. 2001;98:12473–8. doi: 10.1073/pnas.211436698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brylinski M, Kochanczyk M, Konieczny L, et al. Sequence-structure-function relation characterized in silico. In Silico Biol. 2006;6:589–600. [PubMed] [Google Scholar]
- Elcock AH. Prediction of functionally important residues based solely on the computed energetics of protein structure. J Mol Biol. 2001;312:885–96. doi: 10.1006/jmbi.2001.5009. [DOI] [PubMed] [Google Scholar]
- Hetenyi C, van der Spoel D. Blind docking of drug-sized compounds to proteins with up to a thousand residues. FEBS Lett. 2006;580:1447–50. doi: 10.1016/j.febslet.2006.01.074. [DOI] [PubMed] [Google Scholar]
- Oshiro CM, Kuntz ID, Dixon JS. Flexible ligand docking using a genetic algorithm. J Comput Aided Mol Des. 1995;9:113–30. doi: 10.1007/BF00124402. [DOI] [PubMed] [Google Scholar]
- Cummings MD, DesJarlais RL, Gibbs AC, et al. Comparison of automated docking programs as virtual screening tools. J Med Chem. 2005;48:962–76. doi: 10.1021/jm049798d. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Rodrigo J, Muller P, et al. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins. 2004;57:225–42. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- Ewing TJ, Makino S, Skillman AG, et al. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15:411–28. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–48. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T, et al. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261:470–89. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–62. [Google Scholar]
- Ferrara P, Gohlke H, Price DJ, et al. Assessing scoring functions for protein-ligand interactions. J Med Chem. 2004;47:3032–47. doi: 10.1021/jm030489h. [DOI] [PubMed] [Google Scholar]
- Kim R, Skolnick J. Assessment of programs for ligand binding affinity prediction. J Comput Chem. 2008;29:1316–31. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins. 2004;56:235–49. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- Warren GL, Andrews CW, Capelli AM, et al. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–31. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- Battey JN, Kopp J, Bordoli L, et al. Automated server predictions in CASP7. Proteins. 2007;69(Suppl. 8):68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- Das R, Qian B, Raman S, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69(Suppl. 8):118–28. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- Jauch R, Yeo HC, Kolatkar PR, et al. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69(Suppl. 8):57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- Kopp J, Bordoli L, Battey JN, et al. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl. 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- Qiu J, Sheffler W, Baker D, et al. Ranking predicted protein structures with support vector regression. Proteins. 2008;71:1175–82. doi: 10.1002/prot.21809. [DOI] [PubMed] [Google Scholar]
- Read RJ, Chavali G. Assessment of CASP7 predictions in the high accuracy template-based modeling category. Proteins. 2007;69(Suppl. 8):27–37. doi: 10.1002/prot.21662. [DOI] [PubMed] [Google Scholar]
- Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl. 8):108–17. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Pandit SB, Lee SY, et al. Analysis of TASSER-based CASP7 protein structure prediction results. Proteins. 2007;69(Suppl. 8):90–7. doi: 10.1002/prot.21649. [DOI] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–5. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–91. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Iwadate M, Suzuki E, et al. Enlarged FAMSBASE: protein 3D structure models of genome sequences for 41 species. Nucleic Acids Res. 2003;31:463–8. doi: 10.1093/nar/gkg117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki AK, Zhang Y, Skolnick J. Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–96. doi: 10.1093/bioinformatics/bth044. [DOI] [PubMed] [Google Scholar]
- Bissantz C, Bernard P, Hibert M, et al. Protein-based virtual screening of chemical databases. II. Are homology models of G-Protein Coupled Receptors suitable targets? Proteins. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- Enyedy IJ, Ling Y, Nacro K, et al. Discovery of small-molecule inhibitors of Bcl-2 through structure-based computer screening. J Med Chem. 2001;44:4313–24. doi: 10.1021/jm010016f. [DOI] [PubMed] [Google Scholar]
- Evers A, Hessler G, Matter H, et al. Virtual screening of biogenic amine-binding G-protein coupled receptors: comparative evaluation of protein- and ligand-based virtual screening protocols. J Med Chem. 2005;48:5448–65. doi: 10.1021/jm050090o. [DOI] [PubMed] [Google Scholar]
- McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J Med Chem. 2003;46:2895–907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- Erickson JA, Jalaie M, Robertson DH, et al. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem. 2004;47:45–55. doi: 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- Huang SY, Zou X. Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- Ferrari AM, Wei BQ, Costantino L, et al. Soft docking and multiple receptor conformations in virtual screening. J Med Chem. 2004;47:5076–84. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakser IA. Low-resolution docking: prediction of complexes for underdetermined structures. Biopolymers. 1996;39:455–64. doi: 10.1002/(SICI)1097-0282(199609)39:3%3C455::AID-BIP16%3E3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Vakser IA. Protein docking for low-resolution structures. Protein Eng. 1995;8:371–7. doi: 10.1093/protein/8.4.371. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Russell RB, Sasieni PD, Sternberg MJ. Supersites within superfolds. Binding site similarity in the absence of homology. J Mol Biol. 1998;282:903–18. doi: 10.1006/jmbi.1998.2043. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–09. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Arakaki AK, Skolnick J. TASSER: an automated method for the prediction of protein tertiary structures in CASP6. Proteins. 2005;61(Suppl. 7):91–8. doi: 10.1002/prot.20724. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Tertiary structure predictions on a comprehensive benchmark of medium to large size proteins. Biophys J. 2004;87:2647–55. doi: 10.1529/biophysj.104.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11:1046–53. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Xue L, Stahura FL, Bajorath J. Similarity search profiling reveals effects of fingerprint scaling in virtual screening. J Chem Inf Comput Sci. 2004;44:2032–39. doi: 10.1021/ci0400819. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth D, Lehrach H, Hennig S. GOblet: a platform for Gene Ontology annotation of anonymous sequence data. Nucleic Acids Res. 2004;32:W313–7. doi: 10.1093/nar/gkh406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehetner G. OntoBlast function: From sequence similarities directly to potential functional annotations by ontology terms. Nucleic Acids Res. 2003;31:3799–803. doi: 10.1093/nar/gkg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SM, Rosenblum JS, Knutson S, et al. Synergistic computational and experimental proteomics approaches for more accurate detection of active serine hydrolases in yeast. Mol Cell Proteomics. 2004;3:209–25. doi: 10.1074/mcp.M300082-MCP200. [DOI] [PubMed] [Google Scholar]
- Babbitt PC. Definitions of enzyme function for the structural genomics era. Curr Opin Chem Biol. 2003;7:230–7. doi: 10.1016/s1367-5931(03)00028-0. [DOI] [PubMed] [Google Scholar]
- Chen HM, Liu BF, Huang HL, et al. SODOCK: swarm optimization for highly flexible protein-ligand docking. J Comput Chem. 2007;28:612–23. doi: 10.1002/jcc.20542. [DOI] [PubMed] [Google Scholar]
- Lorber DM, Shoichet BK. Flexible ligand docking using conformational ensembles. Protein Sci. 1998;7:938–50. doi: 10.1002/pro.5560070411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taufer M, Crowley M, Price DJ, et al. Study of a highly accurate and fast protein-ligand docking method based on molecular dynamics. Concurrency and Computation: Practice and Experience. 2005;17:1627–41. [Google Scholar]