Abstract
The Duffy binding protein of Plasmodium vivax (DBP) is a critical adhesion ligand that participates in merozoite invasion of human Duffy-positive erythrocytes. A small outbreak of P. vivax malaria, in a village located in a non-malarious area of Brazil, offered us an opportunity to investigate the DBP immune responses among individuals who had their first and brief exposure to malaria. Thirty-three individuals participated in the five cross-sectional surveys, 15 with confirmed P. vivax infection while residing in the outbreak area (cases) and 18 who had not experienced malaria (non-cases). In the present study, we found that only 20% (three of 15) of the individuals who experienced their first P. vivax infection developed an antibody response to DBP; a secondary boosting can be achieved with a recurrent P. vivax infection. DNA sequences from primary/recurrent P. vivax samples identified a single dbp allele among the samples from the outbreak area. To investigate inhibitory antibodies to the ligand domain of the DBP (cysteine-rich region II, DBPII), we performed in vitro assays with mammalian cells expressing DBPII sequences which were homologous or not to those from the outbreak isolate. In non-immune individuals, the results of a 12-month follow-up period provided evidence that naturally acquired inhibitory antibodies to DBPII are short-lived and biased towards a specific allele.
Keywords: allele-specific, antibody response, duffy binding protein, malaria, Plasmodium vivax
Introduction
The Duffy binding protein of Plasmodium vivax (DBP) is a critical adhesion ligand that participates in merozoite invasion of human Duffy/Duffy antigen receptor for chemokines (DARC)-positive erythrocytes [1,2]. DBP belongs to a family of homologous Duffy binding-like erythrocyte binding proteins (DBL–EBP) located within the micronemes of P. vivax and P. knowlesi merozoites [3]. The functional binding domains of DBL–EBP lie in region II, and for P. vivax the critical binding residues have been mapped to a central 170-amino acid stretch that includes cysteines 5–8 [4–6]. The gene encoding the P. vivax DBP region II (DBPII) is highly polymorphic, and this diversity varies geographically from region to region [7–13]. The pattern of excessive polymorphism is consistent with a high selection pressure on the DBP gene and suggests that allelic variation functions as a mechanism of immune evasion [14,15].
Invasive merozoites are believed to sequester microneme proteins until merozoites contact the target erythrocyte, presumably as a mechanism to reduce exposure of DBP to immune inhibition [16]. Currently, available data on humoral immune responses to DBP in human populations demonstrate that anti-DBP antibodies increase with exposure to P. vivax[17–20], and this immune response includes antibody activity that blocks adherence of DBPII to its receptor on erythrocytes [18,21]. The same antibodies that block the DBPII–DARC interaction also inhibit P. vivax erythrocyte invasion [22], which is proof-of-concept that anti-PvDBP antibodies can inhibit merozoite invasion. Of importance, children residing in hyperendemic areas for P. vivax develop anti-DBP inhibitory antibodies that seem to confer protection against blood-stage infection [23].
As most studies on the DBP antibody response reported to date have been carried out in areas where malaria is highly endemic, there is a scarcity of data on the responses to exposure to a single infection and about the persistence of this antibody response in the absence of reinfection. An outbreak of P. vivax malaria, in a village located in a non-malarious area of Brazil, offered us an opportunity to investigate the DBP immune response among individuals who had their first and brief exposure to malaria. In the outbreak area, we hypothesized that a first exposure to P. vivax malaria induces an anti-DBP antibody response that blocks the interaction between DBP and its receptor on erythrocyte. To analyse this neutralizing antibody response, we used an in vitro cytoadherence assay that uses the putative ligand domain of the DBP (region II, DBPII) expressed on the surface of cultivated mammalian cells [18]. To investigate whether neutralizing antibodies recognize DBPII in a strain-specific manner, we analysed polymorphisms within the critical binding motif of P. vivax DBPII from the outbreak isolates, and performed inhibition of cytoadherence assays with DBPII sequences which are homologous or not to that from the outbreak area. In this study, carried out with non-immune individuals, we provide evidence that naturally acquired neutralizing antibodies to DBPII can be strain-specific and are relatively short-lived in the absence of reinfection.
Materials and methods
The P. vivax malaria outbreak
Between April and May 2003, 25 cases of P. vivax malaria were diagnosed for the first time in a small community, Souza, located 70 km from Belo Horizonte, Minas Gerais State, a non-endemic area of Brazil [24,25]. Malaria has never been reported in this area and the Brazilian endemic region, the Amazon area, is 2000 km away. According to the Minas Gerais Department of Health, the source of the infection was a man from the community who had returned from the Amazon, infected by P. vivax, in January 2003. The subsequent outbreak in Souza began in April 2003, and entomological surveys incriminated the vector Anopheles darlingi as responsible for local malaria transmission [24]. The first human malaria case detected in the outbreak area, named S14, remained at the hospital for about 10 days, until a malaria diagnosis could be established. Because malaria infection had never been reported in the outbreak area previously, the physicians failed to consider malaria on presentation of this patient. After the first case, all patients were treated promptly with chloroquine (1·5 g for 3 days) plus primaquine (30 mg daily for 7 days), and a second round of treatment was given in case of relapses and/or recrudescence (3-day course of chloroquine and 15-day course of primaquine). Control activities also included an active search for acute malaria by thick blood smears and outdoor/indoor spraying of residual insecticide (cypermethrine) [25]. The outbreak was considered of short duration (50–60 days), with the last malaria case diagnosed on 21 May 2003; since then, local/regional Departments of Health have maintained entomological and epidemiological surveillance of the area.
Volunteers and blood collection
Cross-sectional surveys were carried out after discussions with the community about the objectives of the project and its protocols. Individuals who had been infected with P. vivax were enrolled in the study if they met the following criteria: (i) informed written consent in accordance with guidelines for human research, as specified by the Brazilian National Council of Health (Resolution 196/96); (ii) residence in the outbreak area; (iii) a minimum age of 15 years; (iv) if women, an indicator of the absence of pregnancy; and (v) a willingness to remain in the outbreak area during the intervening year. As shown in Table 1, a total of 15 individuals met the inclusion criteria (aged 32 ± 13 years). We also included relatives and neighbours who were considered to be exposed to the risk of infection (n = 18; 34 ± 19 years). The latter group had had neither symptoms nor blood parasites by direct examination of Giemsa-stained thick smears. Of the 33 volunteers, 32 did not recall previous history of malaria, temporary residence in malaria-endemic areas or travel to the endemic area during their lifetime. A single volunteer with confirmed malaria in the outbreak area (S1) recalled previous malaria infection, temporary residence in the endemic (gold-mining) area, and travelling to the Amazon during the 6 months preceding the outbreak. We collected 5 ml blood samples (ethylenediamine tetraacetic acid) from all subjects. At the time of blood collection, Giemsa-stained thick blood smears were examined for parasites and nested polymerase chain reaction (PCR) assays for malaria diagnosis were conducted later in our laboratory. Blood samples were used to obtain plasma and for DNA preparation. Three, 6, 9 and 12 months after the first survey, four other identical cross-sectional surveys were carried out. The ethical and methodological aspects of this study were approved by the Ethical Committee of Research on Human Beings from the Centro de Pesquisas René Rachou/FIOCRUZ (Reports 002/2002 and 07/2006), according to the Resolution of the Brazilian Council on Health-CNS 196/96.
Table 1.
Demographic, immunological and genetic data of individuals who had been enrolled in the study carried out in the Plasmodium vivax malaria outbreak area.
| Characteristics | Cases (n = 15) | Non-cases (n = 18) |
|---|---|---|
| Age, years (mean ± s.d.)* | 32 ± 13 | 34 ± 19 |
| Antibody response, n (%)† | ||
| MSP1-19 | 12 (80%) | 0 (0%) |
| DBPII–IV | 3 (20%) | 0 (0%) |
| DARC functional alleles, n (%)‡ | ||
| One (Fy*A or Fy*B) | 6 (40%) | 6 (33%) |
| Two (Fy*A and/or Fy*B) | 9 (60%) | 12 (67%) |
| None (Fy*BES)§ | 0 | 0 |
Difference not significant (t = 0·02, P > 0·05).
Number (%) of individuals with a positive antibody response at the time of first cross-sectional survey.
The frequencies of individuals bearing the functional alleles Fy*A and Fy*B (Fya and Fyb antigens on erythrocytes respectively) were similar between cases and non-cases (P > 0·05).
Homozygosis for the FY*BES (ES, erythroid silent) allele abrogates Duffy antigen receptor for chemokines (DARC) antigen expression on the erythrocyte surface, and designates the DARC negative phenotype. DBP, Duffy binding protein; MSP, merozoite surface protein-1; s.d., standard deviation.
Microscopy and Plasmodium diagnosis by nested PCR
Well-trained microscopists examined 200 fields of Giemsa-stained thick blood smears. DNA was extracted from 300 µl of individual whole-blood samples by using a genomic DNA purification kit (Puregene, Gentra Systems, Minneapolis, MN, USA), according to the manufacturer's protocol. Parasite species identification was performed by nested PCR amplification of the small subunit ribosomal RNA (18S SSU rRNA) genes, as described previously [26].
The DARC genotyping
Extracted DNA was used to detect the three common alleles at the FY locus –FY*A, FY*B, FY*BES(es, erythroid silent) – using real-time PCR with allele-specific primers, essentially as we have described recently [27].
Recombinant proteins and serological assay
Two recombinant P. vivax proteins were used to detect total immunoglobulin G (IgG) antibodies. The recombinant DBP, which includes amino acids 132–771 (regions II–IV, DBPII–IV), was expressed as a soluble glutathione S transferase (GST) fusion protein of 140 kDa, as described previously [17,20]. The recombinant protein representing the 19-kDa C-terminal region of the merozoite surface protein-1 of P. vivax (MSP1-19), which represents amino acids 1616–1704 of the MSP1 of P. vivax, has been described elsewhere [28]. To assess IgG antibodies against DBPII–IV and MSP1-19, an enzyme-linked immunosorbent assay (ELISA) was carried out, as described previously [20], with serum samples at 1 : 100. For the recombinant proteins DBPII–IV (5 µg/ml) and MSP1-19 (1 µg/ml), the final optical density (OD) at 492 nm was calculated by subtracting the OD obtained with GST (antigen control). The results were expressed as an index of reactivity (IR = OD492 values of test sample divided by the value of the cut-off). Cut-off points were set at three standard deviations above the mean OD492 of sera from 30 individuals who had never been exposed to malaria. Values of IR > 1·0 were considered positive.
The P. vivax DBPII amplification and sequencing
Extracted DNA was used as a template in the PCR to amplify the fragment corresponding to nucleotide positions 870–1545 (amino acids 290–515) of the DBPII, as described previously [13]. Platinum high fidelity Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA, USA) was used in PCR to reduce possible nucleotide mis-incorporation. Amplicons were purified using the GFX-96 PCR kit (Amersham Biosciences, Little Chalfont, UK) and sequenced directly using DYEnamic™ ET dye terminator kit (Amersham Biosciences) and MegaBace 500 automated DNA sequencer (Amersham Biosciences). The sequences were analysed using Bioedit sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) to identify DBPII polymorphisms relative to the SAL-1 sequence [29].
The DBP–pEGFP constructs
Region II of DBP (DBPII) from a P. vivax laboratory reference clone (Sal-1) [29] has been subcloned previously into the pEGFP–N1 plasmid (Clontech, Mountain View, CA, USA), with a flanking signal sequence from the herpes simplex virus glycoprotein D1 (HSVgD1) [18]. This targets expression to the surface of the transfected COS cells as a green fluorescent protein (GFP) fusion protein. An additional GFP construct with the DBPII sequence from the outbreak P. vivax isolate was made by subcloning a fragment corresponding to aa 198–522 of region II into pEGFP–HSVgD1 plasmid, using primers described previously [30]. Recombinant plasmids were purified by use of an endotoxin-free plasmid DNA purification system (Qiagen, Valencia, CA, USA).
COS cell transfection and erythrocyte-binding assays
Recombinant plasmids were transfected into COS-7 cells (American Type Culture Collection, Manassas, VA, USA) using lipofectamine and PLUS-reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocols. Briefly, COS-7 cells in six-well culture plates (1·5 × 105 cells/well) were transfected with plasmids (0·5 µg/well)–liposome complexes (5% plus-reagent and 3% lipofectamine) in Dulbecco's modified Eagle medium (DMEM; Sigma, St Louis, MO, USA) without serum. After 6 h of cell exposure to DNA-liposome complexes (37°C, 5% CO2), transfection medium was replaced by DMEM with 10% of fetal bovine serum (Gibco-BRL, Gaithersburg, MD, USA). At 24 h after transfection, the efficiency of transfection was assessed by fluorescence; the recombinant protein expression levels were similar between the Sal-1 and outbreak DBPII variants (data not shown). Forty-eight hours after transfection, the erythrocyte-binding assays were performed as described previously [21]. For this, anti-serum was added at 1 : 20 (1 h at 37°C, 5% CO2) followed by incubation with 10% of human O+ erythrocytes suspension (2 h, room temperature). Unbound erythrocytes were removed by washing and binding was quantified by counting rosettes (10–20 fields, 200×). Positive rosettes were defined as adherent erythrocytes covering more than 50% of the cell surface. For each assay, pooled plasma samples from Souza residents, characterized as non-responders by ELISA, were used as a negative control (100% binding). The percentage inhibition was calculated as 100 × (Rc − Rt)/Rc, where Rc is the average of the number of rosettes in the control wells and Rt is the average of the number of rosettes in the test wells.
Statistical analysis
Statistical analysis was performed using the Epi-Info 2002 software (CDC, Atlanta, GA, USA) or MiniTab statistical software (Minitab Inc., State College, PA, USA). Differences in means were tested by Student's t-test or one-way analysis of variance. Differences in proportions were evaluated by Yates's χ2 or Fisher's exact tests. P-values < 0·05 were considered significant.
Results
Antibody responses to DBPII–IV and MSP1-19 at enrollment
Thirty-three individuals participated in the five cross-sectional surveys, 15 with previously confirmed P. vivax infection in the outbreak area (cases) and 18 who had not experienced malaria infection (non-cases). At the first cross-sectional survey, 20% (three of 15) of malaria cases had antibodies to DBPII–IV (Table 1); in contrast, the MSP1-19 was recognized initially by 80% (12 of 15) of these individuals. The remaining 18 individuals (non-case) did not develop a detectable antibody response against either anti-DBPII–IV or anti-MSP1-19. Although ‘resistance’ to vivax malaria would result from the lack of DARC glycoprotein on red blood cells, none of the individuals studied were homozygous for the allele FY*BES. Therefore, we concluded that absence of DARC on RBCs was not responsible for refractoriness to P. vivax infection in this group (Table 1).
Relationship between malaria status and anti-DBP antibodies during a 12-month follow-up period
Although DBP was recognized initially by 20% of individuals who had had a P. vivax infection, a secondary boosting could be achieved with a new episode of malaria, making 80% into responders at this time (Fig. 1a). Nevertheless, in those individuals the frequency of responders decreased a few months after the clinical attack. By analysing the levels of anti-DBP antibodies during the 12-month follow-up, we observed a wide range of antibody responses among study participants (Fig. 1c), which made the difference between groups without statistical significance (recurrent versus no recurrent infection). Of interest, during the follow-up period, the levels of anti-DBP antibodies were relatively higher in a single individual (Fig. 1c, asterisk in each time-point of the follow-up); this result was not unexpected, because this patient (S14) remained at the hospital for about 10 days until a malaria diagnosis could be established. Despite individual variations, the levels of anti-DBP decreased markedly within the first 6 months of the follow-up.
Fig. 1.
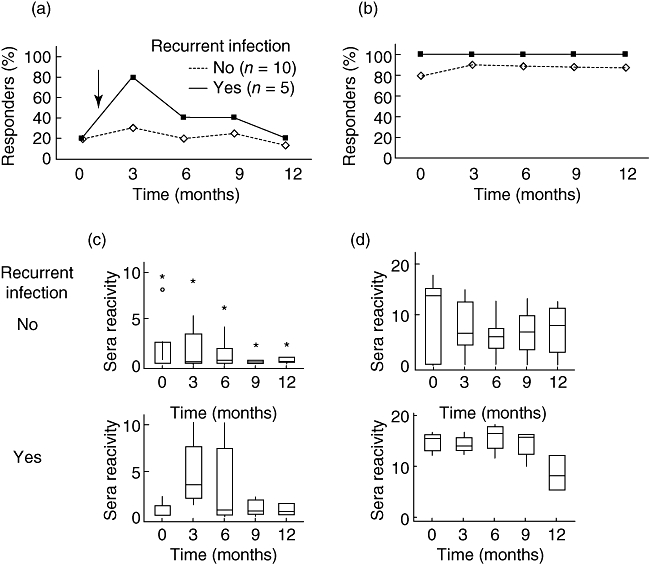
Antibody responses to Duffy binding protein (DBPII–IV) (a,c) and merozoite surface protein-1 (MSP1-19) (b,d) among individuals who had confirmed Plasmodium vivax infection in the outbreak area, and developed (n = 5) or not (n = 10) recurrent P. vivax infections (↓) during the 12-month follow-up period. In (a) and (b), the percentage of responders to DBPII–IV or MSP1-19, respectively, as detected by enzyme-linked immunosorbent assay (ELISA). In (c) and (d), box-plot representations of sera reactivity to DBPII–IV or MSP1-19 respectively; sera reactivity were expressed as index of reactivity (IR) at 492 nm, IR > 1 being considered positive. Box-plots: solid line across the box is the median, and the 25th and 75th percentiles were represented by the bottom and the top of each box respectively; the vertical lines represent the range, with outliers marked by asterisks (S14) or circle (S38).
Altogether, eight of 15 (53%) malaria cases developed anti-DBP antibodies during the follow-up period. The serological response to MSP1-19 was distinctly different (Fig. 1b). Regardless of the occurrence of a relapse and/or recrudescence, the MSP1-19 was a relatively highly immunogenic protein for most individuals who had malaria, with 14 of 15 (93%) positives for anti-MSP1-19 IgG antibodies. However, decreasing levels of antibody reactivity to MSP1-19 was more evident in the group who did not develop a recurrent P. vivax infection (Fig. 1d). No one from the uninfected group (non-cases) developed antibodies against either anti-DBP or anti-MSP1-19 (data not shown).
Because the antibody response against DBP decreased few months after the clinical attack, we also investigated antibodies against another P. vivax apical antigen and vaccine candidate, the apical membrane antigen-1 (AMA-1) [31]. Our results demonstrated that although the profile of AMA-1 immune response was similar to that obtained with DBP, AMA-1 appears to be more immunogenic, with 53% (eight of 15) of responders at the beginning of the study and with all individuals converting into responders at the time of a new episode of malaria (see Supporting information, Fig. S1). However, the frequencies as well as the levels of anti-AMA-1 antibodies were lower in those individuals who did not develop a recurrent P. vivax infection (see Supporting information, Fig. S1b).
The DBPII polymorphisms and inhibitory activity of naturally acquired anti-DBP antibodies
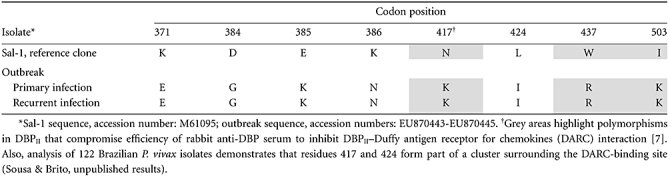
To characterize the P. vivax isolates responsible for the malaria outbreak, we analysed DNA sequences from primary and recurrent infections and identified a single dbp allele in the outbreak area (Table 2). This allele differed, at multiple codons, from the P. vivax laboratory reference clone Sal-1, including differences in three polymorphic codons (417, 437 and 503) suggested to play a synergistic functional effect on DBPII inhibitory binding [7].
Table 2.
Variant amino acids in Duffy binding protein (DBPII) from the Plasmodium vivax outbreak isolates, compared with the P. vivax laboratory reference clone Sal-1.
To investigate the specificities of the anti-DBP outbreak plasmas to inhibit the erythrocyte-binding function of the protein, we performed erythrocyte-binding assays using COS-7 cells expressing sequences of DBPII which are identical or not to those of the outbreak isolate. Previously, this in vitro assay proved to be a suitable alternative tool for the live-cell invasion inhibition assay [22]. For that, plasma samples of those eight individuals who had developed conventional anti-DBP antibodies, at any time-point of the follow-up period, were tested for inhibition of DBPII–DARC binding (Fig. 2). Three months after the first malaria attack, when the majority of responders were detected, seven of eight individuals had developed inhibitory antibodies against the homologous DBPII sequence, while sera of two (S1 and S31) presented inhibitory activity against the heterologous sequence (Fig. 2a). Despite the occurrence of recurrent infections, most of these individuals lost their anti-DBP inhibitory antibody response within 6 months of follow-up. A single exception was an individual (S1) who had had previous malaria infection during frequent trips to the malaria-endemic area, and who developed inhibitory antibodies against homologous and heterologous DBPII sequences. Beyond the frequency of response, the levels of inhibitory antibodies were also related to the DBPII sequence; the greatest levels were observed with COS cells expressing the homologous DBPII sequence, and no cross-reactivity could be detected at 1 : 40 sera dilution (Fig. 2b).
Fig. 2.
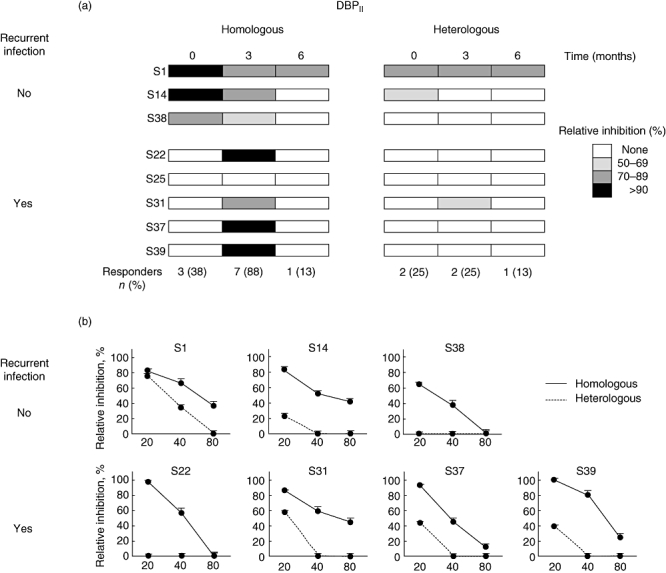
Inhibition of Duffy binding protein–Duffy antigen receptor for chemokine (DBPII–DARC) binding by outbreak plasma samples from eight individuals who developed conventional anti-DBP antibodies during the follow-up period. Erythrocyte binding assays were carried out with COS cells expressing the outbreak (homologous) or Sal-1 (heterologous) DBPII sequences, and individual samples were grouped according to the occurrence (S22, S25, S31, S37, S39) or not (S1, S14, S38) of a recurrent Plasmodium vivax infection. (a) Results of each subject (at 1 : 20 plasma dilution), at enrollment (time zero), and 3 and 6 months later; values at the bottom of the figure represent the overall frequency of responders for each cross-sectional survey. (b) Titration of the inhibitory antibody responses against homologous or heterologous DBPII sequences, using those seven positive samples from the second cross-sectional survey (3-month follow-up). The x-axis represents antibody titers.
Discussion
Naturally occurring antibodies to DBP are prevalent in individuals living in areas where vivax malaria is endemic [17,19,20], and these antibodies can block the DBPII–DARC interaction [18,21,23] and inhibit P. vivax erythrocyte invasion [22]. In previous studies, carried out in malaria-endemic areas, we and others have found strain-transcendent inhibitory responses to DBPII[21,23]. However, those previous studies could not dismiss the possibility that DBPII cross-variant inhibitory activity reflected only an accumulation of antibodies to strain-specific epitopes. Here, we have examined antibody responses of non-immune individuals after a brief initial malaria infection during a malaria outbreak outside the endemic area. Our study demonstrates that DBP has low immunogenicity and induces a short-term humoral immune response that is lost within 6 months of follow-up. In general, this profile of antibody response is similar to that detected to another apical P. vivax vaccine candidate, AMA-1, in which the ELISA positivity rates dropped during the first 9 months of follow-up (Supporting information). Nevertheless, during the follow-up period the frequencies of AMA-1 responders were usually higher than those obtained to DBP, especially among those individuals with recurrent P. vivax infections. The possibility that AMA-1 is more immunogenic than DBP could be explained by those findings demonstrating that AMA-1 is expressed in both pre-erythrocytic (sporozoite) and erythrocytic (merozoite) stages of malaria parasites [32], while DBP is a merozoite-specific protein.
Of importance, single P. vivax exposure appears to induce an anti-DBP inhibitory response that is biased towards a specific DBP allele. The longevity of this DBP inhibitory antibody response is different from that observed among some asymptomatic children residing in a P. vivax hyperendemic area [23], in which the anti-DBP inhibitory antibody response was remarkably stable over the 12-month follow-up period. Together, these findings suggest that vaccines based on DBPII should consider short-term antibody responses in non-immune individuals.
The poorer, unstable antibody responses against DBP during the outbreak follow-up period is in contrast to the stronger, stable response to MSP1-19, which is a much more abundant blood-stage molecule than DBP. Regardless of the presence of recurrent P. vivax infections, the frequency of responders to MSP1-19 was similar at all five time-points, albeit at a lower magnitude in those without recurrent P. vivax infections, as described previously [33]. This longer-term stability of antibodies against P. vivax MSP1 has been well documented [34,35], including its persistence for 30 years after malaria exposure [36]. In none of the study individuals did the absence of DARC on erythrocytes play a role in the anti-DBP or anti-MSP1-19 responses.
In the Souza community, where the outbreak occurred, the period of malaria transmission was short (approximately 50 days), being interrupted by treatment of all patients with anti-malarial drugs (chloroquine and primaquine) and the comprehensive spraying of residual insecticide [25]. Considering that the control intervention of the outbreak was so thorough, the origin of the second attack of P. vivax in five individuals, about 2 months after the first malaria episode, is unclear. Typically, these infections may have had two origins: (i) a recrudescence originating from asexual blood-stage parasites that survived drug treatment; or (ii) a relapse arising from the dormant liver stages, hypnozoites [37]. The recurrences for the P. vivax appear to be more probably relapses, as treatment regimens used in the outbreak area were effective in clearing parasitaemias and there was a long period until the blood-stage infections reappeared. To analyse whether the isolate causing the secondary attack was genetically different from the isolate of initial infections, we compared DNA sequences from primary and recurrent P. vivax infections. Molecular analysis demonstrated that a single dbp allele was detected in the outbreak area (GenBank Accession numbers: EU870443–EU870445). The dbp outbreak allele belongs to allelic family VII, one of the eight DBPII variant families identified in a preliminary analysis of 40 P. vivax Brazilian isolates [13].
Although the activation of heterologous hypnozoite populations seems to be the most common cause of relapse in patients with vivax malaria [38,39], the presence of a single dbp outbreak allele is consistent with either a relapse or a recrudescence. In contrast to previous studies in Asia, the P. vivax transmission in the outbreak area originated from a single patient who had had a P. vivax relapse after returning from the Amazon area [24,25]. In fact, our results are similar to a previous study of P. vivax relapses in Brazil which demonstrated, using the MSP1 molecule as a genetic marker, that parasites from the primary attack were identical to those in relapses [40].
An important finding of our study is the discovery of how parasite genetic diversity relates to naturally acquired neutralizing antibodies against DBP. The results demonstrated that the phylogenetically distant Sal-1 variant was significantly less sensitive to immune inhibition of its DARC binding activity than was the homologous effect against the DBPII allele of the outbreak variant. Significant antibody cross-reactivity was observed in a single individual (S1), a result which was attributed to past cured infections in a gold-mine worker who had a history of previous malaria illnesses in a malaria-endemic area. Although it is not possible at this time to characterize the fine specificity of the inhibitory anti-DBP antibodies, these data demonstrate that variation in few polymorphic residues compromising the inhibitory efficacy of these antibodies. Further work will be necessary to identify the main epitopes recognized by naturally acquired antibodies to DBP in humans.
Altogether, our results indicate that polymorphisms change DBP antigenic character and can compromise immune inhibition, as suggested previously using rabbit immune sera [7]. Of importance, the outbreak and Sal-1 alleles do not share the trio of polymorphic residues (at codons 417, 437 and 503) shown to collectively alter sensitivity to inhibitory antibodies. Overall, these results point towards strain specificity in the natural immune response against DBP. Consistent with this hypothesis, the only individuals in the Amazon area who were observed previously to acquire anti-DBP antibodies that inhibit binding of different DBPII variants to erythrocytes were people who had had long-term exposure [21]. Consequently, it is not surprising that only 9% of asymptomatic children residing in a P. vivax hyperendemic area had acquired a significant anti-DBP inhibitory antibody response that transcended strain-specificity [23].
Even though the current data demonstrate that individuals exposed briefly to P. vivax developed anti-DBP antibodies which exert a receptor-blocking effect, the magnitude of the inhibitory antibody response was very low compared with that from individuals with long-term exposure to malaria; in the outbreak area, inhibitory activity was achieved with immune sera diluted up to 1 : 80, whereas in the Amazon endemic area inhibitory antibodies could still be detected at a 1 : 1280 sera dilution [21]. It is possible that the low levels of immune response in the outbreak area could be due to the short and brief exposure to parasite blood stages. In fact, in this area, a secondary antibody boost was achieved with a recurrent P. vivax infection. Also, our previous data in the Amazon area indicate accumulative exposure to P. vivax as the strongest predictor of the presence of anti-DBP antibodies [21]. None the less, it is currently unclear how effective such natural antibody responses may be in preventing disease in this population. A long-term prospective study in a non-immune population is needed to determine the protective nature of the inhibitory anti-DBPII antibodies in terms of anti-disease immunity.
Recently, it has been predicted that the hypervariable region of DBPII is located on sites remote from the DARC binding site, implying that polymorphism cannot alter the capacity of the protein to bind DARC-positive erythrocytes [41,42]. Another line of evidence suggests that those few polymorphic residues surrounding the DARC binding domain might elude binding of inhibitory antibody [7,16]. The second model seems to explain why antibodies to DBP can inhibit reticulocyte invasion by P. vivax effectively [22]. The results presented here provide strong evidence that the DARC and antibody binding sites have sufficient overlap for antibodies to inhibit binding and provide support for the role of allelic diversity in anti-DBP immune responses.
Acknowledgments
We thank Vania D. A. Cerbino (Minas Gerais Regional Department of Health) and other members of the local malaria control team from the Minas Gerais State Secretariat of Health and Sanitation for their logistic support in the fieldwork. This work is supported by the Brazilian National Research Council (CNPq) and Fundação Oswaldo Cruz, FIOCRUZ, PAPES IV (CNPq-400086/2006-9), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG-CBB-880/06). L. H. C. is supported by a research fellowship from CNPq.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Antibody responses to the apical membrane antigen-1 (AMA-1) among individuals who had confirmed Plasmodium vivax infection in the outbreak area, and developed (n = 5) or not (n = 10) recurrent P. vivax infections (↓) during the 12-month follow-up period. (a) The percentage of responders to AMA-1, as detected by enzyme-linked immunosorbent assay (ELISA); (b) box-plot representations of sera reactivity; sera reactivity were expressed as index of reactivity (IR) at 492 nm, IR > 1 being considered positive. Box-plots: solid line across the box is the median, and the 25th and 75th percentiles were represented by the bottom and the top of each box respectively. The recombinant protein AMA-1 was produced as described previously [31].
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Wertheimer SP, Barnwell JW. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol. 1989;69:340–50. doi: 10.1016/0014-4894(89)90083-0. [DOI] [PubMed] [Google Scholar]
- 2.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 3.Adams JH, Hudson DE, Torii M, et al. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–53. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- 4.Ranjan A, Chitnis CE. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci USA. 1999;96:14067–72. doi: 10.1073/pnas.96.24.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, Singh AP, Pandey S, Yazdani SS, Chitnis CE, Sharma A. Definition of structural elements in Plasmodium vivax and P. knowlesi Duffy-binding domains necessary for erythrocyte invasion. Biochem J. 2003;374:193–8. doi: 10.1042/BJ20030622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.VanBuskirk KM, Sevova E, Adams JH. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc Natl Acad Sci USA. 2004;101:15754–9. doi: 10.1073/pnas.0405421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VanBuskirk KM, Cole-Tobian JL, Baisor M, et al. Antigenic drift in the ligand domain of Plasmodium vivax duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis. 2004;190:1556–62. doi: 10.1086/424852. [DOI] [PubMed] [Google Scholar]
- 8.Tsuboi T, Kappe SH, al-Yaman F, Prickett MD, Alpers M, Adams JH. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect Immun. 1994;62:5581–6. doi: 10.1128/iai.62.12.5581-5586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xainli J, Adams JH, King CL. The erythrocyte binding motif of Plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol. 2000;111:253–60. doi: 10.1016/s0166-6851(00)00315-7. [DOI] [PubMed] [Google Scholar]
- 10.Cole-Tobian J, King CL. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 2003;127:121–32. doi: 10.1016/s0166-6851(02)00327-4. [DOI] [PubMed] [Google Scholar]
- 11.Ampudia EM, Patarroyo A, Patarroyo ME, Murillo LA. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol Biochem Parasitol. 1996;78:269–72. doi: 10.1016/s0166-6851(96)02611-4. [DOI] [PubMed] [Google Scholar]
- 12.Kho WG, Chung JY, Sim EJ, Kim DW, Chung WC. Analysis of polymorphic regions of Plasmodium vivax Duffy binding protein of Korean isolates. Korean J Parasitol. 2001;39:143–50. doi: 10.3347/kjp.2001.39.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa TN, Ceravolo IP, Fontes CJF, Couto A, Carvalho LH, Brito CF. The pattern of major polymorphisms in the Duffy binding protein ligand domain among Plasmodium vivax isolates from the Brazilian Amazon area. Mol Biochem Parasitol. 2006;146:251–4. doi: 10.1016/j.molbiopara.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Cole-Tobian J, King CL. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 2003;127:121–32. doi: 10.1016/s0166-6851(02)00327-4. [DOI] [PubMed] [Google Scholar]
- 15.Martinez P, Suarez CF, Cardenas PP, Patarroyo MA. Plasmodium vivax Duffy binding protein: a modular evolutionary proposal. Parasitology. 2004;128:353–66. doi: 10.1017/s0031182003004773. [DOI] [PubMed] [Google Scholar]
- 16.McHenry AM, Adams JH. The crystal structure of P. knowlesi DBPalpha DBL domain and its implications for immune evasion. Trends Biochem Sci. 2006;31:487–91. doi: 10.1016/j.tibs.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser T, Michon P, Barnwell JW, et al. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect Immun. 1997;65:2772–7. doi: 10.1128/iai.65.7.2772-2777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michon P, Fraser T, Adams JH. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect Immun. 2000;68:3164–71. doi: 10.1128/iai.68.6.3164-3171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xainli J, Cole-Tobian JL, Baisor M, et al. Epitope-specific humoral immunity to Plasmodium vivax Duffy binding protein. Infect Immun. 2003;71:2508–15. doi: 10.1128/IAI.71.5.2508-2515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceravolo IP, Bruña-Romero O, Braga EM, et al. Anti-Plasmodium vivax duffy binding protein antibodies measure exposure to malaria in the Brazilian Amazon. Am J Trop Med Hyg. 2005;72:675–81. [PubMed] [Google Scholar]
- 21.Ceravolo IP, Souza-Silva FA, Fontes CJ, et al. Inhibitory properties of the antibody response to Plasmodium vivax Duffy binding protein in an area with unstable malaria transmission. Scand J Immunol. 2008;67:270–8. doi: 10.1111/j.1365-3083.2007.02059.x. [DOI] [PubMed] [Google Scholar]
- 22.Grimberg BTR, Udomsangpetch J, Xainli A, et al. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 2007;4:e337. doi: 10.1371/journal.pmed.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King CL, Shakri AR, Marcotty A, et al. Naturally acquired Duffy-binding protein protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci USA. 2008;105:8363–8. doi: 10.1073/pnas.0800371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerbino VDA, Zumpano JF, Leopoldo FL, et al. Follow-up of control measures during the P. vivax malaria outbreak in Rio Manso, Minas Gerais. Rev Bras Med Trop. 2004;37:269. [Google Scholar]
- 25.Zumpano JFR, Chaves MOC, Silva RB, et al. Study of P. vivax malaria relapses in Sousa District, Rio Manso municipality, Minas Gerais, Brazil. Rev Bras Med Trop. 2004;37:267. [Google Scholar]
- 26.Snounou GS, Viriyakosol XP, Jarra ZW, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–20. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 27.Sousa TN, Sanchez BA, Ceravolo IP, Carvalho LH, Brito CF. Real-time multiplex allele-specific polymerase chain reaction for genotyping of the Duffy antigen, the Plasmodium vivax invasion receptor. Vox Sang. 2007;92:373–80. doi: 10.1111/j.1423-0410.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 28.Cunha MG, Rodrigues MM, Soares IS. Comparison of the immunogenic properties of recombinant proteins representing the Plasmodium vivax vaccine candidate MSP1(19) expressed in distinct bacterial vectors. Vaccine. 2001;20:385–96. doi: 10.1016/s0264-410x(01)00359-0. [DOI] [PubMed] [Google Scholar]
- 29.Fang XD, Kaslow DC, Adams JH, Miller LH. Cloning of the Plasmodium vivax Duffy receptor. Mol Biochem. Parasitol. 1991;44:125–32. doi: 10.1016/0166-6851(91)90228-x. [DOI] [PubMed] [Google Scholar]
- 30.Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Múfalo BC, Gentil F, Bargieri DY, Costa FT, Rodrigues MM, Soares IS. Plasmodium vivax apical membrane antigen-1: comparative recognition of different domains by antibodies induced during natural human infection. Microbes Infect. 2008;10:1266–73. doi: 10.1016/j.micinf.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 32.Florens L, Washburn MP, Raine JD, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–6. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 33.Barbedo MB, Ricci R, Jimenez MCS, et al. Comparative recognition by human IgG antibodies of recombinant proteins representing three asexual erythrocytic stage vaccine candidates of Plasmodium vivax. Mem Inst Oswaldo Cruz. 2007;102:335–9. doi: 10.1590/s0074-02762007005000040. [DOI] [PubMed] [Google Scholar]
- 34.Braga EM, Fontes CJ, Krettli AU. Persistence of humoral response against sporozoite and blood-stage malaria antigens 7 years after a brief exposure to Plasmodium vivax. J Infect Dis. 1998;177:1132–5. doi: 10.1086/517412. [DOI] [PubMed] [Google Scholar]
- 35.Morais CG, Soares IS, Carvalho LH, Fontes CJ, Krettli AU, Braga EM. IgG isotype to C-terminal 19 kDa of Plasmodium vivax merozoite surface protein 1 among subjects with different levels of exposure to malaria in Brazil. Parasitol Res. 2005;95:420–6. doi: 10.1007/s00436-005-1314-x. [DOI] [PubMed] [Google Scholar]
- 36.Lim KJ, Park JW, Yeom JS, et al. Humoral responses against the C-terminal region of merozoite surface protein 1 can be remembered for more than 30 years in persons exposed to Plasmodium vivax. Parasitol Res. 2004;92:384–9. doi: 10.1007/s00436-003-1009-0. [DOI] [PubMed] [Google Scholar]
- 37.Krotoski WA. The hypnozoite and malarial relapse. Prog Clin. Parasitol. 1989;1:1–19. [PubMed] [Google Scholar]
- 38.Imwong M, Snounou G, Pukrittayakamee S, et al. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–33. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 39.Chen N, Auliff A, Rieckmann K, Gatton M, Cheng Q. Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J Infect Dis. 2007;195:934–41. doi: 10.1086/512242. [DOI] [PubMed] [Google Scholar]
- 40.Kirchgatter K, del Portillo HA. Molecular analysis of Plasmodium vivax relapses using the MSP1 molecule as a genetic marker. J Infect Dis. 1998;177:511–15. doi: 10.1086/517389. [DOI] [PubMed] [Google Scholar]
- 41.Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–4. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- 42.Chitnis CE, Sharma A. Targeting the Plasmodium vivax Duffy-binding protein. Trends Parasitol. 2008;24:29–34. doi: 10.1016/j.pt.2007.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


