Abstract
We have reported that CXCL16, a recently discovered transmembrane chemokine, is expressed in human gingival fibroblasts (HGF). However, it is not known whether HGF express CXCR6, the receptor for CXCL16, or CXCL16 affects HGF biology. We have shown that HGF expressed CXCR6 by reverse transcription–polymerase chain reaction and flow cytometric analysis. Moreover, we elucidated that tumour necrosis factor (TNF)-α and cytosine-guanine dinucleotide (CpG) DNA (Toll-like receptor-9 ligand) treatment enhanced CXCR6 expression by HGF. Interleukin (IL)-4, IL-13 and CpG DNA up-regulated CXCR6 expression by TNF-α-stimulated HGF. On the other hand, IL-1β and interferon-γ inhibited CXCR6 expression on TNF-α-treated HGF. CXCL16 treatment induced HGF proliferation and phosphorylation of extracellular regulated kinase (ERK) and protein kinase B (AKT) in HGF. In conclusion, HGF expressed CXCR6 functionally, because CXCL16 induced HGF proliferation and ERK and AKT phosphorylation in HGF. These results indicate that CXCL16 may play an important role in the pathogenesis and remodelling in periodontally diseased tissues.
Keywords: CXCR6, HGF, periodontal disease
Introduction
CXCL16 is a member of the CXC chemokine subfamily. Unlike other members of this subgroup, CXCL16 is structurally different and has four distinct domains: a chemokine domain tethered to the cell surface via a mucin-like stack, which is attached in turn to transmembrane and cytoplasmic domains [1,2]. Fractalkine (CX3CL1), the only member described so far in the CX3C family [3], has a similar structure to that of CXCL16 and both CXCL16 and fractalkine act as adhesion molecules when expressed on the cell surface, and upon cleavage from the cell surface the soluble chemokines act as chemoattractants [1–3]. CXCL16 attracts CXCR6 expressing T, natural killer and natural killer T cells to the site of inflammation/injury [4–8]. Recently, Nakayama et al.[9] have reported enrichment of CXCR6-expressing plasma cells and myeloma cells in bone marrow and other target tissues via CXCR6–CXCL16 interaction. In addition, CXCL16, via its chemokine domain, facilitates phagocytosis of Gram-positive and Gram-negative bacteria by antigen-presenting macrophages and dendritic cells [10]. CXCL16 was identified originally as a scavenger receptor for phosphatidylserine and oxidized lipoprotein [11]. In human periodontally diseased tissues we have shown that both CXCL16 and CXCR6, expressed in periodontally diseased tissues, and human gingival fibroblasts (HGF) could produce CXCL16 [12]. Our previous report suggests that CXCL16 may play a role in the pathobiology of periodontal disease. However, it is not known whether HGF express CXCR6 and CXCL16 affects HGF biology. In the present study, we aimed to examine CXCR6 expression in HGF, and the effects of cytokines and Toll-like receptor (TLR) ligands on CXCR6 expression by HGF. Moreover, we investigated the effects of CXCL16 on HGF proliferation and phosphorylation on protein kinase B (AKT) and extracellular regulated kinase (ERK), because it is reported that AKT and ERK are involved in HGF proliferation.
Materials and methods
Gingival tissue biopsy and cell culture
We used HGF isolated from three clinically healthy gingiva during routine distal wedge surgical procedure. Gingival specimens were cut into small pieces and transferred to culture dishes. HGF that grew from the gingivae were cultured on uncoated 100 mm2 plastic dishes in Dulbecco's modified Eagle's medium (Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (gibco, Grand Island, NY, USA) and antibiotics (penicillin G, 100 units/ml; streptomycin, 100 mg/ml) at 37°C in humidified air with 5% CO2. Confluent cells were transferred and cultured for use in the present study. After three to four subcultures by trypsinization, cultures contained homogeneous, slim, spindle-shaped cells growing in characteristic swirls. The cells were used for experiments after five passages. Informed consent was obtained from all subjects participating in this study. The study was performed with the approval and compliance of the Tokushima University Ethical Committee.
RNA extraction and reverse transcription–polymerase chain reaction analysis
Total RNA was prepared from biopsied gingival tissue or HGF using the Rneasy total RNA isolation Kit (Qiagen, Hilden, Germany). Single-stranded cDNA for a polymerase chain reaction (PCR) template was synthesized from 48 ng of total RNA using a primer, oligo(dT)12·18 (Invitrogen, Carlsbad, CA, USA), and superscript3 reverse transcriptase (Invitrogen) under the conditions indicated by the manufacturer. Specific primers were designed from the cDNA sequences for CXCL16, CXCR6 and glyceraldehydes-3-phosphate dehydrogenase. Each cDNA was amplified by PCR using Hot star Taq DNA polymerase (Qiagen). The sequences of the primers were as follows: CXCR6-F (5′-CTGGTGGTGTTTGTCTGTGG-3′) and CXCR6-R (5′-GGCTGACAAAGGCATAGAGC-3′). The conditions for PCR were 1× (95°C, 15 min), 35× (94°C, 1 min, 59°C, 1 min, 72°C, 1 min) and 1× (72°C, 10 min). The products were analysed on a 1·5% agarose gel containing ethidium bromide. The expected size of the PCR product for CXCR6 was 762 base pairs.
Flow cytometric analysis
The HGF were stimulated with interleukin (IL)-1β (Peprotech, Rocky Hill, NJ, USA), tumour necrosis factor (TNF)-α (Peprotech), interferon (IFN)-γ (Peprotech), IL-4 (Peprotech), IL-13 (Peprotech), IL-10 (Peprotech), transforming growth factor (TGF)-β1 (Peprotech), Escherichia coli lipopolysaccharide (LPS) (Sigma), Staphylococcus aureus peptidoglycan (Sigma) and cytosine-guanine dinucleotide (CpG) DNA (Hokkaido System Science, Sapporo, Japan) for 24 h. For AKT and ERK analysis, cells were stimulated with CXCL16 (Peprotech) for 0, 15, 30 or 60 min. After culture, cells were washed twice with ice-cold phosphate-buffered saline (PBS). HGF were harvested by incubation with Trypsin/ethylenediamine tetraacetic acid (Sigma). Most cells were rounded up following this treatment and could be removed by gentle agitation. Cells were washed twice with ice-cold PBS and incubated (20 min on ice) in PBS-1% bovine serum albumin (BSA). Cells were incubated with mouse anti-human CXCR6 antibody (R&D Systems, Minneapolis, MN, USA) or isotype control antibody on ice for 30 min. After washing twice with PBS-1% BSA, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse F(ab′)2 fragments (Dako, Kyoto, Japan) or FITC-conjugated swine anti-rabbit F(ab′)2 fragments (Dako) for 30 min on ice. After washing twice with PBS-1% BSA, cells were analysed immediately by flow cytometry (Epics XL-MCL; Coulter, Hialeah, FL, USA). Cells were gated using forward- versus side-scatter to remove any dead cells and cellular debris and thus provide a uniform population of HGF. For each sample, 10 000 cells were analysed.
Cell proliferation assay
Cells were seeded in 96-well plates at a density of 1 × 103 cells/well and then stimulated with CXCL16 (100 ng/ml). After 24 h, we added TetraColor One (Seikkagaku Corporation, Tokyo, Japan) and incubated for an additional 4 h. Afterwards, we measured spectrophotometrically at 450 nm. In selected experiments, HGF were cultured for 1 h in the presence or absence of PD98059 (20 µM; Calbiochem, La Jolla, CA, USA) or LY294002 (20 µM; Calbiochem) prior to incubation with CXCL16 (100 ng/ml).
Western blot analysis
Western blot analysis was performed to confirm CXCL16-induced phosphorylation of signal transduction molecules. HGF stimulated with CXCL16 (100 ng/ml) were washed once with cold PBS, followed by incubation on ice for 30 min with lysis buffer (Cell Signaling Technology, Danvers, MA, USA) supplemented with protease inhibitors (Sigma). After removal of debris by centrifugation, the protein concentrations in lysates were quantified with Bradford protein assay using IgG as a standard. Protein (20 mg) was loaded onto a 4–20% sodium dodecyl sulphate-polyacrylamide gel electrophoresis gel, followed by electrotransfer to a polyvinylidene difluoride membrane. Activation of ERK and AKT was assessed using Phospho-Akt (Ser473) (193H12) rabbit monoclonal antibody (Cell Signaling Technology), Phospho-p44/42 MAP kinase (Thr202/Tyr204) rabbit antibody (Cell Signaling Technology), pan-AKT rabbit antibody (Cell Signaling Technology) or p-44/42 MAP kinase rabbit antibody (Cell Signaling Technology) according to the manufacturer's instructions. Protein bands were visualized by incubation with horseradish peroxidase-conjugated secondary antibody (Sigma), followed by detection using the enhanced chemiluminescence system (GE Healthcare, Uppsala, Sweden).
Statistical analysis
Statistical significance was analysed with Student's t test. P-values < 0·05 were considered significant. We used StatView software to perform the statistical analysis.
Results
CXCR6 expression in HGF
It is not known whether HGF express CXCR6. Using gene-specific primers, we have analysed total RNA isolated from HGF by reverse transcription–PCR. Our results indicate that HGF express CXCR6 at basal conditions (Fig. 1a). These results were confirmed further by flow cytometric analysis that revealed surface expression of CXCR6 on HGF (Fig. 1b).
Fig. 1.
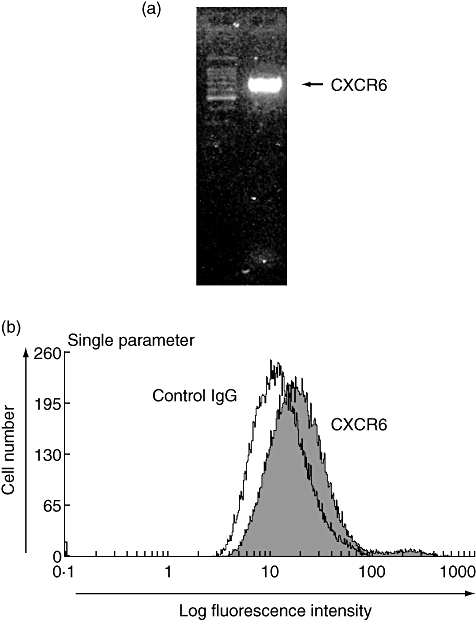
CXCR6 expression in human gingival fibroblasts (HGF). (a) Total RNA was prepared from non-stimulated HGF. The expression of CXCR6 mRNA in non-stimulated HGF was analysed by reverse transcription–polymerase chain reaction, as described in the Methods. (b) Flow cytometric analysis of CXCR6 expression in non-stimulated HGF. The filled line represents CXCR6-specific fluorescence and the open line represents the background level of fluorescence caused by isotype matched antibody.
Effects of cytokine and TLR-ligands on CXCR6 expression by HGF
It has been reported that various cytokines and TLR-ligands exist in periodontally diseased tissues. Therefore, we examined the effects of these factors on CXCR6 expression by HGF and showed that TNF-α enhanced CXCR6 expression on HGF in the cytokines we used in this study (Fig. 2a). Moreover, we elucidated that CpG DNA (TLR-9 ligand) up-regulated CXCR6 expression on HGF (Fig. 2b), although E. coli LPS (TLR-4 ligand) and S. aureus peptidoglycan (TLR-2 ligand) did not modulate CXCR6 expression (Fig. 2c).
Fig. 2.
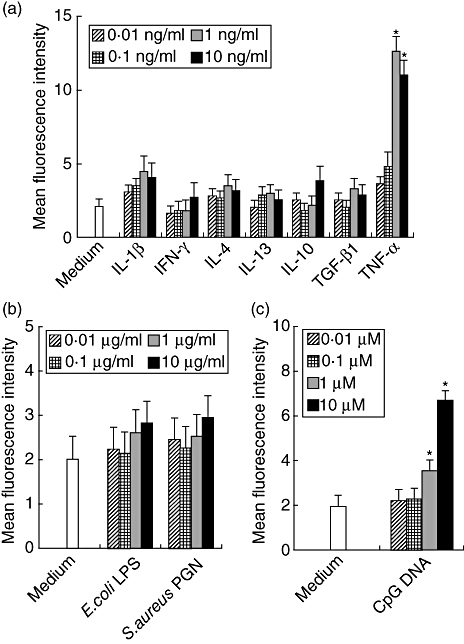
The effects of cytokines and Toll-like receptor ligands on CXCR6 expression by human gingival fibroblasts (HGF). HGF were treated with interleukin (IL)-1β (0·01, 0·1, 1 or 10 ng/ml), interferon-γ (0·01, 0·1, 1 or 10 ng/ml), IL-4 (0·01, 0·1, 1 or 10 ng/ml), IL-13 (0·01, 0·1, 1 or 10 ng/ml), IL-10 (0·01, 0·1, 1 or 10 ng/ml), transforming growth factor-β1 (0·01, 0·1, 1 or 10 ng/ml), tumour necrosis factor-α (0·01, 0·1, 1 or 10 ng/ml), Escherichia coli lipopolysaccharide (0·01, 0·1, 1 or 10 µg/ml), Staphylococcus aureus peptidoglycan (0·01, 0·1, 1 or 10 µg/ml) or cytosine-guanine dinucleotide DNA (0·001, 0·01, 0·1 or 1 µM) and the cells were collected after 24 h. The expression levels of CXCR6 on the surface of HGF were measured using flow cytometry. The data are representative of three different HGF samples from three different donors. The results were calculated as mean and standard deviation (s.d.) of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05 significantly different from the medium.
Cytokines and CpG DNA modulated CXCR6 expression on TNF-α-stimulated HGF
With the exception of TNF-α, the cytokines did not change CXCR6 levels on HGF. We hypothesized that these cytokines might modulate CXCR6 expression enhanced by TNF-α stimulation. Therefore, we examined the effects of IL-1β, IFN-γ, IL-4, IL-13, IL-10 and TGF-β1 on CXCR6 expression on TNF-α-stimulated HGF. IL-1β and IFN-γ inhibited CXCR6 expression on TNF-α-treated HGF. Conversely, IL-4 and IL-13 up-regulated CXCR6 expression enhanced by TNF-α stimulation (Fig. 3a). Moreover, we examined the effects of CpG DNA on TNF-α-enhanced CXCR6 by HGF. CpG DNA enhanced CXCR6 expression synergistically on HGF induced by TNF-α (Fig. 3b).
Fig. 3.
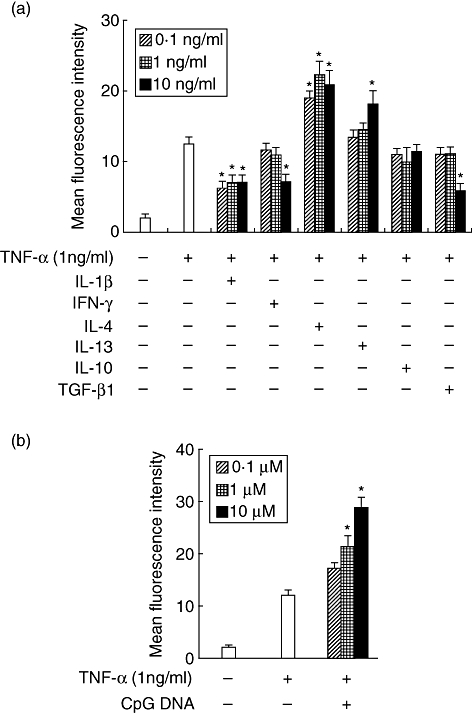
The effects of cytokines and Toll-like receptor ligands on tumour necrosis factor (TNF)-α-induced CXCR6 expression by human gingival fibroblasts (HGF). HGF were treated with TNF-α (1 ng/ml) and interleukin (IL)-1β (0·1, 1 or 10 ng/ml), interferon-γ (0·1, 1 or 10 ng/ml), IL-4 (0·1, 1 or 10 ng/ml), IL-13 (0·1, 1 or 10 ng/ml), IL-10 (0·1, 1 or 10 ng/ml), transforming growth factor-β1 (0·1, 1 or 10 ng/ml) or cytosine-guanine dinucleotide DNA (0·01, 0·1 or 1 µM) and the cells were collected after 24 h. The expression levels of CXCR6 on the surface of HGF were measured using flow cytometry. The data are representative of three different HGF samples from three different donors. The results were calculated as mean and standard deviation (s.d.) of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05 significantly different from the TNF-α single-stimulated HGF.
CXCL16-induced cellular proliferation of HGF
We then investigated the effects of CXCL16 on cell proliferation of HGF. The results showed that CXCL16 induced HGF cell proliferation in a dose-dependent manner (Fig. 4a).
Fig. 4.
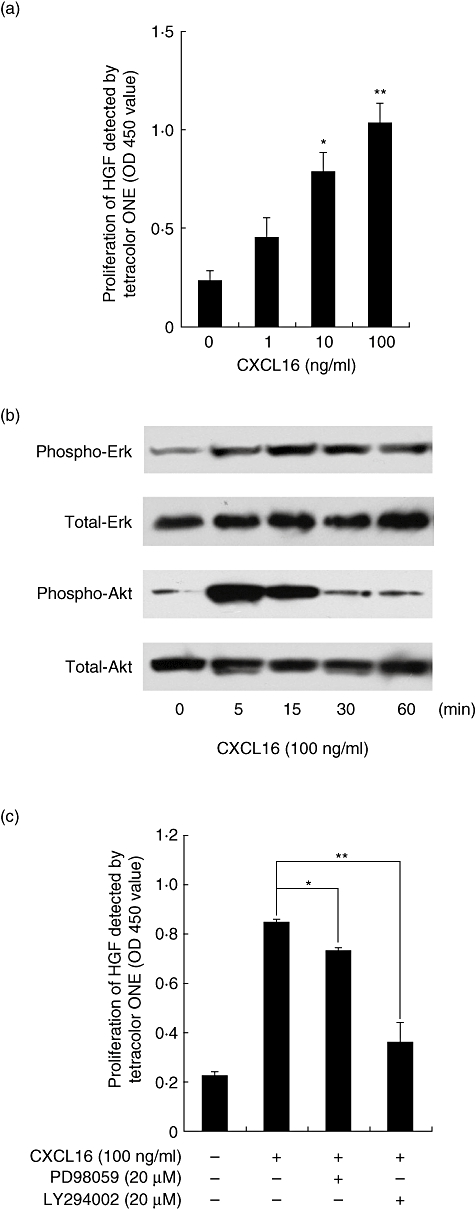
CXCL16 induced human gingival fibroblasts (HGF) proliferation and extracellular regulated kinase (ERK) and protein kinase B (AKT) phosphorylation. (a) HGF were treated with or without CXCL16 (1, 10 or 100 ng/ml) for 24 h. Cell proliferation was assessed using TetraColor One assay. Data are representative of HGF from three different donors. The results were calculated as the mean and standard deviation (s.d.) of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05; **P < 0·01 significantly different from the median. (b) HGF were stimulated with CXCL16 (100 ng/ml) for 5, 15, 30 or 60 min. Extracts were electrophoresed and blotted with phosphor-ERK, total ERK, phospho-AKT or total AKT antibodies. (c) HGF were preincubated with PD98059 (20 µM) or LY294002 (20 µM) for 1 h and then incubated with CXCL16 (100 ng/ml) for 24 h. Cell proliferation was assessed using TetraColor One assay. Data are representative of HGF from three different donors. The results were calculated as the mean and s.d. of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05; **P < 0·01 significantly different from CXCL16-treated HGF without inhibitors.
CXCL16-activated ERK and AKT in HGF
Recent studies have shown that activation of ERK and AKT is involved in HGF proliferation [13]. Therefore, we examined whether CXCL16 can stimulate ERK and AKT in HGF. Subconfluent HGF were exposed to CXCL16 (100 ng/ml) and activation of ERK and AKT was analysed by Western blot analysis, as described in Materials and methods. As shown in Fig. 4, CXCL16 induced ERK and AKT phosphorylation in HGF. The AKT phosphorylation peaked at 5 min after stimulation with CXCL16 and declined thereafter; the ERK phosphorylation peaked at 60 min in this study (Fig. 4b). Moreover, to determine whether activation of ERK and AKT is required for the proliferation of HGF in response to CXCL16, the effects of several inhibitors on the proliferation of HGF were examined (Fig. 4c). PD98059, a specific inhibitor that binds inactive forms of mitogen-activated protein/extracellular signal-related kinase and prevents their activation and phosphorylation, slightly suppressed the proliferation of CXCL16-treated HGF. LY294002, a selective PI3K inhibitor, inhibited almost completely HGF proliferation induced by CXCL16 treatment.
Discussion
Results from the present study indicate for the first time that human fibroblasts express CXCR6. CXCR6 expression is enhanced with TNF-α or CpG DNA treatment, and CXCR6 expression induced by TNF-α is modified with various cytokines. Moreover, treatment with CXCL16 activates the ERK and AKT pathways. Furthermore, CXCL16 increases HGF proliferation.
We have reported previously that CXCL16 is expressed in periodontal diseased tissues, and TNF-α-stimulated HGF could produce CXCL16 [12]. In this study, we show that HGF express CXCR6, and TNF-α can enhance CXCR6 expression on HGF, which might indicate that TNF-α is involved positively in HGF proliferation to induce both CXCL16 and CXCR6 expression by HGF.
We show CpG DNA enhanced CXCR6 expression by HGF, although E. coli LPS (TLR-4 ligand) and S. aureus peptidoglycan (TLR-2 ligand) did not modulate CXCR6 expression. Recently, Mohanonda and colleagues reported that the TLR-2, TLR-3, TLR-4 and TLR-5 ligands enhanced IL-8 production from HGF, although the TLR-9 ligand did not [14]. Moreover, they reported that the TLR-9 ligand suppressed IL-8 production from TLR-3 ligand-stimulated HGF [14]. Their report may explain why the TLR-9 ligand modulated the function of HGF differently compared with other TLR ligands. Their report agrees with ours.
It is reported that both T helper type 1 (Th1) and Th2 cells exist in periodontally diseased tissues [15]. In this report, we reveal that the Th1-type cytokine, IFN-γ, inhibited CXCR6 expression on HGF. Conversely, Th2-type cytokines IL-4 and IL-13 enhanced CXCR6 expression induced with TNF-α treatment. These results might explain that Th1 cells are related to tissue destruction and Th2 cells are involved in tissue repair to modify CXCR6 expression by HGF, because CXCL16 treatment enhanced the proliferation of HGF.
We reveal that CXCL16 treatment led to activation of ERK and AKT. Zhuge and colleagues reported that CXCL16 induced ERK activation in human umbilical vein endothelial cells [16]. Chandrasekar and colleagues also reported that CXCL16 treatment induced AKT activation in human aortic muscle cells [17]. Our results agree with their reports. Recently, Danciu and colleagues reported that the ERK and AKT pathways are related to HGF proliferation and survival [13]. Figure 4c shows that LY294002 inhibited CXCL16-induced HGF proliferation almost completely, although PD98059 showed only a slight suppression. This result suggests that AKT is involved mainly in CXCL16-induced HGF activation.
In conclusion, these results show that cytokines and bacterial components modulate CXCR6 expression in HGF and control HGF proliferation in periodontally diseased tissues. Therefore, CXCR6 and CXCL16 might be targets of periodontal disease treatment, especially periodontal tissue remodelling, because HGF are the major cells in periodontal tissues.
Acknowledgments
This study was supported by a grant-in-aid from the Ministry of Education, Science and Culture of Japan.
Disclosure
The authors have no financial conflict of interest.
References
- 1.Wilbanks A, Zondlo SC, Murphy K, et al. Expression cloning of the STRL33/BONZO/TYMSTR ligand reveals elements of CC, CXC, and CX3C chemokines. J Immunol. 2001;166:5145–54. doi: 10.4049/jimmunol.166.8.5145. [DOI] [PubMed] [Google Scholar]
- 2.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 3.Bazan JF, Bacon KB, Hardiman G, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–4. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 4.Shashkin P, Simpson D, Mishin V, Chesnutt B, Ley K. Expression of CXCL16 in human T cells. Arterioscler Thromb Vasc Biol. 2003;23:148–9. doi: 10.1161/01.atv.0000043906.61088.4b. [DOI] [PubMed] [Google Scholar]
- 5.Heydtmann M, Adams DH. Understanding selective trafficking of lymphocyte subsets. Gut. 2002;50:150–2. doi: 10.1136/gut.50.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unutmaz D, Xiang W, Sunshine MJ, Campbell J, Butcher E, Littman DR. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J Immunol. 2000;165:3284–92. doi: 10.4049/jimmunol.165.6.3284. [DOI] [PubMed] [Google Scholar]
- 7.Kim CH, Kunkel EJ, Boisvert J, et al. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest. 2001;107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CH, Johnston B, Butcher EC. Trafficking machinery of NKT cells: shared and differential chemokine receptor expression among V alpha 24(+)V beta 11(+) NKT cell subsets with distinct cytokine-producing capacity. Blood. 2002;100:11–16. doi: 10.1182/blood-2001-12-0196. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama T, Hieshima K, Izawa D, Tatsumi Y, Kanamaru A, Yoshie O. Cutting edge: profile of chemokine receptor expression on human plasma cells accounts for their efficient recruitment to target tissues. J Immunol. 2003;170:1136–40. doi: 10.4049/jimmunol.170.3.1136. [DOI] [PubMed] [Google Scholar]
- 10.Shimaoka T, Nakayama T, Kume N, et al. Cutting edge: SR-PSOX/CXC chemokine ligand 16 mediates bacterial phagocytosis by APCs through its chemokine domain. J Immunol. 2003;171:1647–51. doi: 10.4049/jimmunol.171.4.1647. [DOI] [PubMed] [Google Scholar]
- 11.Shimaoka T, Kume N, Minami M, et al. Molecular cloning of a novel scavenger receptor for oxidized low density lipoprotein, SR-PSOX, on macrophages. J Biol Chem. 2000;275:40663–6. doi: 10.1074/jbc.C000761200. [DOI] [PubMed] [Google Scholar]
- 12.Hosokawa Y, Hosokawa I, Ozaki K, Nakae H, Matsuo T. CXC chemokine ligand 16 in periodontal diseases: expression in diseased tissues and production by cytokine-stimulated human gingival fibroblasts. Clin Exp Immunol. 2007;149:146–54. doi: 10.1111/j.1365-2249.2007.03398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danciu TE, Gagari E, Adam RM, Damoulis PD, Freeman MR. Mechanical strain delivers anti-apoptotic and proliferative signals to gingival fibroblasts. J Dent Res. 2004;83:596–601. doi: 10.1177/154405910408300803. [DOI] [PubMed] [Google Scholar]
- 14.Mohanonda R, Sa-Ard-Iam N, Montreekachon P, et al. IL-8 and IDO expression by human gingival fibroblasts via TLRs. J Immunol. 2007;178:1151–7. doi: 10.4049/jimmunol.178.2.1151. [DOI] [PubMed] [Google Scholar]
- 15.Takeichi O, Haber J, Kawai T, Smith DJ, Moro I, Taubman MA. Cytokine profiles of T-lymphocytes from gingival tissues with pathological pocketing. J Dent Res. 2000;79:1548–55. doi: 10.1177/00220345000790080401. [DOI] [PubMed] [Google Scholar]
- 16.Zhuge X, Murayama T, Arai H, et al. CXCL16 is a novel angiogenic factor for human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2005;331:1295–300. doi: 10.1016/j.bbrc.2005.03.200. [DOI] [PubMed] [Google Scholar]
- 17.Chandrasekar B, Bysani S, Mummidi S. CXCL16 signals via Gi, phosphatidylinositol 3-kinase, Akt, I kappa B kinase, and nuclear factor-kappa B and induces cell–cell adhesion and aortic smooth muscle cell proliferation. J Biol Chem. 2004;279:3188–96. doi: 10.1074/jbc.M311660200. [DOI] [PubMed] [Google Scholar]


