Summary
MIG-10/RIAM/lamellipodin (MRL) proteins link activated Ras-GTPases with actin regulatory Ena/VASP proteins to induce local changes in cytoskeletal dynamics and cell motility. MRL proteins alter monomeric (G):filamentous (F) actin ratios, but the impact of these changes had not been fully appreciated. We report here that the Drosophila MRL ortholog, pico, is required for tissue and organismal growth. Reduction in pico levels resulted in reduced cell division rates, growth retardation, increased G:F actin ratios and lethality. Conversely, pico overexpression reduced G:F actin ratios and promoted tissue overgrowth in an epidermal growth factor (EGF) receptor (EGFR)-dependent manner. Consistently, in HeLa cells, lamellipodin was required for EGF-induced proliferation. We show that pico and lamellipodin share the ability to activate serum response factor (SRF), a transcription factor that responds to reduced G:F-actin ratios via its co-factor Mal. Genetics data indicate that mal/SRF levels are important for pico-mediated tissue growth. We propose that MRL proteins link EGFR activation to mitogenic SRF signaling via changes in actin dynamics.
Keywords: CELLCYCLE, CELLBIO, DEVBIO
Introduction
The construction of properly sized and functional tissues and organs during animal development requires tight control of cell growth, proliferation, differentiation, and death. Networks of intracellular signal transduction pathways that respond to various secreted ligands and cell surface proteins coordinate these processes. Elucidating the nature of the intracellular signaling networks that connect extracellular stimuli to basic cellular machinery controlling proliferation, growth, and morphology is not only critical for the understanding of tissue size regulation during normal development, but is also important for the identification of aberrant events underlying numerous disease processes, including cancer.
A number of pathways regulating cellular development are initiated by ligation of transmembrane receptor tyrosine kinases (RTKs), such as the epidermal growth factor (EGF) receptor (EGFR). One of the key mediators of RTK signaling is the Ras GTPase, capable of activating proteins harboring Ras association (RA) domains to initiate downstream signaling pathways, such as the mitogen-activated protein kinase (MAPK) cascade, and ultimately resulting in changes in gene transcription. The Ras/MAPK and other canonical RTK signaling pathways have been well characterized, yet they cannot account for all of the observed effects of their respective extracellular signals.
The MIG-10/Rap1-GTP-interacting adaptor molecule (RIAM)/lamellipodin (Lpd) (MRL) proteins are a family of recently identified molecular adaptors, harboring an RA, pleckstrin homology (PH), and several proline-rich domains (Krause et al., 2004; Lafuente et al., 2004). Several lines of evidence indicate that MRL proteins act downstream of Ras-like GTPases and transduce extracellular signals to changes in the actin cytoskeleton, cell motility, and adhesion. In particular, Lpd interacts with active Ras and RIAM with active Rap1. Consistent with this, only RIAM is required for Rap1-induced cell adhesion (Lafuente et al., 2004; Rodriguez-Viciana et al., 2004). Lpd also binds to PI(3,4)P2 via its PH domain, which is sufficient for membrane targeting after platelet-derived growth factor stimulation (Krause et al., 2004). Both Lpd and RIAM utilize their proline-rich motifs to directly interact with the Enabled (Ena)/vasodilator-stimulated phosphoprotein (VASP) actin regulators, known to regulate lamellipodia formation and cell migration (Jenzora et al., 2005; Krause et al., 2004; Lafuente et al., 2004). In addition, Lpd knockdown impairs lamellipodia formation, whereas Lpd overexpression increases speed of lamellipodia protrusion in an Ena/VASP-dependent manner (Krause et al., 2004). Finally, both Lpd and RIAM have been shown to alter the cellular ratio between monomeric (G) and filamentous (F) actin (Krause et al., 2004; Lafuente et al., 2004), suggesting a wider role in regulating cell metabolism. Indeed, control of the G:F actin ratio is an essential way for cells to regulate gene transcription via the transcription factor serum response factor (SRF), and has been linked to changes in proliferation, migration, and differentiation (Miralles et al., 2003).
Here we report the characterization of the Drosophila MRL ortholog, which we have named pico on the basis of the retarded growth phenotype resulting from pico knockdown or loss-of-function mutant. Reduction in pico levels results in reduced rates of cell growth and proliferation, whereas ectopic expression of pico promotes coordinated cell growth and proliferation, leading to tissue overgrowth. pico's effect on cell proliferation is conserved in its mammalian ortholog, Lpd. We present evidence that pico and Lpd link extracellular signaling to tissue growth via changes in actin dynamics and SRF activation. To our knowledge, this is the first time that MRL proteins have been implicated in controlling cell proliferation and tissue growth.
Results
Pico Encodes the Only MRL Protein in Drosophila
Phylogenetic analysis has shown that pico (CG11940) encodes the only member of the MRL family of proteins in Drosophila (Krause et al., 2004; Lafuente et al., 2004). The organization of the pico transcription unit, located on the first chromosome at cytological position 18F2-4 (Consortium, 2003), is shown in Figure 1A. We identified two transcripts that are generated from alternative transcription start sites of the pico transcription unit: pico and pico-L. pico-L encodes a 1159 amino acid protein that is identical to the protein encoded by pico, except for the presence of an additional 128 N-terminal residues. Both pico proteins contain RA and PH domains and proline-rich Ena/VASP binding sites characteristic of the MRL proteins (Krause et al., 2004; Lafuente et al., 2004; see Supplemental Data and Figure S1 available online).
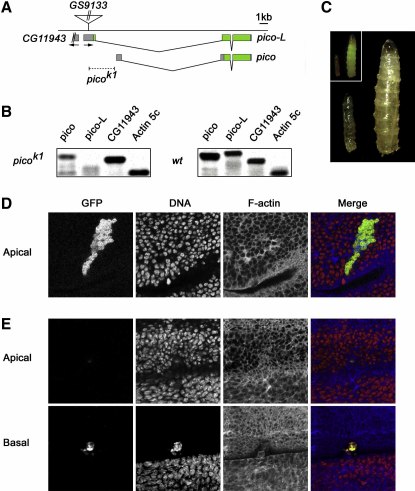
Figure 1.
pico Is Essential for Growth and Viability
(A) Genomic organization of the pico locus. pico encodes two transcripts: pico and pico-L. Untranslated regions are shown in gray; coding regions are in green. Orientation of pico and CG11943 transcription units is indicated with arrows. The insertion site of the P-element, GS9113, used to generate picok1, is marked. The deletion in picok1 is indicated with a dashed line.
(B) RT-PCR analysis showing reduction in levels of pico and pico-L expression in picok1 mutant larvae.
(C) Hemizygous picok1 mutant larva to the left, showing arrested growth compared to pico+ siblings of the same age to the right. Inset: mutant larvae were identified on the basis that they lacked a GFP balancer chromosome.
(D and E) pico mutant clones fail to divide and appear to be displaced from the wing imaginal disc epithelium. Compared to wild-type control clones (D), which are located in apical sections and contain, on average, about 45 positively marked cells, picok1 mutant clones (E) contain, on average, fewer than five cells and appear to be located at the basal surface of the wing epithelium. Merged images show cells positively marked with GFP in green, DNA stained with propidium iodide in red, and F-actin labeled with phalloidin in blue.
Pico Is Essential for Organismal Growth and Viability
To determine the in vivo function of pico, we generated a mutant allele, picok1, by imprecise excision of a viable P element transposon, which we found inserted in the pico 5′ untranslated region. picok1 showed little or no pico-L expression, reduced levels of pico, but wild-type levels of the neighboring gene CG11943 mRNA (Figure 1B), consistent with molecular analysis revealing a 2.81 kb deletion removing the 5′ end of pico-L and a large region upstream of the predicted pico transcription start site in this mutant (Figure 1A). Hemizygous picok1 animals were larval lethal and displayed phenotypes reminiscent of mutations in positive regulators of growth and proliferation: mutant larvae were dramatically reduced in size, with severely reduced endoreduplicated tissue and little or no detectable imaginal disc tissue; mutant larvae eventually died following an extended larval period of up to 2 weeks (Figure 1C, and data not shown). Heat shock-induced co-overexpression of pico and pico-L rescued the hemizygous lethality of picok1, indicating that the lethality of picok1 is due to disruption of the pico locus (data not shown). The presence of food in the guts of picok1 mutant larvae verified that they had eaten, and suggests that the inhibition of larval growth may be caused by a cellular growth defect. To assess this, we examined the behavior of picok1 mutant cells randomly generated in the wing imaginal disc by mitotic recombination. Homozygous picok1 cells could not readily be detected, but were occasionally observed to achieve clone sizes of up to 15 cells in clones positively marked with GFP. Their predominantly basal localization indicated that they might be displaced from the basal surface of the disc epithelium (Figures 1D and 1E; Figure S2). This phenotype is reminiscent of cells that have sustained inappropriate cell cycle arrest. However, unlike mutants that prevent passage through the cell cycle, but allow continued growth (Neufeld et al., 1998), pico mutant cells did not become enlarged, suggesting that pico loss-of-function also results in a cellular growth defect.
Knockdown of pico Results in Cell Growth and Proliferation Defects
To further assess the cellular requirement for pico, we used a heritable double-stranded (ds) RNA interference (RNAi) approach to flexibly knockdown pico and pico-L levels. We generated stable lines of transgenic flies carrying an inverted repeat construct (picoIR) capable of expressing intron-spliced hairpin dsRNA for a sequence common to pico and pico-L under GAL4-UAS control. Sequence analysis predicted minimal off-targets for picoIR (0 off-targets from 459 possible 19 mers). Flies expressing this construct ubiquitously under the control of tubulin-GAL4 (tub > picoIR) were semilethal; survivors were small compared to tubulin-GAL4 siblings (tub >) (Figures 2A and 2B). Levels of pico and pico-L mRNA were greatly reduced in tub > picoIR larvae compared with control animals, indicating knockdown of endogenous pico expression (Figure 2C). We also found levels of ectopic Myc-tagged Pico were severely reduced by coexpression of picoIR (Figures 2D and 2E). Ectopic overexpression of picoIR in the developing wing using MS1096-GAL4 (MS1096 > picoIR) resulted in a significant reduction in wing area (p < 0.001; Figures 2F and 2G). MS1096-GAL4 is expressed at higher levels in the dorsal half of the developing wing pouch. Accordingly, MS1096 > picoIR wings were curled upwards slightly, indicating that, relative to the ventral wing blade, the dorsal wing blade was somewhat reduced in size (data not shown). As a single wing hair marks each adult wing cell, we measured the wing hair density to gauge cell size. The wing hair density in MS1096 > picoIR wings was significantly increased relative to wild type (p < 0.01; Figures 2F and 2G), indicating that the reduction in wing area was at least in part due to a reduction in cell size. An independent inverted repeat construct (National Institute of Genetics: 11940R), showed qualitatively similar effects to picoIR, suggesting that the growth defects we observed are not due to off-target effects (data not shown).
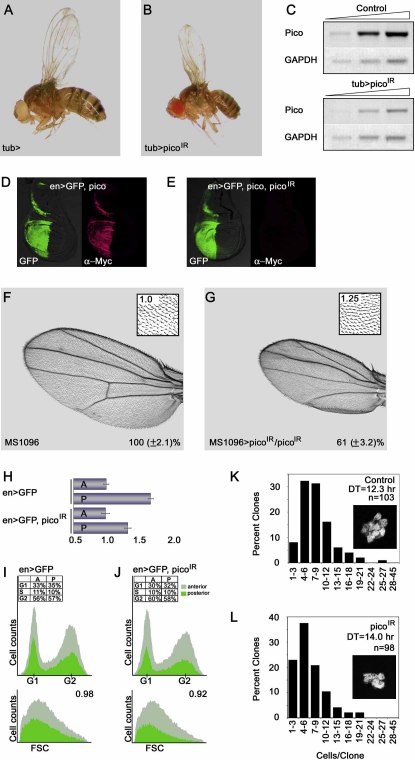
Figure 2.
Knockdown of pico by RNAi Reduces Tissue Size
Moderate ubiquitous expression of picoIR results in a reduction in overall body size.
(A and B) A tub-GAL4 fly and a tub-GAL4 UAS-picoIR (tub > picoIR) fly are shown for comparison. Quantitative measurements of female flies (n = 50 per genotype) show a 23% reduction in body weight in tub > picoIR flies (p < 0.01).
(C) Representative RT-PCR analysis showing reduction in pico and pico-L expression in tub-GAL4 and UAS-picoIR flies using primers recognizing both transcripts. A range of cDNA concentrations normalized against GAPDH was used to assess transcript levels.
(D and E) Expression of picoIR knocks down levels of Myc-tagged pico in wing imaginal discs. (D) High levels of ectopic pico in the posterior compartment marked by GFP (in green) were detected by Myc staining (in red) in en-GAL4, UAS-Myc-pico discs. (E) No Myc staining was detected in discs coexpressing picoIR.
(F and G) Compared with control (F), expression of two copies of picoIR (G) results in a significant reduction in adult wing size (p < 0.01). Male wings are shown. Wing area is expressed as a percentage of control (±SD). The reduction in wing size is due to fewer and smaller cells, as revealed by relative bristle density measurements (see insets).
(H–J) Consequences of picoIR overexpression in the posterior compartment of the wing under the control of en-GAL4. (H) Compared to control, expression of picoIR with en-GAL4 results in a reduction in size of the posterior compartment of the adult wing. A, anterior; P, posterior. Numerical scale is in arbitrary units. Error bars indicate 1 SD. (I and J) picoIR-induced growth retardation is not due to aberrant cell cycle phasing. Flow cytometry was performed on dissociated wing disc cells overexpressing GFP alone (I) or picoIR and GFP (J). GFP-positive experimental populations are marked in green; GFP-negative internal controls are marked in gray. Comparison of representative cell cycle profiles shows that picoIR does not alter cell cycle phasing. Percentage of cells in G1, S, and G2 phases of the anterior and posterior compartments is shown in insets. Graphs of forward scatter (FSC; bottom panels) show that picoIR results in a modest reduction in cell size at the third instar larval stage. The ratio of the mean FSC value of GFP-positive verses GFP-negative cells is shown in the top right corner of the bottom panels.
(K and L) The distribution of clonal cell number for control (K) and picoIR-expressing clones (L). Cell-doubling time (DT) is markedly increased by picoIR. Insets show representative clones of each genotype as visualized by nuclear GFP.
To study the effects of changes in steady-state levels of pico in more detail in a defined cell population, we used en-GAL4 to continuously drive picoIR expression in the posterior compartment of the wing disc from the earliest stages of disc formation. Adult wing measurements showed that en-GAL4, UAS-picoIR (en > picoIR) flies exhibited a specific reduction in the area of the posterior wing compartment (Figure 2H). The ratio of posterior to anterior area of en > picoIR wings (1.33:1) was significantly reduced compared to that of control wings (1.68:1; p < 0.01). To determine the effect of pico on cell cycle progression, we obtained cell cycle profiles using flow cytometry on live cells from dissociated en > picoIR wing imaginal discs. DNA profiles revealed that cells expressing picoIR had normal cell cycle phasing, while forward scatter analysis confirmed that cells were slightly smaller than controls (Figures 2I and 2J). To assess in vivo cell division rates, we generated clones coexpressing GFP and picoIR using the flip-out technique (Neufeld et al., 1998) and counted the number of GFP-expressing cells per clone 38 hr after induction. Clones expressing picoIR had significantly fewer cells than control clones expressing GFP alone (p < 0.01), and therefore contained cells that divided at a slower rate (Figures 2K and 2L).
Ectopic pico Promotes Coordinated Cell Growth and Proliferation
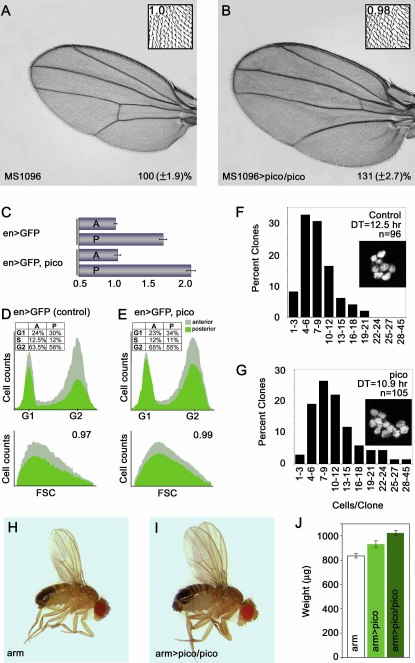
To determine whether pico is limiting for tissue growth, we examined the effect of ectopically expressing pico in the wing. Ectopic pico resulted in significant wing overgrowth with little or no disruption of patterning (p < 0.01; Figures 3A and 3B). MS1096 > pico wings were curled downwards slightly, indicating that, relative to the ventral wing blade, the dorsal wing blade was enlarged (data not shown). Ectopic pico had no effect on wing hair density, suggesting that increased tissue growth driven by pico results from an increased rate of cell division with a matching increase in growth rate. When pico was overexpressed together with picoIR, the phenotypic effects of each alone were nullified, resulting in wings that were of wild-type size and appearance (data not shown). Ectopic expression of pico in the posterior compartment of the developing wing resulted in an expansion of the posterior tissue (Figure 3C). The posterior to anterior area ratio of wings from en-GAL4, UAS-pico flies (2.03:1) was significantly increased compared to that of control wings (1.65:1; p < 0.01). Flow cytometric analysis revealed that cells overexpressing pico had normal cell cycle phasing and were of a normal size (Figures 3D and 3E). Clones expressing pico had significantly more cells than control clones expressing GFP alone (p < 0.01), and therefore contained cells that divided at a faster rate (Figures 3F and 3G). Therefore, in the context of the developing wing, ectopic pico induces a coordinated increase in cell cycle and cell growth rates, leading to substantial tissue overgrowth. Moderate ectopic expression of pico throughout the fly resulted in dose-dependent increase in body mass, indicating that pico functions as a general growth promoter in multiple tissues (Figures 3H–3J). These data show that the loss-of-function phenotype of pico is complementary to its gain-of-function phenotype. The gain-of-function phenotype appears to reflect the overactivation of the natural function of pico, which is to promote cell growth and proliferation.
Figure 3.
Ectopic Expression of pico Promotes Tissue Overgrowth
(A) Male adult MS1096-GAL4 wing resembling wild-type.
(B) Overexpression of pico in the developing wing pouch (MS1096 > pico/pico) results in significant wing overgrowth (p < 0.01). Wing area is expressed as a percentage of control (±SD). Insets are magnified images of wings showing the relative wing bristle density.
(C–E) Consequences of pico overexpression in the posterior compartment of the wing under the control of en-GAL4. (C) Adult male wings expressing pico and GFP (en > GFP, pico) show an increase in the size of the posterior compartment compared to control (en > GFP). A, anterior; P, posterior. Numerical scale is in arbitrary units. Error bars indicate ±1 SD.
(D and E) Results of flow cytometry on dissociated wing disc cells overexpressing (D) GFP alone or (E) pico and GFP. GFP-positive experimental populations are in green; GFP-negative internal controls are in gray. Comparison of representative cell cycle profiles shows that ectopic pico does not alter cell cycle phasing. Percentage of cells in G1, S, and G2 phases of the A and P compartments is shown in the top left corners of the upper panels. Graphs of forward scatter (FSC; bottom panels) show that cell size is relatively unaffected by ectopic pico. The ratio of the mean FSC value of GFP-positive verses GFP-negative cells is shown in the top right corner of the lower panels.
(F and G) The distribution of clonal cell number for control and pico-expressing clones. Cell doubling time (DT) is markedly decreased by pico. Insets show representative clones of each genotype as visualized by nuclear GFP.
(H–J) Moderate ubiquitous expression of pico results in an increase in organism size; arm-GAL4 male fly (H) and arm-GAL4, UAS-pico/UAS-pico male fly (I) showing size difference. (J) Quantitative representation of adult weight in micrograms from flies of different genotypes, as indicated.
Pico Partially Disrupts Epithelial Architecture but Does Not Induce Cell Death
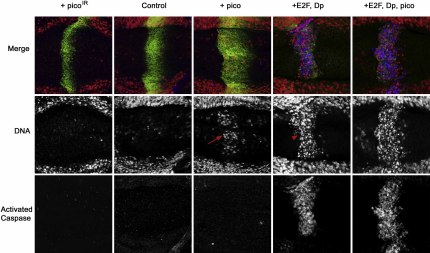
Mutations that slow the rate of cell proliferation can show intrinsic survival defects. Conversely, strong proliferative stimuli, such as the oncogenes E2F and Dp, can induce apoptosis, and net proliferation only occurs when apoptosis is simultaneously prevented. To determine whether cells expressing different levels of pico undergo cell death, we examined the effect of expressing pico or picoIR in positively marked (GFP-positive) cells with ptc-GAL4. Cells undergoing apoptosis were identified by activated caspase 3 staining. Compared to controls expressing GFP alone, the zone of GFP-positive cells in discs ectopically expressing picoIR was narrower and contained fewer cells (Figure 4). We did not observe elevated activated caspase 3 staining, and all nuclei appeared normal, suggesting that cells with reduced pico levels did not have an intrinsic survival defect. Ectopic expression of pico under control of ptc-GAL4 resulted in an expansion of the number of ptc > GFP cells. Pico expressing cells often exhibited an abnormal distribution toward the basal side of the wing disc epithelium, despite appearing morphologically normal (Figure 4). The effect of pico overexpression on adult wing size was not enhanced when apoptosis was suppressed by coexpression of the caspase inhibitor p35 (data not shown). Therefore, stimulation of growth by pico was not associated with an increase in apoptosis. Studies of the miRNA bantam have shown that genes stimulating cell proliferation can simultaneously suppress apoptosis (Brennecke et al., 2003). Therefore, we wondered whether pico could suppress proliferation-induced apoptosis caused by ectopic E2F and Dp. As previously reported, cells overexpressing E2F and Dp under the control of ptc-GAL4 showed pyknotic nuclei and elevated levels of activated caspase 3 in basal optical sections of wing discs, indicative of apoptosis (Figure 4) (Brennecke et al., 2003). Coexpression of pico with E2F/Dp had no effect on the levels of activated caspase (Figure 4). Therefore, stimulation of growth and proliferation by pico is not associated with an increase in apoptosis, and pico is not capable of suppressing apoptosis induced by proliferative stimuli from E2F and DP oncogenes.
Figure 4.
Reduction or Elevation of pico Levels Does Not Induce Apoptosis
The basal view of wing disc epithelia is shown. Discs expressing picoIR with ptc-GAL4 have a reduced zone of GFP-labeled cells compared with discs expressing GFP alone (control). Conversely, discs expressing ectopic pico displayed an expanded zone of GFP-labeled cells. Some cells were located basally, but did not show elevated caspase staining and did not have pyknotic nuclei (arrow). Expression of E2F and Dp resulted in high levels of caspase staining and nuclei were pyknotic (arrowhead). Coexpression of pico was unable to suppress E2F/Dp-induced cell death. Merged images show activated caspase staining in blue to visualize apoptotic cells, ptc-GAL4-expressing cells visualized by GFP in green, and propidium-labeled nuclei in red. All images were taken with identical settings to permit comparison of the intensity of activated caspase.
Pico Appears to Act in a Noncanonical EGFR-Dependent Pathway
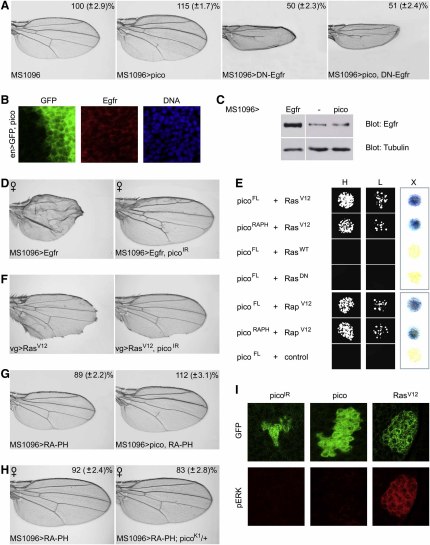
MRL proteins have been postulated to link activated growth factor receptors, such as the EGFR via interactions with Ras GTPases to changes in actin dynamics (Krause et al., 2004). The Drosophila EGFR is critical for imaginal disc growth, as well as for patterning and differentiation (Diaz-Benjumea and Garcia-Bellido, 1990). Ectopic expression of dominant negative Egfr (EgfrDN), which is thought to interfere with signaling by forming inactive heterodimers with the wild-type receptor (Kashles et al., 1991), results in dramatically reduced, narrow wings and loss of wing veins (L3 and distal parts of L4 and L5) (Guichard et al., 1999). Wings coexpressing pico and EgfrDN resembled those expressing EgfrDN alone (Figure 5A), indicating that pico failed to promote cell growth and proliferation when EGFR activity was compromised. This might indicate that pico is either upstream of Egfr or, alternatively, that pico needs to be activated by Egfr signaling to have its effect. Ectopic pico appeared to act cell autonomously, did not phenocopy Egfr-induced wing venation defects (Figure 3), and did not affect EGFR levels or distribution (Figures 5B and 5C), suggesting that pico is unlikely to be upstream of Egfr or its ligands. Conversely, picoIR suppressed the effect of Egfr overexpression on wing size, but not venation (Figure 5D), indicating that pico may be an effector of Egfr-mediated tissue growth.
Figure 5.
pico Appears to Act in a Noncanonical EGFR-Dependent Pathway
(A) The effect of MS1096 > pico is completely abrogated by coexpression of EgfrDN. Wing area is expressed as a percentage of MS1096-GAL4-only control (±SD).
(B) Ectopic pico does not affect EGFR distribution or levels in wing discs.
(C) EGFR levels on immunoblots of wing disc extracts are unaffected by ectopic pico.
(D) Wing overgrowth, but not aberrant wing venation, induced by ectopic Egfr is partially suppressed by co-overexpression of picoIR.
(E) Pico full-length and picoRA-PH bind RasV12 and Rap1V12 in the yeast two-hybrid system, but not controls: wild-type Ras (RasWT), dominant negative Ras (RasDN), or an unrelated control (NIPP1). Interaction is indicated by blue X-gal coloration (X) and growth on auxotrophic media at high (H) and low (L) density.
(F) The effect of overexpressing RasV12 along the presumptive wing margin is partially suppressed by cooverexpression of picoIR.
(G and H) Reduced tissue growth resulting from ectopic expression of picoRA-PH can be (G) suppressed by co-overexpression of full-length pico or (H) enhanced in females by pico loss of function. Wing area is expressed as a percentage of MS1096-GAL4 only control (±SD).
(I) pico does not activate MAPK. Ectopic expression of picoIR or pico in flip-out clones marked with GFP (in green) do not affect levels of activated MAPK (dpERK, in red), whereas ectopic RasV12 leads to elevated levels of activated MAPK and rounded clones.
EGFR is known to activate Ras-like GTPases capable of binding to mammalian MRL proteins (Rodriguez-Viciana et al., 2004). We found that full-length Pico and a fragment of Pico containing the RA-PH domain (PicoRA-PH) bound to constitutively activated Ras and Rap1 (RasV12 and Rap1V12, respectively) in the two-hybrid system. Pico did not bind to wild-type or dominant-negative Ras, suggesting that pico may be an effector of Ras GTPases (Figure 5E). In support of this, we found that ectopic picoIR partially suppressed the effect of ectopic overexpression of RasV12 in the wing (Figure 5F). In addition, wings overexpressing picoRA-PH were significantly smaller than controls (89 ± 2.2%; p < 0.01). This effect could be overcome by cooverexpression of full-length pico or enhanced by loss of one copy of pico (Figures 5G and 5H), suggesting that ectopic PicoRA-PH may interfere with pico function by competing for its binding partners, and that other regions of the pico protein are required for its growth-promoting effect. To determine whether pico contributes to canonical Ras effector signaling, we stained tissues ectopically expressing pico in flip-out clones with an antibody that recognizes the diphosphorylated (activated) form of MAPK (Gabay et al., 1997). Ectopic pico had no detectable effect on activated MAPK staining relative to controls (Figure 5I). Taken together, these data suggest that pico acts downstream of EGFR and may act as a Ras or Rap1 effector, but does not induce the canonical MAPK pathway.
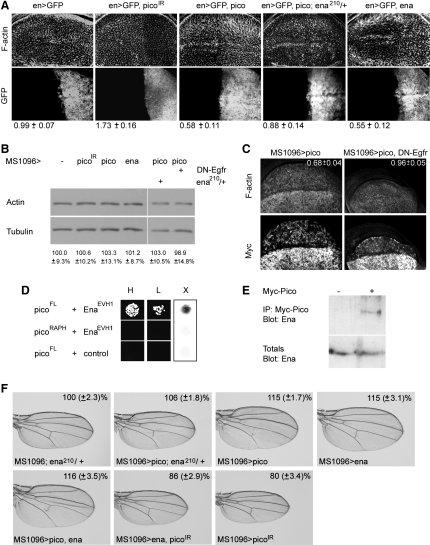
Pico Interacts with ena and Modifies Actin Dynamics
Mammalian MRL proteins have been shown to stimulate F-actin formation without influencing total actin content (Krause et al., 2004; Lafuente et al., 2004). We found that, in the context of the wing imaginal disc, ectopic pico promoted F-actin formation, and ectopic picoIR reduced F-actin levels (Figure 6A). Total actin content was unaffected in extracts (Figure 6B), suggesting that pico regulates the ratio of G:F-actin content. EgfrDN suppressed pico-mediated changes in G:F actin ratio (Figures 6B and 6C), suggesting that this aspect of pico function is dependent on EGFR signaling.
Figure 6.
pico Interacts with ena and Modifies Actin Dynamics
(A) Representative wing discs of the indicated genotypes stained for F-actin. Ectopic expression of pico or ena results in elevated F-actin, whereas ectopic picoIR reduces F-actin. pico-induced F-actin formation is dominantly suppressed by ena210. Ectopic expression, driven by en-GAL4, is limited to the posterior compartment marked by GFP. Quantitation of ratios of anterior:posterior F-actin levels is shown at the bottom of (A) (±SD).
(B) Immunoblots showing levels of total actin normalized to tubulin in extracts from wing discs of the indicated genotypes. Mean actin levels from six independent experiments are expressed below as percentage of the control ± SD.
(C) Ectopic expression of Myc-pico using MS1096-GAL4 results in elevated F-actin staining in the dorsal region of the wing disc where Myc-pico levels are highest. This effect is suppressed by coexpression of DN-Egfr. Quantitation of ratios of ventral:dorsal F-actin levels (±SD) is shown in the upper right corners of the upper panels.
(D) Pico full-length, but not picoRA-PH, binds the Ena EVH1 domain, but not an unrelated control (NIPP1), in the yeast two-hybrid system. Interaction is indicated by blue X-gal (X) and growth on auxotrophic media at high (H) and low (L) density.
(E) Ena coimmunoprecipitates with Myc-tagged pico from MS1096 > Myc-pico wing disc extracts. Lower panel shows protein immunoblot analyses of total cell lysates to control for loading.
(F) Wing images of the indicated genotypes showing functional interactions between pico and ena in the wing. Wing from ena210 heterozygote resembles wild-type. ena210 dominantly suppresses the effect of ectopic pico on tissue overgrowth; compare with the effect of one copy of pico alone. Ectopic ena drives tissue overgrowth. The effect of ectopic ena is not enhanced by coexpression of pico, but can be suppressed by one copy of picoIR. The effect of one copy of picoIR alone is shown for comparison. Mean wing area is expressed as a percentage of control (±SD).
MRL proteins alter actin dynamics through their interactions with proteins that can regulate the length and branching density of actin filaments, such as Ena/VASP (Krause et al., 2004; Lafuente et al., 2004). MRL-Ena/VASP interactions are mediated by the Ena/VASP homology 1 domain (EVH1) domain in Ena/VASP. In support of this, we found that the EVH1 domain of Drosophila Ena bound directly to full-length Pico in the two-hybrid system, but not PicoRA-PH, which lacks canonical EVH1 binding sites (Figure 6D). Endogenous, full-length Ena coimmunoprecipitated with Pico from Drosophila tissue extracts (Figure 6E), suggesting that Ena is complexed to Pico in vivo.
Given the conserved ability of Pico to bind Ena and promote F-actin formation, we tested the involvement of ena in pico-mediated F-actin accumulation and growth. For this analysis we used a mutant of ena (ena210), which fails to interact with EVH1-binding partners (Ahern-Djamali et al., 1998). Wings from ena210 heterozygotes resembled wild-type (Figure 6F). ena210 dominantly suppressed pico-mediated F-actin accumulation (p < 0.01, Figure 6A) and wing overgrowth (60%, p < 0.01, Figure 6F), suggesting that ena is limiting for pico function. We also examined the effect of ena overexpression. Like ectopic pico, ena overexpression resulted in a decreased G:F actin ratio (Figures 6A and 6B) and significant wing overgrowth (p < 0.01; Figure 6F). Wings coexpressing ena and picoIR resembled those expressing picoIR alone (Figure 6F), indicating that ena is largely dependent on pico for its growth-promoting effect. Co-overexpression of ena and pico phenotypically resembled the effect of overexpressing either gene alone (Figure 6F). The lack of an additive effect makes it unlikely that ena and pico act in parallel pathways to drive tissue overgrowth. Together, these data suggest that the effects of pico on tissue growth may be linked to specific, ena-mediated changes in actin dynamics.
MRL Proteins Facilitate Mal/SRF Activation and Cell Proliferation
The SRF is a mitogen-responsive transcription factor that is inhibited by binding of cellular G-actin to the SRF cofactor Mal (Posern and Treisman, 2006). Ena/VASP has been reported to induce SRF activity via a region of Ena/VASP that mediates F-actin assembly (Grosse et al., 2003; Sotiropoulos et al., 1999). Given that the MRL proteins share the ability to bind Ena/VASP and modify actin dynamics, we wondered whether Mal/SRF could be a conserved downstream target of the MRL proteins.
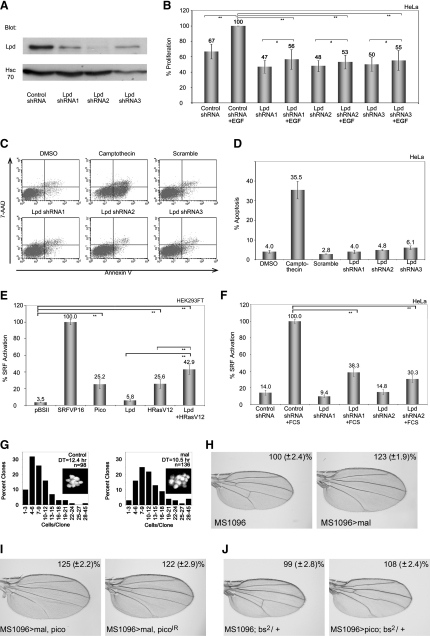
First, we tested the ability of human Lpd to promote cell proliferation in HeLa cells, which only express Lpd and not RIAM. We generated three clonal Lpd knockdown HeLa cell lines using Lpd-specific or scrambled control short hairpin RNA (shRNA) expression. We quantified effects on cell proliferation by measurement of viable cell numbers using a tetrazolium dye (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide [MTT])-based metabolic assay. Knockdown of Lpd resulted in a reduction in cell proliferation (Figures 7A and 7B). Levels of apoptotic markers were not affected in Lpd knockdown cell lines (Figures 7C and 7D), indicating that reduction in cell number was not due to increased cell death. Importantly, there was no significant difference in proliferation of Lpd knockdown cell lines treated with or without EGF, unlike control cells, which overproliferated when treated with EGF (Figure 7B). Taken together, these data suggest that Lpd is required for EGF-induced proliferation, and that the effect of pico on proliferation is conserved in human Lpd.
Figure 7.
MRL Proteins Facilitate Mal/SRF Activation and Cell Proliferation
(A) Immunoblot analysis of Lpd expression in HeLa cell lines expressing Lpd-specific or scrambled shRNA. Hsc70 staining serves as loading control.
(B) Lpd knockdown reduces cell proliferation and abrogates cell proliferation in response to EGF. Cell proliferation was measured using an MTT assay after 5 days. The increase in proliferation compared to Day 1 was calculated; the scrambled control with EGF was set to 100%. The mean values (±SEM) of four independent experiments are shown. ∗∗p < 0.05; #p > 0.05; one-way ANOVA.
(C and D) Lpd knockdown does not induce apoptosis. (C) Dot plots showing Annexin-V-PE/7-AAD staining of HeLa cell lines examined by flow cytometry. Camptothecin treatment (positive control, 20 μM for 24 hr) induced apoptosis. DMSO treatment acted as negative control. (D) Percentage of apoptotic cells as measured by Annexin-V staining for each of the indicated treatments. The mean values (±SEM) of three independent experiments are shown.
(E and F) Transient expression of pico and coexpression of Lpd and HRasV12 significantly induce SRF-reporter gene activity in serum-starved 293FT cells. (E) Graph showing SRF activation for each of the indicated treatments, expressed as a percentage of the effect of constitutive active SRF (SRFVP16) activity.(F) Lpd knockdown abrogates serum-induced SRF activation. SRF activity is expressed as percentage relative to the serum-induced reporter activity in control cells. The mean values (±SEM) of three independent experiments are shown. ∗∗Statistically significant results (p < 0.05, one-way ANOVA).
(G) The distribution of clonal cell number for control and mal-expressing clones. Cell-doubling time (DT) is markedly reduced by mal. Insets show representative clones of each genotype as visualized by nuclear GFP.
(H) Overexpression of mal in the developing wing pouch (MS1096 > mal) results in significant wing overgrowth (p < 0.01).
(I) The effect of ectopic mal is not significantly modified by either coexpression of pico or picoIR.
(J) Wing from bs2 heterozygote resembling wild-type. bs2 dominantly suppresses the effect of ectopic pico on tissue overgrowth.
Second, we used an established cell culture system (Sotiropoulos et al., 1999) to analyze the effect of MRL proteins on SRF activity. Ectopic expression of pico induced a 7.2-fold increase in SRF-responsive gene expression (Figure 7E), similar to the effect of activated H-Ras (H-RasV12). Lpd also appeared to increase SRF activation. The effect was much less pronounced than pico, most likely because there is tight negative regulation of ectopic Lpd expression in mammalian cells. However, the effect of Lpd was significantly enhanced in the presence of H-RasV12. The effect of Lpd and H-RasV12 co-overexpression was significantly higher than that of H-RasV12 alone (Figure 7E). When we tested whether Lpd was required for SRF activation, we found that serum-induced SRF activation was abrogated in Lpd knockdown cell lines compared with control cell lines (p < 0.05; Figure 7F). These data show that MRL proteins are capable of inducing SRF activation, and that Lpd is required for efficient serum-induced SRF activation in mammalian cells.
Finally, we examined the role of mal in pico-mediated overproliferation. Overexpression of wild-type or activated mal or blistered (bs), which encodes Drosophila SRF, has been previously reported to cause overproliferation of the adult wing (Han et al., 2004), similar to the phenotype resulting from ectopic pico. We found that ectopic mal reduced cell-doubling time (Figure 7G) without inducing apoptosis (as determined by activated caspase 3 staining; data not shown), and resulted in a significant increase in wing area (p < 0.01; Figure 7H). Co-overexpression of mal and pico in flies phenotypically resembled the effect of overexpressing mal alone. Furthermore, mal-mediated overgrowth could not be suppressed by ectopic picoIR (Figure 7I). Pico-mediated wing overgrowth was dominantly suppressed (47%; p < 0.01) by a hypomorphic mutation in bs (Figure 7J), indicating that bs is limiting for pico-mediated growth. Collectively, these data indicate that MRL proteins activate SRF-dependent gene expression and that mal/SRF mediate pico-induced tissue overgrowth.
Discussion
MRL Proteins Are Positive Regulators of Cell Proliferation and Tissue Growth
Here we show that pico, which encodes the only Drosophila member of the MRL family of proteins, and its mammalian ortholog, Lpd, have a conserved role in the regulation of cellular proliferation. Reduced pico or Lpd levels result in reduced rates of cellular proliferation, but do not impair cell survival. Too much pico promotes coordinated growth and proliferation, leading to larger tissues with more normal-sized cells. In this respect, the effect of pico is distinct from that of many known Drosophila growth drivers. Growth regulators, such as Drosophila S6K, cause cells to accumulate mass faster than they can divide, primarily due to effects on translation, leading to cellular hypertrophy. Other regulators, such as E2F, can drive cell division without stimulating cell growth, leading to hyperplastic cellular hypotrophy and/or apoptosis.
MRL Proteins Link EGFR Activation to Changes in Actin Dynamics and Cellular Proliferation
We found that attenuating EGFR signaling abrogates the effect of ectopic pico on both F-actin accumulation and tissue growth. pico acts cell autonomously and is therefore unlikely to act upstream of Egfr by affecting the level of EGFR ligands. To rule out that pico regulates levels of EGFR, we examined receptor levels and distribution in wing imaginal discs overexpressing pico or picoIR. EGFR levels and distribution in these genetic backgrounds resembled wild-type. Another possibility is that pico regulates EGFR activity. Although suitable reagents were not available to directly monitor EGFR activity levels in wing discs, we analyzed effects on extracellular signal-regulated kinase (ERK) activation, which provides a molecular readout for EGFR/Ras/Raf signaling. Diphosphorylated (dp) ERK levels were not affected by ectopic pico. These data suggest that, rather than being upstream of EGFR, Pico needs to be activated by EGFR or a downstream component of EGFR signaling, such as activated Ras. Consistently, both Lpd and Pico bind to activated, but not wild-type, Ras. Furthermore, we found that pico knockdown partially suppresses the effects of ectopic Egfr and activated Ras; in addition, Lpd knockdown impairs the EGF-induced increase in proliferation. Taken together, these data suggest that pico and Lpd are downstream effectors of EGFR.
Ena/VASP has been reported to act downstream of MRL proteins (Krause et al., 2004; Lafuente et al., 2004). Correspondingly, we found that pico-mediated wing overgrowth and F-actin accumulation are sensitive to the levels of ena. Importantly, ena is also sufficient to cause overgrowth and F-actin accumulation when overexpressed. Changes in actin dynamics induced by Ena/VASP proteins can activate SRF-dependent gene expression in mammalian cells. Similarly, we found that Pico and Lpd can activate SRF activity. Like pico, ectopic mal or bs/SRF in flies (Han et al., 2004) are sufficient to cause wing overgrowth. Pico-mediated overgrowth is sensitive to the levels of bs/SRF, but mal-induced overgrowth could not be suppressed by pico knockdown, suggesting that Mal/SRF may act downstream of pico in flies. Collectively, these data suggest that MRL proteins may exert their mitogenic effects by specifically interacting with Ena/VASP proteins and inducing SRF-responsive transcription. Interactions between EGFR, MRL proteins, Ena/VASP, and Mal may provide a mechanism linking growth factor signaling and Mal-mediated SRF activation.
Are MRL proteins uniquely able to stimulate Mal/SRF-mediated tissue growth? Although other actin regulators are known to activate Mal/SRF (Posern and Treisman, 2006), there is currently little data to indicate that they play a role in proliferation control. This might be explained if different transcriptional responses occur at different Mal-dependent SRF activation thresholds, leading to diverse cellular outcomes. Alternatively, other actin regulators might influence processes that limit net tissue growth. For instance, Rho activates Mal/SRF in mammalian cells (Posern and Treisman, 2006), but increased Rho activity in flies is associated with loss of epithelial integrity and cell extrusion (Speck et al., 2003), which may negate any potential mal-mediated growth-promoting effects. These issues warrant further study in both flies and mammals. Future studies are also needed to characterize transcriptional targets of Drosophila SRF and resolve the contribution of SRF targets to MRL-mediated growth and proliferation.
MRL Proteins and Cancer
Lpd expression appears to be differentially regulated in cancer compared to normal tissues (Dahl et al., 2005; Eppert et al., 2005; Ginestier et al., 2006). Our data, showing a conserved role for MRL proteins in proliferation control, may provide a potential mechanistic explanation for these observations. In this regard, it is interesting that loss of pico or Lpd can abrogate the effects of EGFR/Erb signaling, deregulation of which has also been implicated in cancer progression. Collectively, these data suggest that MRL proteins might play a role in the pathogenesis of certain cancers and may therefore represent novel molecular targets for therapeutic intervention.
Experimental Procedures
Mutational Analysis
Mutant pico alleles were generated by mobilization of an isogenic line of GS9133 (Toba et al., 1999) using delta 2-3 transposase, and deletions were mapped by Southern blotting. FM7i, Act-GFP was used to identify hemizygous mutant animals and determine the lethal phase. RT-PCR with gene-specific primers verified levels of pico and pico-L expression in hemizygous mutant larvae. Genomic PCR and sequencing confirmed the breakpoints of picok1 following Southern blotting. Expression of UAS-pico alone, or together with UAS-pico-L, with hs-GAL4 and heat shock rescued the lethality of picok1 mutant males. Mosaic analysis of picok1 clones was performed using FRT-mediated recombination (see Supplemental Data for further details).
UAS-Constructs for Heritable RNAi and Ectopic Expression
To make a dsRNAi construct targeting both pico and pico-L, a 477 bp DNA fragment corresponding to 17–493 bp of the pico coding sequence was subcloned into EcoRI/AvrII and NheI/XbaI sites of pWIZ (Lee and Carthew, 2003). This created a UAS-responsive element carrying a tail-to-tail pico inverted repeat flanking the second intron of the white gene. Off-targets were analyzed using dsCheck (http://dscheck.rnai.jp/; Naito et al., 2005). Full-length pico and pico-L open reading frames were amplified by RT-PCR and subcloned into pUAS-HM (Parker et al., 2001) and pPFMW (Drosophila Genome Resource Center [DGRC]) for expression in flies with an N-terminal Myc tag. A fragment encoding the RA-PH domain of pico (picoRA-PH) was subcloned into pPGW (DGRC) for expression in flies with an N-terminal GFP tag. For each construct, at least 10 stable transgenic lines were generated by Genetic Services Inc. (Sudbury) using P element-mediated germline transformation into a w1118 strain. Different transgenes gave qualitatively similar phenotypes.
Fly Stocks and Genetics
Information about the transgenes and mutations used in this study and the genotypes of flies examined are provided in the Supplemental Data. Positively marked, flip-out clones for cell-doubling time analysis and activated MAPK staining were generated in hsFLP122; Act > CD2 > GAL4, UAS-GFP animals. Upon hatching, 50 staged larvae were transferred to vials containing yeast paste and raised at 25°C. Clones were induced at 77 hr for 20 min at 37°C, producing 5–10 clones/disc, and analyzed at 115 hr. Each experiment was performed at least twice.
Weight and Area Analysis
Body weight was the average of at least 40 flies, 3 days after eclosion. Adult wings were mounted in Canada Balsam and examined by light microscopy. Cell density was assessed by counting number of wing hairs on the dorsal wing surface as described by Böhni et al. (1999) (n = 25 per genotype). The area of wing, exclusive of the alula and the costal cell, was measured using NIH ImageJ (n = 25 per genotype).
Cell Cycle Phasing and FCS
Wing imaginal discs were dissected from larvae at wandering third instar stage 125 hr after egg deposition and flow cytometry was performed essentially as described by Neufeld et al. (1998) using Hoechst 33342 to stain the DNA of trypsinated cells. Approximately 30 wing discs were examined per experiment. At least three experiments were performed for each genotype. Data were collected on a Dako (Cytomation) MoFlo flow cytometer and analyzed using Summit V4.0 software.
Immunostaining
Wing discs were fixed with Brower's Fix (three parts buffer: 0.15 M PIPES pH 6.9, 3 mM MgSO4, 1.5 mM EGTA, 1.5% NP-40; one part fix: 8% formaldehyde) for 2 hr at 4°C before staining with Myc antibody (9E10 mouse monoclonal supernatant). For caspase and EGFR staining, wing discs were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature before staining with cleaved human caspase 3 antibody (Yu et al., 2002) or Drosophila EGFR antibody (Santa Cruz Biotechnology), respectively. For activated-MAPK staining, wing discs dissected in 10 mM Tris-Cl, pH 6.8, 180 mM KCl, 50 mM NaF, 1 mM NaVO4, 10 mM β-glycerophosphate and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature were immunostained using dpERK antibody (Sigma Aldrich). Cy3- or Cy5-conjugated secondary antibodies were used for immunofluorescent detection (Jackson ImmunoResearch Labs, Inc.). F-actin was visualized with Alexa Fluor-633 phalloidin (Invitrogen). DNA was stained with propidium iodide (Sigma Aldrich). Images were collected on a scanning confocal microscope, imported to Photoshop (Adobe), and adjusted for brightness and contrast uniformly across entire fields.
Yeast Two-Hybrid and Immunoprecipitation Assays
Full-length pico and picoRA-PH were cloned into the GAL4 DNA-binding domain vector pGBKT7 and fragments encoding NIPP1, wild-type Ras, dominant negative Ras, RasV12, Rap1V12, and the EVH1 domain of ena (aa 1–113) were subcloned into the GAL4 activation domain vector pACT2 for yeast two-hybrid binding assays. Yeast two-hybrid assays were performed as described by Bennett and Alphey (2007). Immunoprecipitation assays were performed as described by Vereshchagina et al. (2004) from MS1096-GAL4 or MS1096-GAL4 UAS-Myc-pico third instar wing disc extracts. Ena antibody (5G2; Developmental Studies Hybridoma Bank, Iowa City, IA), was used for Western Blotting.
MTT Assay in HeLa Cells
HeLa cells were transfected with Lpd-specific shRNA or the corresponding scrambled sequence (Krause et al., 2004) using Lipofectamine 2000 (Invitrogen). Single clones were selected in DMEM, 10% FBS, penicillin/streptomycin, 2 mM L-glutamine, 2 μg/ml puromycin, screened for Lpd expression/knockdown, and expanded. Proliferation of these cells lines was assessed in four independent experiments using the MTT cell proliferation assay (American Type Culture Collection) according to the manufacturer's instructions. For each experiment, cell lines were plated in triplicate and left growing in the absence or presence of 100 ng/ml EGF (Sigma Aldrich) for 5 days. The increase in proliferation compared to Day 1 was calculated, and cell lines expressing Lpd shRNA were compared to the control, which was set to 100%. The percentage of apoptosis in these cell lines was assessed using the Annexin-V-PE apoptosis detection kit (BD Biosciences). Flow cytometry was performed on a FACSCalibur cytometer (BD); 12,000 cells each were analyzed using the Cell Quest Pro software (BD).
SRF Assay in HEK293FT Cells
HEK293FT cells were transfected with p3D.ALuc (100 ng) (Geneste et al., 2002), pRL-TK (100 ng), and expression plasmids (1.8 μg), as indicated in the legend for Figure 7E, using Lipofectamine 2000 (Invitrogen). Cells were maintained in 0.5% FCS for 24 hr before lysis. HeLa cell lines were transfected with p3D.ALuc (100 ng), pRL-TK (100 ng) using Fugene HD (Roche). Cells were maintained in 0.5% FCS for 16 hr before 8 hr stimulation with 15% FCS prior to lysis.
Acknowledgments
We thank the Bloomington, Kyoto, Szeged, and Japanese National Institute of Genetics stock centers, Buzz Baum, Steven Cohen, Bruce Edgar, Laura Johnston, Julian Ng, Pernille Rorth, and Clive Wilson for fly strains, the DGRC, Jacques Camonis, Oskar Laur, Sally Leevers, and Richard Treisman for DNA constructs and vectors, Susan Zusman for help with generating transgenic lines, Nigel Rust for assistance with flow cytometry, and Karen Clifton for technical assistance. This work was supported by Cancer Research UK (grant C20691/A6678) to D.B., with additional support from the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and the Royal Society. Work in the laboratory of M.K. is supported by a Wellcome Trust University Award (grant 077429/Z/05/Z) and an Medical Research Council studentship. E.L. was funded by an Oxford University European Scatcherd Scholarship and a Phizackerley Senior Scholarship at Balliol College, Oxford. D.B. was Todd-Bird Research Fellow at New College, Oxford.
Published: November 10, 2008
Footnotes
Supplemental Data include Supplemental Experimental Procedures and two figures and can be found with this article online at http://www.developmentalcell.com/cgi/content/full/15/5/680/DC1/.
Supplemental Data
References
- Ahern-Djamali S.M., Comer A.R., Bachmann C., Kastenmeier A.S., Reddy S.K., Beckerle M.C., Walter U., Hoffmann F.M. Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol. Biol. Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D., Alphey L. Yeast two-hybrid screens to identify Drosophila PP1-binding proteins. Methods Mol. Biol. 2007;365:155–179. doi: 10.1385/1-59745-267-X:155. [DOI] [PubMed] [Google Scholar]
- Böhni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., Andruss B.F., Beckingham K., Hafen E. Autonomous control of cell, and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- The FlyBase Consortium The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2003;31:172–175. doi: 10.1093/nar/gkg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl E., Sadr-Nabavi A., Klopocki E., Betz B., Grube S., Kreutzfeld R., Himmelfarb M., An H.X., Gelling S., Klaman I. Systematic identification and molecular characterization of genes differentially expressed in breast and ovarian cancer. J. Pathol. 2005;205:21–28. doi: 10.1002/path.1687. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea F.J., Garcia-Bellido A. Behaviour of cells mutant for an EGF receptor homologue of Drosophila in genetic mosaics. Proc. Biol. Sci. 1990;242:36–44. doi: 10.1098/rspb.1990.0100. [DOI] [PubMed] [Google Scholar]
- Eppert K., Wunder J.S., Aneliunas V., Tsui L.C., Scherer S.W., Andrulis I.L. Altered expression and deletion of RMO1 in osteosarcoma. Int. J. Cancer. 2005;114:738–746. doi: 10.1002/ijc.20786. [DOI] [PubMed] [Google Scholar]
- Gabay L., Seger R., Shilo B.Z. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Geneste O., Copeland J.W., Treisman R. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J. Cell Biol. 2002;157:831–838. doi: 10.1083/jcb.200203126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C., Cervera N., Finetti P., Esteyries S., Esterni B., Adelaide J., Xerri L., Viens P., Jacquemier J., Charafe-Jauffret E. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin. Cancer Res. 2006;12:4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- Grosse R., Copeland J.W., Newsome T.P., Way M., Treisman R. A role for VASP in RhoA-Diaphanous signalling to actin dynamics and SRF activity. EMBO J. 2003;22:3050–3061. doi: 10.1093/emboj/cdg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard A., Biehs B., Sturtevant M.A., Wickline L., Chacko J., Howard K., Bier E. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development. 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- Han Z., Li X., Wu J., Olson E.N. A myocardin-related transcription factor regulates activity of serum response factor in Drosophila. Proc. Natl. Acad. Sci. USA. 2004;101:12567–12572. doi: 10.1073/pnas.0405085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenzora A., Behrendt B., Small J.V., Wehland J., Stradal T.E. PREL1 provides a link from Ras signalling to the actin cytoskeleton via Ena/VASP proteins. FEBS Lett. 2005;579:455–463. doi: 10.1016/j.febslet.2004.10.110. [DOI] [PubMed] [Google Scholar]
- Kashles O., Yarden Y., Fischer R., Ullrich A., Schlessinger J. A dominant negative mutation suppresses the function of normal epidermal growth factor receptors by heterodimerization. Mol. Cell. Biol. 1991;11:1454–1463. doi: 10.1128/mcb.11.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Leslie J.D., Stewart M., Lafuente E.M., Valderrama F., Jagannathan R., Strasser G.A., Rubinson D.A., Liu H., Way M. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Lafuente E.M., van Puijenbroek A.A., Krause M., Carman C.V., Freeman G.J., Berezovskaya A., Constantine E., Springer T.A., Gertler F.B., Boussiotis V.A. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Carthew R.W. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.I., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Naito Y., Yamada T., Matsumiya T., Ui-Tei K., Saigo K., Morishita S. dsCheck: highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res. 2005;33:W589–W591. doi: 10.1093/nar/gki419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz A.F., Johnston L.A., Edgar B.A. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Parker L., Gross S., Alphey L. Vectors for the expression of tagged proteins in Drosophila. Biotechniques. 2001;31:1280–1286. doi: 10.2144/01316st01. [DOI] [PubMed] [Google Scholar]
- Posern G., Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Sabatier C., McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos A., Gineitis D., Copeland J., Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Speck O., Hughes S.C., Noren N.K., Kulikauskas R.M., Fehon R.G. Moesin functions antagonistically to the Rho pathway to maintain epithelial integrity. Nature. 2003;421:83–87. doi: 10.1038/nature01295. [DOI] [PubMed] [Google Scholar]
- Toba G., Ohsako T., Miyata N., Ohtsuka T., Seong K.H., Aigaki T. The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics. 1999;151:725–737. doi: 10.1093/genetics/151.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereshchagina N., Bennett D., Szoor B., Kirchner J., Gross S., Vissi E., White-Cooper H., Alphey L. The essential role of PP1beta in Drosophila is to regulate nonmuscle myosin. Mol. Biol. Cell. 2004;15:4395–4405. doi: 10.1091/mbc.E04-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.Y., Yoo S.J., Yang L., Zapata C., Srinivasan A., Hay B.A., Baker N.E. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.