Abstract
Pollination triggers not only embryo development but also the differentiation of the ovule integuments to form a specialized seed coat. The mucilage secretory cells of the Arabidopsis thaliana seed coat undergo a complex differentiation process in which cell growth is followed by the synthesis and secretion of pectinaceous mucilage. A number of genes have been identified affecting mucilage secretory cell differentiation, including MUCILAGE-MODIFIED4 (MUM4). mum4 mutants produce a reduced amount of mucilage and cloning of MUM4 revealed that it encodes a UDP-L-rhamnose synthase that is developmentally up-regulated to provide rhamnose for mucilage pectin synthesis. To identify additional genes acting in mucilage synthesis and secretion, a screen for enhancers of the mum4 phenotype was performed. Eight mum enhancers (men) have been identified, two of which result from defects in known mucilage secretory cell genes (MUM2 and MYB61). Our results show that, in a mum4 background, mutations in MEN1, MEN4, and MEN5 lead to further reductions in mucilage compared to mum4 single mutants, suggesting that they are involved in mucilage synthesis or secretion. Conversely, mutations in MEN2 and MEN6 appear to affect mucilage release rather than quantity. With the exception of men4, whose single mutant exhibits reduced mucilage, none of these genes have a single mutant phenotype, suggesting that they would not have been identified outside the compromised mum4 background.
Keywords: Arabidopsis thaliana, cell wall, germination, MEN, mucilage, MUM4, pectin, RHM2, rhamnogalacturonan I, seed coat
Introduction
Pollination in flowering plants leads not only to the initiation of embryogenesis and endosperm development, but also to differentiation of the ovule integuments to form the seed coat. The seed coat layers are derived from maternal tissue and can undergo a number of specializations that aid embryo nutrition, seed dispersal, germination, and seed longevity (Esau, 1977; Fahn, 1982; Boesewinkel and Bouman, 1984). One such specialization is the production of a hydrophilic polysaccharide slime, known as mucilage, in the seed coat epidermis. This trait, known as myxospermy, is found in a number of species, including the Brassicaceae, Solanaceae, Linaceae, and Plantaginaceae. Seed coat mucilage has been suggested to play a number of roles, including the promotion of seed hydration and germination, the prevention of gas exchange, and attachment to soil substrates and animal vectors (Esau, 1977; Fahn, 1982; Grubert, 1981).
The seed coat mucilage secretory cells of the model genetic plant, Arabidopsis thaliana (Arabidopsis), undergo a complex differentiation process, including separable stages of pectinaceous and cellulosic cell wall production, making them an excellent model in which to study the regulation of cell wall biosynthesis in a developmental context (Haughn and Chaudhury, 2005; Western, 2006). Epidermal cells of the seed coat first grow by vacuolar expansion. This growth phase is followed by the biosynthesis of a large quantity of pectinaceous mucilage, which is secreted to the apical tangential regions of the cell, forming a doughnut shaped mucilage pocket between the plasma membrane and primary cell wall (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). Mucilage production is accompanied by an increase in the number of Golgi stacks, consistent with the synthesis of pectins in the Golgi apparatus (Western et al., 2000; Young et al., 2008). Concurrent with mucilage synthesis and accumulation, the vacuole contracts towards the bottom of the cell and the cytoplasm is constricted to a volcano shape in the centre of the cell. Pectin biosynthesis and secretion is succeeded by the production of a cellulosic secondary cell wall that fills in the remaining cytoplasm to form a volcano-shaped columella in the centre of the cell (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). Programmed cell death is followed by seed desiccation and the shrinking of the mucilage around the columella to reveal hexagonal-shaped cells with thickened radial cell walls surrounding the columella. Seed wetting leads to the almost instantaneous hydration of the hydrophilic mucilage, followed by rupture of the primary cell wall and the release of mucilage to form a gel capsule surrounding the seed (Western et al., 2000; Windsor et al., 2000). Arabidopsis seed mucilage is primarily composed of an unbranched form of the pectin rhamnogalacturonan I (RG I), with smaller quantities of pectic side chains (arabinans, galactans), homogalacturonan, hemicellulose, and cellulose (Penfield et al., 2001; Willats et al., 2001a; Western et al., 2004; Macquet et al., 2007a).
Mutations in several genes have been identified to have pleiotropic effects on Arabidopsis mucilage secretory cell differentiation. These include the developmental regulator, APETALA2 (AP2), the epidermal cell differentiation factors TRANSPARENT TESTA GLABRA1 (TTG1), TTG2, GLABRA2 (GL2), TRANSPARENT TESTA2 (TT2), TT8, ENHANCER OF GLABRA3 (EGL3), MYB5, and the transcription factor MYB61 (Koornneef, 1981; Bowman and Koornneef, 1993; Jofuku et al., 1994; Rerie et al., 1994; Penfield et al., 2001; Johnson et al., 2002; Zhang et al., 2003; Gonzalez et al., 2009; Li et al., 2009). Loss of function mutants of each of these regulators result in a reduced amount of mucilage and flattened columellae. The most severe are ap2 mutants, which completely lack mucilage and columellae. Reduced mucilage has also been observed in mutants for the KANADI family transcription factor ABERRANT TESTA SHAPE (ATS), the putative glucosidase II, RADIAL SWELLING3, MICROTUBULE ORGANIZATION1, the abscisic acid biosynthetic gene ABSCISIC ACID1, and the gibberellin biosynthetic gene GIBBERELLIN-3 OXIDASE4 (Karssen et al., 1983; Léon-Kloosterziel et al., 1994; Burn et al., 2002; Kim et al., 2005; Messmer McAbee et al., 2006; McFarlane et al., 2008). A screen for mucilage-specific genes led to the identification of MUCILAGE-MODIFIED1–5 (MUM1–5) (Western et al., 2001). mum4 mutants have reduced mucilage and flattened columellae, while mum3 and mum5 mutants have mucilage of altered composition. By contrast, mum1, mum2, and the recently identified subtilase1.7 (Atsbt1-7) mutants have defects in mucilage release (Western et al., 2001; Rautengarten et al., 2008). Both MUM2 and MUM4 have been cloned. MUM2 encodes a β-galactosidase, which, along with a putative pectin methylesterase target of SBT1.7, appears to be required to modify pectin structure in the mucilage and/or primary cell wall to facilitate mucilage release (Dean et al., 2007; Macquet et al., 2007b; Rautengarten et al., 2008). Conversely, MUM4 encodes a UDP-L-rhamnose synthase (also known as RHM2) required for the production of the primary mucilage pectin RG I (Usadel et al., 2004a; Western et al., 2004; Oka et al., 2007). Expression of MUM4 is specifically up-regulated at the time of mucilage synthesis. AP2 and a TTG1–EGL3–TT8–MYB5–TT2 transcription factor complex activate GL2, which, in turn, regulates MUM4 gene expression (Western et al., 2004; Gonzalez et al., 2009; Li et al., 2009). Alternate pathways of mucilage production appear to be regulated by TTG2, also downstream of AP2 and the TTG1–EGL3–TT8–MYB5–TT2 complex, and MYB61, which may be acting indirectly on mucilage production through a role in sugar allocation (Johnson et al., 2002; Zhang et al., 2003; Newman et al., 2004; Western et al., 2004). Thus, while many regulatory genes, and even some cell wall modification genes, have been identified for roles in mucilage secretory cell differentiation, only one biosynthetic gene has yet been identified.
An enhancer mutant screen of the reduced mucilage mutant mum4 was performed to identify additional downstream genes in the mucilage production pathway. In addition to isolating new alleles of mum2 and myb61, six new mucilage secretory cell differentiation genes, MUM ENHANCER1–6 (MEN1–6) were identified and characterized. men1–6 mutants demonstrate varying degrees of enhancement of the mum4 phenotype, with three having significant loss of mucilage, suggesting direct roles in mucilage biosynthesis or secretion.
Materials and methods
Plant lines, mutagenesis, and growth conditions
Lines of Arabidopsis thaliana used were mum4-1 (Col-2 ecotype) (Western et al., 2004) and ttg1-1 (Ler ecotype; Arabidopsis Biological Resource Centre, Columbus, OH). Seeds were planted on AT minimal medium plates (Haughn and Somerville, 1986) or directly on soil (Sunshine Mix No. 5, SunGro Horticulture), stratified for 3–4 d at 4 oC and then transferred to growth chambers at 22 oC under continuous light (90–120 μE m−2 s−1 photosynthetically active radiation), unless otherwise specified. Flower staging for days post anthesis (DPA) was performed as in Western et al. (2001).
For mutagenesis, 0.33 g mum4-1 seeds (∼15 000) were treated for 12 h with 0.25% (v/v) ethyl methanesulphonate. After rinsing, mutagenized seeds were planted in 80 batches of ∼150 plants (M1) and bulk-harvested. For screening, seeds from individual M2 plants were isolated and stained with ruthenium red after pretreatment with EDTA as described below. The nine mutants described were isolated from screening approximately 5000 M3 lines from 10 batches (∼1500 parental lines). men2 mum4 and men3 mum4 were isolated from a common parental batch, as were men6-2 mum4 and myb61-6 mum4; the rest were single isolates from separate parental batches. Prior to study, all double mutant lines were backcrossed at least twice to mum4-1.
In accordance with journal policy on the distribution of novel materials, the men mutants will be made available by the authors upon request.
Sequencing of mum2-13 and myb61-6
The coding regions of MUM2 and MYB61 were PCR amplified from the mum2-13 mum4-1 and myb61-6 mum4-1 double mutants, respectively, using the overlapping primer sets presented in Supplementary Table S1 at JXB online. Sequencing was performed at the McGill-Genome Quebec Innovation Centre sequencing facility and alignments were performed against the wild-type sequences using DNAMAN (Lynnon Corporation).
Microscopy
For ruthenium red staining, seeds were either placed directly in 0.01% (w/v) ruthenium red without shaking, shaken directly in ruthenium red for 90 min, or pre-hydrated with shaking in 0.05 M EDTA for 90 min followed by ruthenium red stain, as indicated. For the seeds stained with shaking, samples were rinsed in dH2O prior to visualization. Seeds were observed on a Leica MZ-16F stereomicroscope and imaged with a Micropublisher 3.3 camera (Qimaging) operated via Openlab 5 (Perkin Elmer).
Developing seeds were prepared for brightfield microscopy, sectioned, and stained with toluidine blue O as described in Western et al. (2001). Samples were examined using a Leica DM 6000B compound microscope and images captured with a Qimaging Retiga CCD camera operated through Openlab. Scanning electron microscopy of dry seeds was performed as described in Western et al. (2001).
To test for mucilage release after extraction with ammonium oxalate, intact seeds were incubated in 0.2% (w/v) ammonium oxalate with vigorous shaking for 2 h at 30 °C. Seeds were then either shaken in 0.01% (w/v) ruthenium red for 90 min, mounted on a depression slide and observed with a Leica DM 6000B compound microscope, or air-dried before mounting on stubs and observed by scanning electron microscopy.
Seed coat permeability was determined using tetrazolium salts as described by Debeaujon et al. (2000). In short, seeds were incubated in 1% (w/v) tetrazolium red for 2 d in the dark at 37 oC and the percentage of red seeds calculated as a measure of permeability.
Chemical analysis
To quantify neutral sugars in crude mucilage extracts, 50 mg of intact seeds were incubated in 0.2% (w/v) ammonium oxalate with vigorous shaking for 2 h at 30 °C. 1 μmol of myo-inositol was added to the supernatant and samples were precipitated with 5 vols ethanol, directly hydrolysed with 2 M trifluroacetic acid, and derivatized to alditol acetates. Derivatization to alditol acetates and gas chromatography were performed as in Gibeault and Carpita (1991), but with an HP-23 glass capillary column (30 m×0.25 mm i.d.; Agilent Technologies). Seeds used for chemical analyses were collected from mutant and control plants cultivated together.
Germination time-course
Two 70 mm diameter Whatman No. 1 filter papers (Whatman) were placed in the lid of a 100 mm plastic Petri dish. To these were added 2 ml of water and 40–80 seeds of each mutant line. The plates were sealed with parafilm and stratified in the dark at 4 °C for 72 h. Seeds were incubated at 22 °C under 16/8 h light/dark, following which they were counted every day for 6 d and germination was scored by the presence of open green cotyledons. The plates were counted again after 9 d to confirm that all lines had reached approximately 100% germination. Seeds used for germination analyses were collected from mutant and control plants cultivated together and stored as distinct seed sets for 6 or 8 months, depending on the set. Seeds were stored in microfuge tubes with holes in the lids at room temperature under ambient humidity and light conditions. Each time-course was done in triplicate and the whole was performed twice using two independent sets of seeds with similar results.
Results
Identification of mum enhancers
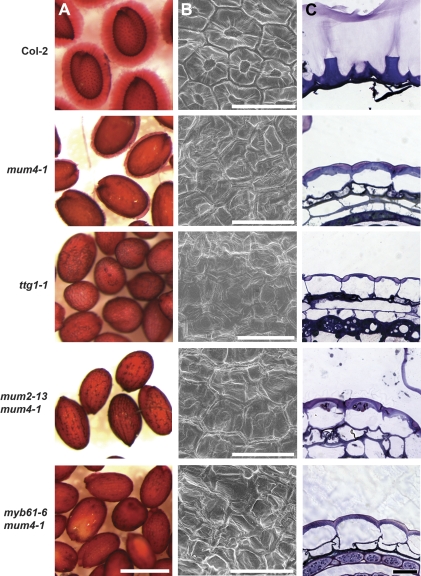
When Arabidopsis seeds are hydrated, the seed coat mucilage swells rapidly, leading to the bursting of the primary cell wall and the release of mucilage to surround the seed in a gel-like capsule (Fig. 1A) (Western et al., 2000; Windsor et al., 2000). mum4 mutants make a significantly reduced amount of mucilage (Western et al., 2001, 2004) that remains within the cells when seeds are hydrated. Addition of a heavy metal chelator such as EDTA or EGTA, however, leads to the release of mum4 mucilage. This is probably due to the withdrawl of Ca2+ ions from the cell wall pectins, leading to weakening of the cell wall and/or permitting increased swelling of the mucilage present. mum4 seeds shaken in EDTA prior to ruthenium red staining reveal a thin layer of stained mucilage around the seeds, consistent with their reduced mucilage production (Fig. 1A) (Western et al., 2004). By contrast, mutants for TTG1, which acts upstream of both the GL2 and TTG2 pathways of mucilage production, make very little mucilage and show no obvious mucilage release when EDTA-treated (Fig. 1A). The moderate level of mucilage release found for mum4 mutants, as well as the ability to differentiate mum4 mutants from mutants with further reduced mucilage, allowed us to perform a genetic screen for phenotypic enhancers of mum4. mum4-1 seeds were mutagenized with ethyl methanesulphonate and seeds from individual M2 plants (M3 lines) were collected and screened for reduced levels of mucilage compared with mum4-1 as observed with ruthenium red staining after EDTA pretreatment. Over 5000 M3 lines derived from ten parental M1 batches (1000–1500 M1 parents) were screened, leading to the identification of nine mum4-1 enhancers [named mum enhancers (men)] that have no visible mucilage release when treated with EDTA (Figs 1A, 2A).
Fig. 1.
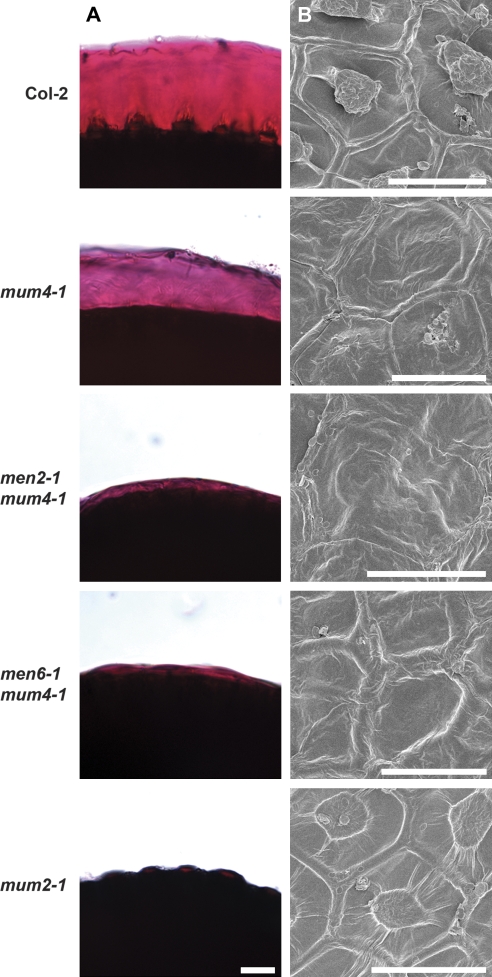
Mucilage release and seed coat structure of controls and mum4 enhancer lines resulting from mutations in MUM2 and MYB61. (A) Seeds pretreated in EDTA with shaking and stained with ruthenium red. Note the thick layer of mucilage surrounding wild-type Col-2 seeds and the very thin layer around mum4-1 seeds. (B) Scanning electron microscopy of dry seeds. Note the prominent hexagonal cell walls and central volcano-shaped columella in the centre of Col-2 cells. (C) Toluidine blue-stained resin sections of 13 DPA seeds. Wild-type mucilage cells have burst open in the aqueous fixative, leaving tall, blue-stained, volcano-shaped columellae with some cell wall material attached to the centre of the columella. mum4-1 cells do not burst, and contain small pockets of purple-stained mucilage in the upper apical corners, subtended by a blue dome of secondary cell wall. ttg1-1 cells, similar to mum4-1, do not burst, however, the mucilage pockets are smaller and the secondary cell wall is thinner in appearance. Scale bars: (A) 500 μm, (B) 50 μm, (C) 10 μm.
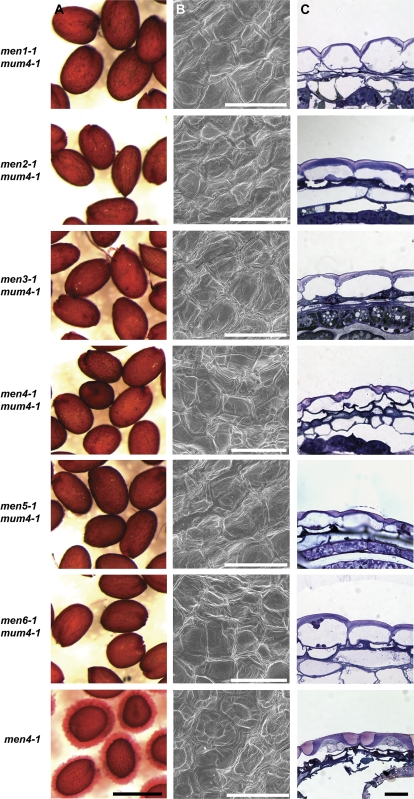
Fig. 2.
Mucilage release and seed coat structure of mum4 enhancer lines. (A) Seeds pretreated in EDTA with shaking and stained with ruthenium red. (B) Scanning electron microscopy of dry seeds. (C) Toluidine blue-stained resin sections of 13 DPA seeds. Scale bars: (A) 500 μm, (B) 50 μm, (C) 10 μm.
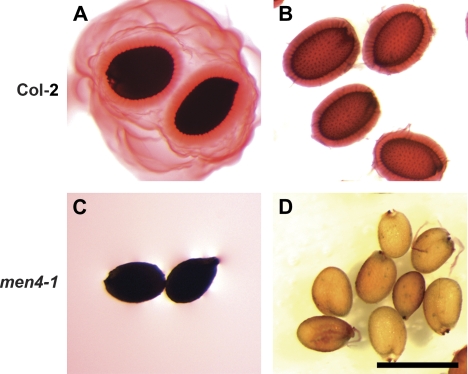
Backcrosses to mum4-1 plants revealed, in each case, that the seed phenotype was the result of a recessive mutation to a single locus (Table 1). Complementation tests were also performed between the nine men mum4-1 lines. Only one pair of mutants did not complement each other, revealing the identification of eight mutant loci. To determine if any of the men mum4-1 lines represented known mucilage mutant loci beyond MUM4, several assays were performed. First, no changes in seed shape were observed as in ap2 and ats mutants (Léon-Kloosterziel et al., 1994). Second, their identity as new alleles of GL2, TTG1, and TTG2 was tested through an examination of seed coat colour and trichome presence (Koornneef, 1981; Rerie et al., 1994; Johnson et al., 2002). All men mum4-1 lines had trichomes, and none had obviously yellow seeds, suggesting that they are different genes. Third, to eliminate tt mutants that were not obviously yellow, tetrazolium red staining was used to detect the increased permeability to solutes found for most tt mutants, including ttg1, tt2, and tt8 that are known to affect mucilage production (Debeaujon et al., 2000). One line that appeared to be wild-type seed colour (named men3-1 mum4-1) showed significant staining with tetrazolium red (data not shown). However, closer examination of men3-1 mum4-1 seeds revealed that they were slightly paler than wild-type seeds. The seed colour phenotype was found to segregate away from the mucilage phenotype, suggesting a background mutation in a tt or related gene that is unlikely to significantly affect mucilage release (data not shown). Fourth, complementation tests were performed with myb61, another reduced mucilage mutant (Penfield et al., 2001). One line was found not to complement myb61-1 and sequencing confirmed that it is a new allele of MYB61, which has been named myb61-6 (G to A transition leading to the conversion of Trp at position 252 to a stop codon). Fifth, the remaining men mum4-1 lines were backcrossed to wild-type Columbia-2 (Col-2) plants to determine if there was a mutant phenotype in the absence of mum4-1, as all mucilage mutants identified to date other than egl3, tt2, and tt8 have detectable mucilage release phenotypes in a wild-type background. Only two mutant lines had detectable single mutant phenotypes where no mucilage was released when shaken in ruthenium red stain without EDTA pretreatment (Fig. 3; data not shown). Since they were already shown not to be allelic to known reduced mucilage mutants, both of these lines were backcrossed to the mucilage release mutants mum1-1, mum2-1, and patchy (Western et al., 2001; AA Arsovski, TL Western, unpublished results). One line was found to complement all three mutants, while the other only complemented mum1-1 and patchy. While sequencing did not reveal an obvious mutation in the coding sequence of MUM2 for the non-complementing line, regulatory region or intron-related mutations cannot be ruled out. This line did, however, map to the MUM2 region, which, in combination with multiple complementation tests using both the double and isolated single mutants, and similar results between the single mutant and mum2 in all further assays (data not shown), suggest that this is, indeed, a new allele of MUM2 that has been named mum2-13. The other line represents a different gene that has been named MEN4. EDTA pretreatment of men4 single mutants revealed the release of a reduced amount of mucilage compared to wild-type seeds (compare Fig. 1A with Fig. 2A).
Table 1.
Segregation analysis and Chi-square test results for mum4 enhancer lines backcrossed to mum4-1
| Mutant line | mum4:no mucilagea | Chi squareb |
| men1-1 mum4-1 | 68:17 | 1.1333, P >0.1 |
| men2-1 mum4-1 | 60:17 | 0.3506, P >0.5 |
| men3-1 mum4-1 | 85:27 | 0.0476, P >0.5 |
| men4-1 mum4-1 | 57:16 | 0.3699, P >0.5 |
| men5-1 mum4-1 | 60:21 | 0.0370, P >0.5 |
| men6-1 mum4-1 | 80:19 | 1.7811, P >0.1 |
F3 seed phenotype (seed of F2 plants), shaken for 90 min in EDTA followed by staining in 0.01% ruthenium red.
Null hypothesis of 3:1 mum4:no mucilage; degrees of freedom=1; cutoff at P=0.05.
Fig. 3.
Mucilage release of men4-1 versus wild-type seeds in ruthenium red with and without shaking. (A, B) Col-2 wild-type seeds. (A) Seeds put directly into ruthenium red without shaking are surrounded by an outer, diffuse layer of mucilage and an inner, dense layer of mucilage, while those shaken in dye lose the soluble outer layer (B). (C, D) men4-1 seeds lack mucilage release when treated directly with ruthenium red, with or without agitation. Scale bar: 500 μm. (This figure is available in colour at JXB online.)
Phenotypic characterization of men seed coats
Wild-type epidermal seed coat cells, when observed by scanning electron microscopy (SEM), are shown to be roughly hexagonal in shape with thickened radial cell walls and a narrow, volcano-shaped columella in the centre of each cell (Fig. 1B) (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). A characteristic of reduced mucilage synthesis mutants, such as mum4 and ttg1, is a flattened columella that is subtly visible or missing when observed with SEM (Fig. 1B) (Koornneef, 1981; Western et al., 2001, 2004). To determine if an exacerbated phenotype was apparent in the mum4 enhancer lines, dry seeds of each double mutant plus the men4-1 single mutant were subjected to SEM. In each case for the double mutants, no significant enhancement of the mum4 phenotype was obvious (Figs 1B, 2B). men4-1 epidermal seed coat cells, however, have apparent columellae, but they are much broader and less prominent than those of wild-type seeds (Fig. 2B).
A more detailed view of the presence of mucilage and the shape of the columella can be gained through the use of sectioning and staining of seed coats with toluidine blue. Toluidine blue is a polychromatic dye that stains various cell components different colours. For example, acidic polysaccharides such as pectins stain pink-purple, and cell walls purple-blue (O'Brien et al., 1964). The timing of mucilage and columella production has been extensively studied, demonstrating that both are generally complete by 13 d post anthesis (DPA) (Western et al., 2000). Wild-type mucilage secretory cells at 13 DPA tend to release their mucilage upon wetting in aqueous fixative, leaving only the tall, volcano-shaped columellae and empty spaces where the mucilage accumulated prior to hydration (Fig. 1C) (Western et al., 2000). mum4-1 mucilage secretory cells, by contrast, remain intact, with a small amount of pink-purple staining mucilage found in the apical cell corners above a dome-shaped secondary cell wall (columella), all found above a large vacuole (Fig. 1C) (Western et al., 2004). ttg1-1 mucilage cells have a more severe phenotype than that of mum4-1, with less mucilage and a very thin secondary cell wall (Fig. 1C) (Penfield et al., 2001; Western et al., 2001). All of the mum4 enhancer lines resemble mum4-1 or ttg1-1 to varying degrees (Figs 1C, 2C). mum2-13 mum4-1 and, men2-1 mum4-1 resemble mum4-1, while men1-1 mum4-1, men4-1 mum4-1, and men5-1 mum4-1 are similar to ttg1-1. myb61-6 mum4-1, men3-1 mum4-1 (with background tt mutation), and men6-1 mum4-1 all appear to have an intermediate phenotype.
Quantitative analysis of men mucilage
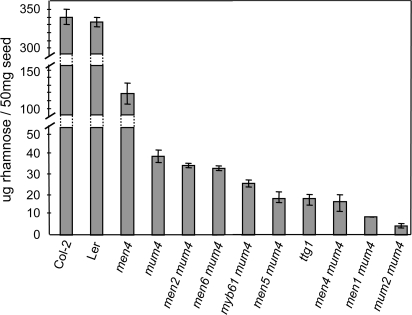
To quantify the amount of mucilage produced by the different mum4 enhancers, ammonium oxalate soluble mucilage was extracted from intact seed samples, hydrolysed, derivatized to alditol acetates, and subjected to gas chromatography. Alditol acetate derivatization allows for the production of a complete neutral sugar profile of cell wall material (fucose, rhamnose, arabinose, xylose, mannose, galactose, and glucose) (Chaplin, 1986). However, the soluble cell wall material from these mutants could be derived from the cell wall and/or the mucilage. Arabidopsis mucilage is primarily comprised of unbranched RG I, a pectin whose backbone is composed of alternating residues of rhamnose and galacturonic acid (Penfield et al., 2001; Western et al., 2004). Comparison between ground wild-type seeds and those of an ap2 mutant, which makes little or no mucilage, suggests that approximately 80% of the rhamnose of Arabidopsis seeds is found in the mucilage (Western et al., 2001). Thus, to focus more specifically on changes to mucilage levels, only rhamnose was considered for comparison between mutants (Fig. 4). Wild-type Col-2 and Ler extracted mucilage contained 339.6±9.7 and 332.9±6.3 μg of rhamnose per 50 mg seed, respectively (SE, n=3). By contrast, mum4-1 seeds have approximately one-tenth that amount (38.90±3.5 μg per 50 mg seed) and ttg1-1 seeds have approximately half as much rhamnose as mum4-1 mutants (17.5±2.6 μg per 50 mg seed) (Fig. 4).
Fig. 4.
Rhamnose levels of soluble mucilage extracted from mum4 enhancers and controls. Ammonium oxalate extracts of concurrently-grown seed batches were hydrolysed with trifluoroacetic acid and derivatized to alditol acetates, followed by gas chromatography. Extractions were done in triplicate, error bars=SE.
men2-1 mum4-1 and men6-1 mum4-1 were found to have approximately the same amount of rhamnose as mum4-1 single mutants (Fig. 4). While this correlates with the similar appearance of mucilage and columella between these two double mutants and mum4-1 observed with toluidine blue-stained sections (Figs 1C, 2C), it is intriguing that the double mutant seeds pretreated with EDTA do not appear to release mucilage (Fig. 2A). In order to determine if this resulted from the differential release of mucilage with ammonium oxalate at 30 oC versus EDTA at room temperature, ammonium oxalate-treated seeds were stained with ruthenium red. Similar to the EDTA results, mum4-1 seeds showed substantial mucilage release with ammonium oxalate treatment, while men2-1 mum4-1 and men6-1 mum4-1 seeds had only slight puffing of the cell walls (Fig. 5). No mucilage release or cell wall puffing was seen for mum2-1 mutants that cannot release mucilage due to mucilage hydration defects (Fig. 5A; Dean et al., 2007; Macquet et al., 2007b).
Fig. 5.
Seed coat phenotype of men2 mum4, men6 mum4, and control seeds following extraction with ammonium oxalate. (A) Seeds extracted with ammonium oxalate with shaking at 30 oC, then stained with ruthenium red. Note substantial mucilage release for Col-2 seeds. mum4-1 seeds release less mucilage, with the outer wall appearing to remain largely intact. (B) Scanning electron microscopy of air-dried ammonium oxalate extracted seeds. Only Col-2 seeds show obvious rupture of outer primary cell wall. Scale bars: (A) 10 μm, (B) 25 μm. (This figure is available in colour at JXB online.)
myb61-6 mum4-1 has an approximately 30% drop compared with mum4-1, confirming the enhanced phenotype seen with toluidine blue-stained sections. This was also the case for men4-1 mum4-1 and men5-1 mum4-1, which are similar to ttg1-1, both in terms of a 50% reduction in rhamnose compared with mum4-1 and in their phenotype in their cross-sections. men1-1 mum4-1, which also appears similar to ttg1-1 in toluidine blue-stained sections, has a further 40–50% drop in soluble mucilage compared to ttg1-1. The lowest amount of rhamnose observed was for mum2-13 mum4-1, reflecting the lack of mucilage release observed in mum2 single mutants (Fig. 5; Dean et al., 2007; Macquet et al., 2007b). The rhamnose level was also determined for the men4-1 single mutant and found to be approximately 35% of wild-type rhamnose levels in its extracted mucilage, consistent with the ruthenium red and toluidine blue section results.
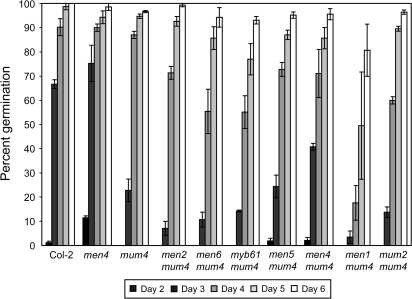
Germination of men lines
Altered seed germination responses have been correlated with changes in seed coat structure, including mucilage quantity and release (Léon-Kloosterziel et al., 1994; Debeaujon et al., 2000; Penfield et al., 2001; Rautengarten et al., 2008). To determine the effect of reduced mucilage levels in the mum4 single mutant, as well as in the men lines, a time-course of germination was performed (Fig. 6). mum4-1 germination lagged significantly behind that of wild-type seeds at 3 d (23% for mum4-1 versus 67% for Col-2), but reached approximately wild-type levels by 4 d (Fig. 6). Similar, or even more severe delays, at 3 d were detected for the set of men mum4-1 lines plus myb61-6 mum4-1 and mum2-1 mum4-1 double mutants, all of which continued to stay significantly below wild-type germination levels at day four, with the exception of men4 mum4 (Fig. 6). All lines reached 95–100% germination within 9 d (data not shown). Together, these results suggest not only that the reduction of mucilage in mum4-1 has an effect on the speed of germination, but also that this delay may be enhanced by further defects in both mucilage release and quantity. Conversely, men4-1, which has approximately three times more mucilage than mum4-1 (Fig. 4), shows no delay in germination. Interestingly, both men4-1 mum4-1 and men5-1 mum4-1 seeds demonstrate precocious germination relative to mum4-1 at 2 d. This is reflected in men4-1, which germinates faster than the wild type at 2 d. This ‘early germination’ may explain the similar if not faster germination exhibited by men4-1 versus wild type, and men4-1 mum4-1 and men5-1 mum4-1 versus mum4-1 exhibited at 3 d. It is possible that, in these lines, there is a germination phenotype beyond that resulting from reduced mucilage levels.
Fig. 6.
Time-course of germination of mum4 enhancers and controls. Genotypes are organized in the same order as Fig. 4 for comparison. Seeds were stratified for 3 d at 4 °C, followed by germination at 22 °C under 16/8 h light/dark. 40–80 seed of each genotype were sowed on filter paper with water, error bars=SE. Similar results were obtained in a separate experiment using seed from an independent set of plants.
The mum4 enhancer lines were also tested for gross changes in whole plant developmental phenotypes by following the Arabidopsis Gantlet Project protocol (http://thale.biol.wwu.edu/). In short, seeds were plated with wild-type and mum4-1 control seeds side-by-side, grown vertically on plates for 14 d, and then transplanted to soil, with regular observation across all stages (daily while on plates and weekly once in soil). No gross developmental phenotypes were observed for any of the lines.
Discussion
The mucilage secretory cells of the Arabidopsis seed coat are a useful model for the identification and study of genes involved in cell wall production and metabolism. In particular, they have started to allow the dissection of genes involved in the regulation of pectin synthesis and in pectin modification (Penfield et al., 2001; Johnson et al., 2002; Zhang et al., 2003; Western et al., 2004; Dean et al., 2007; Macquet et al., 2007b; Rautengarten et al., 2008; Gonzalez et al., 2009; Li et al., 2009). However, to date, only one gene involved directly in mucilage synthesis has been identified, and no genes have been directly implicated in polar secretion of mucilage (Usadel et al., 2004a; Western et al., 2004). Here, six new genes involved in mucilage production have been identified as enhancers of the mum4 reduced mucilage mutant. Three of these genes appear to have further reductions in mucilage production compared with mum4, making them promising candidates for roles in mucilage synthesis and/or secretion.
MEN genes affect mucilage production
Mutations in six new genes (MEN1–6) affecting mucilage secretory cell differentiation were identified, along with new alleles of two known genes: MUM2 and MYB61. The finding of new alleles of these two genes validated the screen in its ability to find genes acting in parallel with MUM4 for mucilage production (MYB61) (Penfield et al., 2001), as well as genes acting in pectic mucilage modifications that are required for mucilage swelling and release (MUM2) (Dean et al., 2007; Macquet et al., 2007b). Further, this screen revealed the utility of such a sensitized screen, as only one of the new genes identified (men4) had an obvious single mutant phenotype. Characterization of the mum4 enhancers revealed two phenotypic categories: reduced mucilage production and lack of mucilage release.
Three of the men mum4 double mutants identified in this screen (men1 mum4, men4 mum4, and men5 mum4) appear to make reduced amounts of mucilage compared to mum4 as determined by both their cell structure and their soluble rhamnose levels (Figs 1, 2, 4). men1 mum4 double mutants have the most significant reduction of mucilage, while men4 mum4 and men5 mum4 have slightly more mucilage. The interpretation of these mutants as being affected in mucilage production is supported by their shared phenotypes with myb61 mum4 and ttg1. TTG1 regulates both the GL2 and TTG2 pathways of mucilage production (Johnson et al., 2002; Zhang et al., 2003; Western et al., 2004), thus, ttg1-like mutants may be expected to be affected in both pathways downstream of TTG1. myb61 mutants have reduced mucilage resulting from disruption of a TTG1-independent pathway (Penfield et al., 2001; Western et al., 2004), so the myb61 mum4 double mutant serves as a control for the disruption of the two independent pathways of mucilage production, as MUM4 acts downstream of TTG1. In addition, the single mutant for men4 has significantly reduced mucilage compared to wild-type seeds (Figs 1, 3, 4), confirming a role in mucilage production for one member of this class of mum4 enhancers.
As mucilage primarily comprises the pectin RG I, genes involved in its manufacture and transport, or the regulation of these processes, would be the most obvious candidates for the men genes affected in mucilage quantity. While our phenotypic and complementation analyses have ruled out most mucilage regulatory mutants, it is possible that one of the men genes could encode a new allele of EGL3 or a weak allele of MYB5 without an obvious single mutant phenotype (Zhang et al., 2003; Gonzalez et al., 2009; Li et al., 2009). Our preliminary mapping data for MEN1, MEN4, and MEN5, however, suggest that this is not the case (AA Arsovski, M Wang, N Martin, J Schafhauser, TL Western, unpublished results). MUM4 is a member of a small gene family that encodes three full-length, trifunctional UDP-L-rhamnose synthase proteins (RHM1, MUM4/RHM2, RHM3), and a protein that catalyses only the latter part of the conversion of UDP-D-glucose to UDP-L-rhamnose (UER) (Reiter and Vanzin, 2001; Usadel et al., 2004a; Watt et al., 2004; Western et al., 2004; Oka et al., 2007). All members are expressed throughout the plant, allowing for genetic redundancy in rhamnose synthesis and the production of some mucilage in mum4 seeds. A mutation in RHM1, RHM3 or UER could result in a further reduction in mucilage production. By contrast, one of the men genes could encode a member of the UDP-D-GLUCOURONATE 4-EPIMERASE (GAE) family, which are required for the synthesis of UDP-D-galacturonic acid, the other sugar comprising the backbone of RG I, as well as the backbone of homogalacturonan (Willats et al., 2001b; Usadel et al., 2004b). Pectins are synthesized in the Golgi apparatus through the activity of glycosyltransferases (GT) that use nucleotide sugars as substrates. Members of GT family 8 have been implicated in pectin synthesis through both mutant studies and enzyme isolation (Scheller et al., 1999; Sterling et al., 2001, 2006; Willats et al., 2001b; Bouton et al., 2002; Lao et al., 2003; Shao et al., 2004; Mohnen, 2008), so it is possible that one of the men genes could encode a GT8 protein required either for RG I backbone synthesis or for the synthesis of one of the other pectins found in Arabidopsis mucilage. Indeed, a gene encoding a GT8 family protein that has a mild mucilage production phenotype has been identified as being up-regulated in seed coats at the time of mucilage synthesis, similar to MUM4 (J Schafhauser, A Abdeen, TL Western, unpublished results). Alternately, a men mutant could be affected in a gene required for secretion of mucilage from the Golgi apparatus to the apoplast. These could include vesicle trafficking factors such as small G-proteins, their effectors or activators, or the multi-subunit exocytosis complex known as the exocyst (Cole and Fowler, 2006; Hála et al., 2008; Nielsen et al., 2008; Rojo and Denecke, 2008; Yalovsky et al., 2008).
Both men2 mum4 and men6 mum4 double mutants did not release mucilage after pretreatment with EDTA, but appear to make a similar amount of mucilage to mum4 as assessed by both cellular appearance and rhamnose levels in soluble mucilage (Figs 2, 4). Specific mucilage release in ammonium oxalate used for mucilage extraction was tested by staining seeds after shaking in ammonium oxalate and revealed only slight puffing of the cell wall for men2 mum4 and men6 mum4 (Fig. 5). It is possible that some primary cell wall breakage occurred in parallel with this puffing, allowing mucilage extraction through fissures in the wall. Conversely, chelator treatment may have extracted pectins from the primary cell wall, allowing further extraction of highly soluble mucilage through the weakened wall. Extraction of only primary cell wall pectins in the case of men2 mum4 and men6 mum4 is unlikely since both have higher levels of soluble rhamnose than other non-releasing mutants that have little or no mucilage (e.g. ttg1) or that have been shown to affect mucilage hydration (mum2 mum4) (Figs 2, 4, 5) (Western et al., 2000; Dean et al., 2007; Macquet et al., 2007b).
The mum4-like phenotypes make it difficult to assess the cause of the lack of mucilage release in men2 mum4 and men6 mum4. One possibility is that there is a decrease in mucilage production that is sufficient to prevent release, but not large enough to be detected though observation of secretory cell structure or rhamnose levels. Alternately, there could be a defect in the cell wall or mucilage structure that prevents mucilage release. While the extremely low soluble rhamnose levels of mum2 mum4 double mutants would seem to argue against this latter hypothesis, mum2 mutants are characterized by a lack of mucilage swelling that probably results from reduced hydration capacity, explaining the mum2 mum4 double mutant phenotype (Fig. 5; Dean et al., 2007; Macquet et al., 2007b). It is possible for the men2 mum4 and men6 mum4 double mutants that the mucilage and/or outer cell wall structural changes are such that mucilage solubility is less significantly affected than for mutations in MUM2. Candidate genes for men2 mum4 and men6 men2, therefore, may be new cell wall modification factors. While preliminary mapping places MEN2 away from known mucilage release genes (MM Villota, TL Western, O Rowland, R Subramaniam, unpublished data), it cannot be ruled out that the two men6 alleles are weak alleles of MUM1, MUM2 or SBT1.7 that lack obvious phenotypes in the absence of mum4.
Effect of mucilage changes on seed germination
A time-course of germination revealed that mum4-1 has delayed germination compared with wild-type seeds (Fig. 6). This is consistent with previous studies that demonstrated that reduced mucilage mutant seeds (myb61-1, ttg1-1, gl2-1) had a decreased ability to germinate under conditions of limited water supply (as exerted by increasing concentrations of polyethylene glycol) compared to wild-type seeds (Penfield et al., 2001). A similar reduction in germination in the presence of polyethylene glycol was seen for Atsbt1.7 mutants, which are defective in mucilage release (Rautengarten et al., 2008). Due to its significant hydrophilicity, Arabidopsis mucilage has been proposed to promote seed hydration, and thus germination, through the attraction and retention of water surrounding the seed (Penfield et al., 2001). In mum4-1, seed hydration, and thus imbibition, could be slowed by either the reduced quantity of mucilage or the lack of release of mucilage to form a hydrated gel around the seed. While pleiotropic effects cannot be ruled out, the enhanced delay seen in both myb61 mum4 and mum2 mum4 double mutants, as well as men mum4 double mutants (Fig. 6), strongly suggests that both mucilage quantity and release are important for efficient seed hydration and germination, even under moist conditions.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table 1. Primers used for the sequencing of mum2-13 and myb61-6.
Acknowledgments
The authors gratefully acknowledge a number of undergraduate research assistants for their help in screening for mum4 enhancer lines and genetic analysis, including Phoenix Bouchard-Kerr, Sonia Rehal, Amin Osmani, Victoria Bond, Marina Gerbin, Michelle Wang, Natalie Martin, Deidre Clark, Sana Ghani, Nicolay Hristozov, and Faizal Kassam. Our thanks also go to Dr Ashraf Abdeen and the anonymous reviewers for comments on the manuscript. Funding for this project was provided by a Natural Sciences and Engineering Research Council Discovery Grant to TLW and an Agriculture and Agri-Foods Canada Network Grant to RS and OR.
Glossary
Abbreviations
- DPA
days post anthesis
- GT
glycosyltransferase
- RG I
rhamnogalacturonan I
- SEM
scanning electron microscopy
References
- Beeckman T, De Rycke R, Viane R, Inzé D. Histological study of seed coat development in Arabidopsis thaliana. Journal of Plant Research. 2000;113:139–148. [Google Scholar]
- Boesewinkel FD, Bouman F. The seed: structure. In: Johri BM, editor. Embryology of Angiosperms. New York: Springer-Verlag; 1984. pp. 567–610. [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker M-T, Talbotek J, Granier F, Lahaye M, Höfte H, Truong H-N. QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. The Plant Cell. 2002;14:2577–2590. doi: 10.1105/tpc.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Koornneef M. Seed morphology. In: Bowman JL, editor. Arabidopsis: an atlas of morphology and development. New York: Springer-Verlag; 1993. pp. 398–401. [Google Scholar]
- Burn JE, Hurley UA, Birch RJ, Arioli T, Cork A, Williamson RE. The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. The Plant Journal. 2002;32:949–960. doi: 10.1046/j.1365-313x.2002.01483.x. [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, et al. The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. The Plant Cell. 2007;19:4007–4021. doi: 10.1105/tpc.107.050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin M. Monosaccharides. In: Chaplin M, Kennedy J, editors. Carbohydrate analysis: a practical approach. Washington DC: IRL Press; 1986. pp. 1–36. [Google Scholar]
- Cole RA, Fowler JE. Polarized growth: maintaining focus on the tip. Current Opinion in Plant Biology. 2006;9:579–588. doi: 10.1016/j.pbi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiology. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Anatomy of seed plants. Toronto: Wiley; 1977. The seed; pp. 455–473. [Google Scholar]
- Fahn A. Plant anatomy. Toronto: Pergamon Press; 1982. The seed; pp. 479–496. [Google Scholar]
- Gibeaut DM, Carpita NC. Cleanup procedure for partially methylated alditol acetate derivatives of polysaccharides. Journal of Chromatography. 1991;587:284–287. [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. TTG1 complex MYBs, MYB5, and TT2, control outer seed coat differentiation. Developmental Biology. 2009;325:412–421. doi: 10.1016/j.ydbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Grubert M. Mucilage or gum in seeds and fruits of Angiosperms: a review. Munich: Minerva Press; 1981. [Google Scholar]
- Hála M, Cole R, Synek L, et al. An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. The Plant Cell. 2008;20:1330–1345. doi: 10.1105/tpc.108.059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Chaudhury A. Genetic analysis of seed coat development in Arabidopsis. Trends in Plant Science. 2005;10:472–477. doi: 10.1016/j.tplants.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville CR. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Molecular and General Genetics. 1986;204:430–434. [Google Scholar]
- Jofuku KD, den Boer BGW, Montagu MV, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. The Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. The Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DLC, Breekland AE, Koornneef M. Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heyhn. Planta. 1983;157:158–165. doi: 10.1007/BF00393650. [DOI] [PubMed] [Google Scholar]
- Kim Y-C, Nakajima M, Nakayama A, Yamaguchi I. Contribution of gibberellins to the formation of Arabidopsis seed coat through starch degradation. Plant and Cell Physiology. 2005;46:1317–1325. doi: 10.1093/pcp/pci141. [DOI] [PubMed] [Google Scholar]
- Koornneef M. The complex syndrome of TTG mutants. Arabidopsis Information Service. 1981;18:45–51. [Google Scholar]
- Lao NT, Long D, Kiang S, Coupland G, Shoue DA, Carpita NC, Kavanagh TA. Mutation of a family 8 glycosyltransferase gene alters cell wall carbohydrate composition and causes a humidity-sensitive semi-sterile dwarf phenotype in Arabidopsis. Plant Molecular Biology. 2003;53:687–701. doi: 10.1023/B:PLAN.0000019074.60542.6c. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Keijzer CJ, Koornneef M. A seed shape mutant of Arabidopsis that is affected in integument development. The Plant Cell. 1994;6:685–692. doi: 10.1105/tpc.6.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. The Plant Cell. 2009;21:72–89. doi: 10.1105/tpc.108.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet M-C, Kronenberger J, Marion-Poll A, North HM. In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant and Cell Physiology. 2007a;48:984–999. doi: 10.1093/pcp/pcm068. [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Oudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. A naturally occurring mutation in an Arabidopsis accession affects a β-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. The Plant Cell. 2007b;19:3990–4006. doi: 10.1105/tpc.107.050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H, Young R, Wasteneys G, Samuels A. Cortical microtubules mark the mucilage secretion domain of the plasma membrane in Arabidopsis seed coat cells. Planta. 2008;227:1363–1375. doi: 10.1007/s00425-008-0708-2. [DOI] [PubMed] [Google Scholar]
- Messmer McAbee J, Hall TA, Skinner DJ, Izhaki A, Hauser BA, Meister RJ, Reddy GV, Meyerowitz EM, Bowman JL, Gasser CS. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. The Plant Journal. 2006;46:522–531. doi: 10.1111/j.1365-313X.2006.02717.x. [DOI] [PubMed] [Google Scholar]
- Mohnen D. Pectin structure and biosynthesis. Current Opinion in Plant Biology. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Newman LJ, Perazza DE, Juda L, Campbell MM. Involvement of the R2R2-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. The Plant Journal. 2004;37:239–250. doi: 10.1046/j.1365-313x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Cheung AY, Ueda T. The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiology. 2008;147:1516–1526. doi: 10.1104/pp.108.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Feder N, McCully M. Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma. 1964;59:366–373. [Google Scholar]
- Oka T, Nemoto T, Jigami Y. Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-d-glucose to UDP-l-rhamnose conversion. Journal of Biological Chemistry. 2007;282:5389–5403. doi: 10.1074/jbc.M610196200. [DOI] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. The Plant Cell. 2001;13:2777–2791. doi: 10.1105/tpc.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. The Plant Journal. 2008;54:466–480. doi: 10.1111/j.1365-313X.2008.03437.x. [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks DM. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes and Development. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Vanzin GF. Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Molecular Biology. 2001;47:95–113. [PubMed] [Google Scholar]
- Rojo E, Denecke J. What is moving in the secretory pathway of plants? Plant Physiology. 2008;147:1493–1503. doi: 10.1104/pp.108.124552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Doong RL, Ridley BL, Mohnen D. Pectin biosynthesis: a solubilized α-1,4-galacturonosyltransferase from tobacco catalyzes the transfer of galacturonic acid from UDP-galacturonic acid onto the non-reducing end of homogalacturonan. Planta. 1999;207:512–517. [Google Scholar]
- Shao M, Zheng H, Hu Y, Liu D, Jang J-C, Ma H, Huang H. The GAOLAOZHUANGREN1 gene encodes a putative glycosyltransferase that is critical for normal development and carbohydrate metabolism. Plant and Cell Physiology. 2004;45:1453–1460. doi: 10.1093/pcp/pch168. [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kolli VSK, Quigley HF, Hahn MG, Mohnen D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proceedings of the National Academy of Sciences. USA. 2006;103:5236–5241. doi: 10.1073/pnas.0600120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Quigley HF, Orellana A, Mohnen D. The catalytic site of the pectin biosynthetic enzyme α-1,4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiology. 2001;127:360–371. doi: 10.1104/pp.127.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Kuchinsky AM, Rosso MG, Eckermann N, Pauly M. RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiology. 2004a;134:286–295. doi: 10.1104/pp.103.034314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Schlüter U, Mølhøj M, Gipmans M, Verma R, Kossman J, Reiter W-D, Pauly M. Identification and characterization of a UDP-d-glucuronate 4-epimerase in Arabidopsis. FEBS Letters. 2004b;569:327–331. doi: 10.1016/j.febslet.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Watt G, Leoff C, Harper AD, Bar-Peled M. A bifunctional 3,5-epimerase/4-keto reductase for nucleotide-rhamnose synthesis in Arabidopsis. Plant Physiology. 2004;134:1337–1346. doi: 10.1104/pp.103.037192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL. Changing spaces: the Arabidopsis mucilage secretory cells as a novel system to dissect cell wall production in differentiating cells. Canadian Journal of Botany. 2006;84:622–630. [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW. Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiology. 2001;127:998–1011. [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiology. 2000;122:345–355. doi: 10.1104/pp.122.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW. MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiology. 2004;134:296–306. doi: 10.1104/pp.103.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Knox JP. In situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta. 2001a;213:37–44. doi: 10.1007/s004250000481. [DOI] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Molecular Biology. 2001b;47:9–27. [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AL. Arabidopsis seed coat development: morphological differentiation of the outer integument. The Plant Journal. 2000;22:483–493. doi: 10.1046/j.1365-313x.2000.00756.x. [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B. Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiology. 2008;147:1527–1543. doi: 10.1104/pp.108.122150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL. Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. The Plant Cell. 2008;20:1623–1638. doi: 10.1105/tpc.108.058842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.