Abstract
Non-mycorrhizal Hakea actites (Proteaceae) grows in heathland where organic nitrogen (ON) dominates the soil nitrogen (N) pool. Hakea actites uses ON for growth, but the role of cluster roots in ON acquisition is unknown. The aim of the present study was to ascertain how N form and concentration affect cluster root formation and expression of peptide transporters. Hydroponically grown plants produced most biomass with low molecular weight ON>inorganic N>high molecular weight ON, while cluster roots were formed in the order no-N>ON>inorganic N. Intact dipeptide was transported into roots and metabolized, suggesting a role for the peptide transporter (PTR) for uptake and transport of peptides. HaPTR4, a member of subgroup II of the NRT1/PTR transporter family, which contains most characterized di- and tripeptide transporters in plants, facilitated transport of di- and tripeptides when expressed in yeast. No transport activity was demonstrated for HaPTR5 and HaPTR12, most similar to less well characterized transporters in subgroup III. The results provide further evidence that subgroup II of the NRT1/PTR family contains functional di- and tripeptide transporters. Green fluorescent protein fusion proteins of HaPTR4 and HaPTR12 localized to tonoplast, and plasma- and endomembranes, respectively, while HaPTR5 localized to vesicles of unknown identity. Grown in heathland or hydroponic culture with limiting N supply or starved of nutrients, HaPTR genes had the highest expression in cluster roots and non-cluster roots, and leaf expression increased upon re-supply of ON. It is concluded that formation of cluster roots and expression of PTR are regulated in response to N supply.
Keywords: Cluster root, heathland, organic nitrogen, peptide transporters, Proteaceae, roots
Introduction
Organic nitrogen (ON) is emerging as a potentially important soil-derived N source for plants, although the contribution of ON to plant N supply is not well understood (reviewed by Näsholm et al., 2009). Better knowledge of the strategies of plants for acquiring ON is important for understanding nutrient cycles, conservation of biodiversity, and design of sustainable bioproduction systems. The heathland plant Hakea actites grows in soil dominated by ON (Schmidt and Stewart, 1997) and uses amino acids, peptides, and proteins as N sources (Schmidt and Stewart, 1999; Schmidt et al., 2003; Paungfoo-Lonhienne et al., 2008). Hakea actites was chosen as a model to examine how ON is acquired and metabolized, because in contrast to most heathland plants, which rely on fungal and/or bacterial symbioses to access N (Read, 1991), H. actites does not form symbioses but produces cluster roots. Cluster roots are formed by a number of species in selected plant families, but occur in most species of the Southern hemisphere Proteaceae family.
Cluster roots are a strategy for nutrient acquisition in extremely oligotrophic habitats (Lambers et al., 2008) and are formed in response to low nutrient supply (reviewed by Dinkelaker et al., 1995; Neumann and Martinoia, 2002; Shane and Lambers, 2005). Cluster roots enhance the plant's access to phosphorus and other soil nutrients (Dinkelaker et al., 1995; Neumann and Martinoia, 2002) and may have a role for accessing N (Pate and Jeschke, 1993; Schmidt et al., 2003). It has been shown that cluster roots exude a range of inorganic and organic compounds, as well as enzymes, including acid phosphatases (Dinkelaker et al., 1995) and proteases (Schmidt et al., 2003; Paungfoo-Lonhienne et al., 2008).
There is evidence that plants use di- and tripeptides as an N source (Schmidt et al., 2003; Komarova et al., 2008). Transport of peptides across membranes is mediated by transporters of three gene families: members of the NRT1/PTR (nitrate transporter 1/peptide transporter) family which facilitate transport of di- and tripeptides; members of the OPT (oligopeptide transporter) family transporting tetra- and pentapeptides; and members of the ABC (ATP-binding cassette) family which mediate transport of larger peptides, although the latter has not been confirmed in plants (reviewed by Rentsch et al., 2007). While a large number of putative PTR genes exist in plants, functionality has been confirmed for comparatively few transporters (Rentsch et al., 2007; reviewed by Tsay et al., 2007). In Arabidopsis, peptide transporters AtPTR1 and AtPTR5 facilitate uptake of di- and tripeptides into roots and germinating pollen, respectively, and overexpression of AtPTR5 increased growth of Arabidopsis with peptides as the sole N source (Komarova et al., 2008). Thus, the presence and expression of PTRs in roots may determine the plant's ability to use peptides for growth. It is currently not known whether the main function of PTRs is uptake of peptides from the growth medium into the root and/or inter- and intracellular transport of peptides.
A partial cDNA fragment of a putative PTR was previously cloned from H. actites (HaPepT1); this cDNA had increased levels of expression during cluster root development (Schmidt et al., 2003). The partial HaPepT1 cDNA is identical to HaPTR4, one of the transporters characterized here. The aim of this study was to examine formation of cluster roots as well as functionality and expression of peptide transporters from H. actites in the natural habitat and in controlled conditions with different concentrations and forms of N.
Materials and methods
Hydroponic plant culture
In the first experiment, the aim was to determine how cluster roots are formed and peptide transporters are expressed. Hakea actites (W.R. Barker) seedlings were initially cultivated without addition of an N source, and then transferred to nutrient solution with limiting or adequate N supply (inorganic N, ON). Plants were grown from seed in water–vermiculite for 3 months (April to June 2005) in a naturally lit glasshouse (60% full sunlight, maximum temperature 28 °C). Subsequently, plants were cultivated in hydroponic culture containing 4.0 l of nutrient solution (200 μM CaCl2, 100 μM MgSO4, 50 μM KH2PO4, 50 μM K2SO4, 2.5 μM H3BO3, 0.25 μM MnSO4, 0.025 μM ZnSO4, 0.025 μM CuSO4, 0.025 μM NH4Mo7O24, 5 μM Fe-EDTA, pH adjusted to 6.0) and grown for a further 7 months. Nutrient solutions were supplemented with 15 μM or 150 μM N as ammonium nitrate, glycine, or bovine serum albumin (BSA). The control received no N. Each pot contained three seedlings, and each treatment consisted of three pots. The nutrient solution was changed weekly. Tissues of 7-month-old plants were sampled, and immediately submersed in liquid N2 and stored at –80 °C. Additional plants were divided into shoots, non-cluster roots, and cluster roots for biomass analysis (dried for 48 h at 60 °C).
In the second experiment, the aim was to investigate the effect of the plant's original nutrient status on cluster root formation. Four-month-old H. actites from a commercial nursery (Wallum Nurseries Pty, Cleveland, Australia) were grown in University of California (UC) potting mix (Matkin and Chandler, 1957) and watered daily for 4 months with tap water. Roots were washed free of soil, and seedlings of uniform size were selected and transplanted to 4.0 l plastic pots. Nutrient solution and growth conditions were the same as for the plants initially cultivated without N described above. Plants were grown for an additional 6 months (June 2005 to January 2006).
The third experiment was to determine expression of HaPTR genes under conditions of protein re-supply after starvation. Hakea actites seeds were germinated in April 2004, grown in UC potting mix for 2 months, and grown hydroponically for 10 months with low nutrient conditions supplied only with tap water. Seedlings were grown for an additional 7 months under ‘nutrient starvation’ conditions with distilled water amended with 10 μM CaSO4, and replaced weekly. Three pots were re-supplied with 150 μM N in the form of protein (BSA) for 3 d.
Hydroponic pots were covered with aluminium foil to prevent algal growth, and pots were rotated frequently. Young and mature cluster roots, non-cluster roots, and leaves were taken for RNA isolation from the first and third experiments; plants in the second experiment (grown initially at high N) did not produce cluster roots.
Field-grown plants
Hakea actites seedlings were collected in October 2006 from wallum heathland [Beerwah Scientific Area No. 1 (26.9ºS, 152.9ºE)] where mature H. actites shrubs grow to ∼2 m height. During regular fires, imposed every 7 years, mature plants die and release seeds, which germinate in the ash bed. Small plants of up to 50 cm in size were collected from an area which that had been burnt 2 years previously. Hakea actites produces cluster roots at 10–20 cm intervals along lateral roots in the upper 10 cm of soil (Schmidt and Stewart, 1999). Roots were carefully excavated, and adhering soil was removed by gently shaking the roots and washing them in deionized water. Ten young cluster roots were placed in microcentrifuge tubes and stored immediately in liquid N2 for RNA extraction. Roots and leaves were collected from the same plants. Tissues were sampled from five individual plants.
Uptake of the dipeptide Gly-Gly
Axenic Hakea plants (six seeds per plate) were grown for 4 weeks on N-free nutrient medium (see hydroponic culture) on agar plates (0.3% phytagel, Phytotechnology Laboratories, Lenexa, KS, USA) and individual roots of similar size were supplied with 25 μmol of Gly-Gly (99%, Sigma). Three-quarters of each cotyledon was removed when Gly-Gly was added to reduce the seedlings’ reliance on stored N. Roots were extracted after 5 min, 4 h, or 15 h, and rinsed three times in 0.5 mM CaCl2. Roots were immediately placed in liquid N2. Frozen roots were homogenized and resuspended in 250 μl of methanol (20 %). Samples were shaken on a vortex and stored at 4 °C overnight, centrifuged at 23 000 g for 10 min at 4 °C to condense the tissue into a pellet, and 40 μl of supernatant was mixed with 140 μl of borate buffer and 20 μl of AccQTag™ reagent (AccQTag™ derivitization kit), and analysed for peptide and amino acids (UPLC, Waters, Milford, MA, USA, equipped with a BEH C18 1.7 μm 2.1×100 mm column and a tunable UV detector at 254 nm). A solvent gradient from a H2O-based solution to a 55% acetonitrile solvent separated compounds in a 40 min run. Pellets were dried (55 °C, 48 h) and dry weights recorded.
Isolation of HaPTRs
Total RNA from Hakea roots was isolated as previously described (Mason and Schmidt, 2002). DNA was removed with 2 U of DNase (Turbo DNA-free™; Ambion, Austin, TX, USA, 30 min at 37 °C). Total RNA was quantified using a NanoDrop ND-1000 spectrophotometer and separated using agarose gel electrophoresis to monitor RNA degradation. Only RNA preparations with A260/A280 ≥1.8 were used. HaPTR genes were isolated using 3′ and 5′ rapid amplification of cDNA ends (RACE) PCR (FirstChoice RLM-RACE; Ambion) following the manufacturer's instructions. 3′ RACE PCR was accomplished using a degenerate primer 5′-GTGTNWSNWSNTTTGGNGCNGATCAGTTTGATG-3′ designed on conserved regions of di-/tripeptide transporters of the NRT1/PTR family. Subsequently, 5′ ends of the cDNA of HaPTR4, HaPTR5, and HaPTR12 were obtained by 5′ RACE PCR using cDNA-specific primers (outer/inner), respectively: 5′-AACGAAAACCTGGCACATTCTC-3′/5′-CCCTTCTGCACTCTCTCTACTGGATCAG-3′, 5′-CAAACACTTGCCCAATTCTTCTG-3′/5′-TTGGACTTGCATTCCTCTGGATGTC-3′, 5′-AAACACCTGCCCAACTCTTCTG-3′/5′-AAGAGCTCCTAGATTTTATTTCCTCTGGATG-3′.
Full-length cDNAs were amplified by using specific primers designed against regions before the ATG and after the stop codon of each gene: HaPTR4, 5′-CGACCAATCCACCCAGATCTTTAG-3′ and 5′-AGTGGTAACACATCCTACTTGGGAGATCC-3′; HaPTR5, 5′-AGCTTTAATATAATTTGCAGGAGTGGTGATC-3′ and 5′-CCGGGTAAATGCCCATATAAAGGC-3′; HaPTR12, 5′-CTTCAGCGGAAGAAGGAAGAAAATTG-3′ and 5′-TTCTGTGCAATCTCCACAATAAAATGG-3′, respectively. High Fidelity polymerase (Roche) was used in all PCRs to ensure high accuracy. Amplified full-length cDNAs were cloned into pGEM-T Easy vector. Sequencing data of three independent PCR products were analysed and organized by the ContigExpress program from Vector NTI Suite 6.0 (Informax Inc., North Bethesda, MD, USA).
RNA expression analysis
For quantitative real-time-PCR, total RNA from non-cluster roots, cluster roots, and leaves was converted to cDNA using SuperScript™ III Reverse Transcriptase (Invitrogen, La Jolla, CA, USA) following the manufacturer's instructions. A control reaction (absence of reverse transcriptase) was used to check for residual genomic DNA. Primers used for HaPTR4, 5, and 12 were, respectively: 5′-CATCGACCAGAGGCTGTTCAC-3′, 5′-GCAGTCATTGATAGAATTGATATGAAGAG-3′; 5′-CAATCCTCCACATTTTTCACTAAGC-3′, 5′-TTTGTAAGGAGGCAGGTGGTATTT-3′, 5′-GGAGCCACCATGGAGAGAACTA-3′, 5′-TGAAAAGAACTATGGAGAGGTTGATAAA-3′. For quantification of Hakea 18S small subunit nuclear rRNA, primers 5′-CGGCGGCCCTTGAAA-3′ and 5′-CATCGACCAGAGGCTGTTCAC-3′ were used. 18S rRNA was the internal control of transporter expression after demonstrating that its expression was similar in all tissues (data not shown). Quantitative real-time PCR was performed using the 7900 HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR® Green PCR Master Mix (Applied Biosystems). Ten minutes of enzyme activation was used followed by 45 cycles of 15 s at 95 °C, and 15 s at 60 °C. Elongation was not required because PCR products were very short (∼100 bp) and amplification was completed before the denaturation step. After amplification, melting curve analysis was performed to verify the formed product. The measured Ct values were converted to relative copy numbers using the ΔCt method. The values were normalized by comparing them with 18S rRNA.
HaPTR constructs
PCR fragments corresponding to the open reading frame (ORF) of HaPTR4, HaPTR5, and HaPTR12 cDNA were amplified using primers with restriction sites for all vectors used, i.e. for complementation of Saccharomyces cerevisiae mutants, transient protoplast expression, and cRNA synthesis (Xenopus oocyte expression). The following primers were used: HaPTR4, 5′-GGCCCGGGCTAGCATGGGTTCTCTGGAGGAGAGATTGCT-3′, 5′-GGGCTAGCTCGAGCTAGGATCCAGATGCTTTCTTGCATTTGTACCTTGC-3′; HaPTR5, 5′-GGCCATGGGGCCCGGGCTAGCATGGAAGCTCCCTTGTTAAGTGACACTG-3′, 5′-CCGCATGCTCGAGCTAGGATCCCATTATATTTGCCCTATTATATACATAGGATTTTGAG-3′; HaPTR12, 5′-GGCCCGGGCTAGCATGACTTCATCAGCAAAAATGGAGGAG-3′, 5′-CCCTGCAGTTAGGATCCTGTATTATATTCCTTGTTATATACATATGTTTTTGCAAAATAC-3′. Each amplified HaPTR ORF was cloned into the pGEM-T Easy vector.
For expression in yeast, HaPTR4, HaPTR5, and HaPTR12 were excised from the pGEM-T Easy vector and cloned into the XmaI/XhoI, XmaI/XhoI, and XmaI/PstI sites of yeast expression vector pDR196 (Rentsch et al., 1995). For localization of fusion proteins of HaPTR with green fluorescent protein (GFP) in protoplasts, the ORFs of HaPTR4, HaPTR5, and HaPTR12 cDNAs were isolated from the pGEM-T Easy vector and cloned into pUC18-spGFP6 (XmaI/BamHI) and pUC18-GFP5Tsp (NheI/NheI, NheI/SphI and NheI/PstI) for C- and N-terminal fusion proteins, respectively (M Suter-Grotemeyer and D Rentsch, unpublished data). For expression of HaPTRs in Xenopus oocytes, HaPTR4, HaPTR5, and HaPTR12 were excised from the vector and cloned into the oocyte expression vector pBF1 (Baukrowitz et al., 1999) at XmaI/XhoI, XmaI/XhoI, and XmaI/PstI sites, respectively.
Transient expression in protoplasts
Tobacco (Nicotiana tabacum cv. SRI) protoplasts were isolated and transformed (Di Sansebastiano et al. 1998). Images were obtained after 16–28 h with a confocal laser microscope (DMR, Leica Microsystems, Wetzlar, Germany; TCS 4D operating system). GFP and chlorophyll epifluorescence were detected with the filter set for fluorescein isothiocyanate and trimethylrhodamine isothiocyanate, respectively. Digital images were pseudocoloured as red or green images using Photoshop 7.0 (Adobe Systems, Mountain View, CA, USA) in correspondence to the real green or red colour.
Yeast transformation and complementation
Functional expression of HaPTR genes was performed by complementation of S. cerevisiae strain LR2 (MATαhip1-614 his4-401 can1 ino1 ura3-52 ptr2Δ::hisG) (Rentsch et al., 1995), deficient in di- and tripeptide transport. The yeast strain was transformed (Dohmen et al. 1991) and transformants were selected on synthetic complete medium (SC) containing 20 mM histidine. To test for peptide transport activity, transformants were selected on SC medium containing 1 mM His-Ala as the sole source of histidine. Alternatively, minimal medium containing 200 μM histidine, 20 μg ml−1 inositol, and 1 g l−1 of a peptide as the sole N source was used. cDNAs of the HaPTR genes were also expressed in S. cerevisiae strain BY4730 (MATα leu2Δ0 met15Δ0 ura3Δ0) (EUROSCARF, Germany). Peptide transport was analysed by examining the ability of leucine-containing peptides to support growth. Growth assays were carried out as described previously with slight modifications (Hauser et al., 2000): yeast transformants were selected on proline medium containing 1.7 g l−1 yeast N base without amino acid and ammonium sulphate, 20 g l−1 glucose, 1 g l−1 proline (as a nitrogen source), 191 μM methionine, supplemented with 100–200 μM leucine-containing peptides (LLLL, KLLHG, or SFLLRN).
GenBank accession numbers
Sequences have been submitted to GenBank under the accession numbers EF608213 (HaPTR4), EF608214 (HaPTR5), and EF608215 (HaPTR12).
Statistical analysis
Data were analysed using STATISTICA (Statsoft, Tulsa, OK, USA). Significant differences between treatments were determined by analysis of variance (ANOVA) followed by least significant difference (LSD) post hoc test.
Results
N form and concentration affect biomass allocation and cluster root formation
To assess effects of N concentration and form on cluster root formation, plants were grown hydroponically in nutrient solution with 15 μM (low) or 150 μM (adequate) N supplied as NH4NO3, glycine, or protein (BSA). BSA was chosen as the treatment to represent ON of higher molecular mass than glycine, and microbial conversion of BSA in the non-axenic growth conditions was intentional. Plants supplied with 150 μM N produced significantly (P <0.05) more biomass than plants in no-N or 15 μM N treatments. Biomass production with 15 μM or 150 μM N was in the order glycine>NH4NO3>BSA (Table 1). Plants grown with 150 μM glycine had the lowest allocation of biomass to roots (Table 1).
Table 1.
Dry weight and number of cluster roots of hydroponically grown Hakea actites
| N source | Concentration (μM) | Dry weight |
No. of cluster roots per pot |
|||||||||
| Shoot (g) | Roots (g) | Cluster roots (mg) | Cluster root (% of root dry weight) | Total biomass (g) | 13 weeks | 22 weeks | 23 weeks | 24 weeks | 25 weeks | 26 weeks | ||
| No N | 0 | 1.8±0.14a | 0.75±0.08a | 100±66b | 11.4±6.2 | 2.7±0.2a | 1.3±0.9 | 3.0±0.6 | 3.3±0.9 | 7.7±0.3 | 13.0±2.6 | 17.7±2.7 |
| NH4NO3 | 15 | 6.2±0.27b,c | 2.1±0.4b | 37a | 1.9 | 8.4±0.3a,b | 0 | 0.3* | 0.3 | 1.3 | 1.3 | 1.3 |
| 150 | 47.0±4.0d | 14.9±1.3d | 0 | 0 | 62.1±5.2d | 0 | 0 | 0 | 0 | 0 | 0 | |
| Glycine | 15 | 9.3±1.5c | 3.2±0.39b | 35±28a | 1.1±1.0 | 12.7±1.8b | 0 | 0.3±0.3* | 0.3±0.3 | 0.3±0.3 | 1.3±0.9 | 3.3±2.8 |
| 150 | 60.9±6.5e | 15.1±1.7d | 0 | 0 | 76.1±8.2e | 0 | 0 | 0 | 0 | 0 | 0 | |
| BSA | 15 | 3.9±0.2a,b | 1.7±0.2a,b | 49±27a,b | 2.8±1.7 | 5.8±0.4a,b | 0 | 1±1* | 1±1 | 8.0±1.5 | 13.7±2.3 | 23.3±2.3 |
| 150 | 33.9±3.0f | 9.6±0.2c | 0 | 0 | 43.5±3.2c | 0 | 0 | 0 | 0 | 0 | 0 | |
Plants were grown without N for 3 months and grown for an additional 7 months with different N treatments. Low-N-treated plants commenced producing cluster roots 13 weeks after commencement of hydroponic culture with different N treatments, while plants grown in adequate N did not produce cluster roots. Phosphorus was omitted from the growth media after week 22. The average of three pooled seedlings in three pots ±SD is shown. Means marked with different letters are significantly different (P <0.05, ANOVA LSD post hoc test). The dry weight of plants was determined in week 26.
Cluster roots were produced by plants in one pot only.
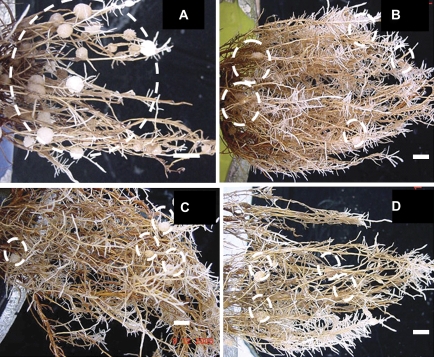
Plants grown with adequate N (150 μM N) did not produce cluster roots (Fig. 1, Table 1). At 13 weeks, no-N plants produced on average 1.3 cluster roots per pot (three plants per pot) (Table 1). At 15 weeks, cluster roots were produced by every plant in the no-N treatment (data not shown). At 22 weeks, three cluster roots were produced on average in each pot in the no-N treatment, while in low N treatments (BSA, glycine, and NH4NO), cluster roots were only produced in one pot per treatment (Table 1). To increase cluster root production, phosphorus was omitted in the nutrient solutions from week 22 in all treatments. In week 24, all plants in the low BSA treatment had cluster roots, and the numbers of cluster roots had increased in no-N and low BSA treatments. Most cluster roots were produced by plants grown in low BSA and no-N treatment 4 weeks after transfer to phosphorus-free medium (Table 1). Cluster roots were smaller in the BSA treatment than in the other treatments.
Fig. 1.
Roots of 9-month-old hydroponically grown Hakea actites initially cultivated without N and then grown for 26 weeks with different N forms. No-nitrogen (A) and a low N concentration (15 μM N) of NH4NO3, glycine, or BSA (B, C, D). Plants grown with a higher N concentration (150 μM N) did not produce cluster roots. Cluster roots are highlighted by white circles. Scale bar = 20 mm. (This figure is available in colour at JXB online.)
Intact dipeptide is taken up into roots
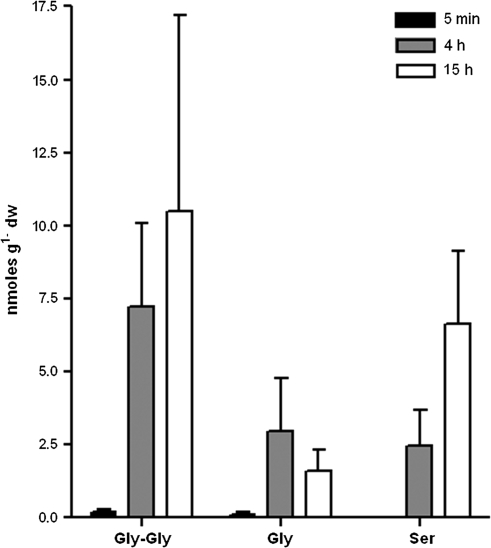
Gly-Gly was offered as the sole N source to axenic H. actites seedlings to determine if intact dipeptides are taken up into the root. Soluble Gly-Gly in roots increased 43- and 63-fold after 4 h and 15 h, respectively (Fig. 2). Glycine increased 46- and 25-fold after 4 h and 15 h, respectively. Serine increased 1520- and 4150-fold after 4 h and 15 h incubation with Gly-Gly, respectively (Fig. 2). Concentrations of 19 amino acids remained constant in the soluble pool of root tissue over the incubation period (data not shown). Low levels of ammonium were detected in the tissue after 15 h incubation (8±3 pmol g−1 dry weight, data not shown).
Fig. 2.
Concentration of soluble Gly-Gly, glycine, and serine in roots of Hakea actites seedlings grown in axenic culture. Roots were analysed 5 min, 4 h, and 15 h after addition of 25 μmol of Gly-Gly to the medium. Bars represent the average of three independent seedlings ±SD.
HaPTR cDNAs belong to subgroups II and III of the PTR/NRT1 transporter family
To investigate the role of peptide transporters in cluster roots, HaPTRs were isolated. Full-length cDNAs were named HaPTR4, HaPTR5, and HaPTR12. Hydropathy analysis of HaPTRs (SVMtm, Yuan et al., 2004; ConPredII, Arai et al., 2004; HMMTOP, Tusnády and Simon, 2001) predicted 10–12 transmembrane domains, which is in agreement with predictions for other PTRs (Steiner et al., 1995).
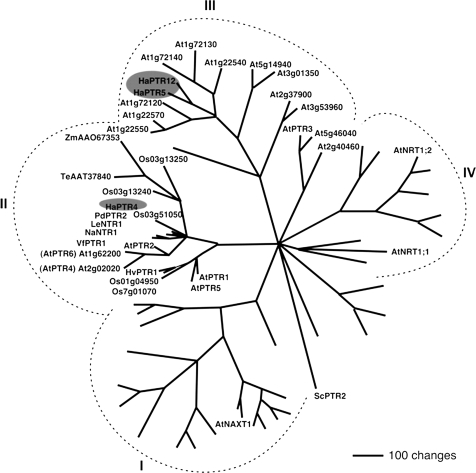
According to the four subgroup nomenclature of the NRT1/PTR family, which contains functionally characterized nitrate, peptide, and carboxylate transporters of Arabidopsis, faba bean (Vicia faba), barley (Hordeum vulgare), and alder (Alnus glutinosa) (Tsay et al., 2007), HaPTR4 belongs to subgroup II, whereas HaPTR5 and HaPTR12 belong to subgroup III (Fig. 3). Phylogenetic analysis revealed that HaPTR4 was identical to the partial cDNA of HaPepT1 (Schmidt et al., 2003) and, with ∼80% identity, highly homologous to (putative) PTRs of almond (Prunus dulcis: PdPTR2, AF213936), tomato (Solanum lycopersicum: LeNTR1, AF016713), faba bean (V. faba: VfPTR1) (Miranda et al., 2003), and Arabidopsis (AtPTR2) (Frommer et al., 1994; Rentsch et al., 1995). HaPTR5 shared the highest similarity with HaPTR12 (72% identity), Arabidopsis At1g22540 (60% identity), and At1g72140 and At1g72130 (51% identity, each). The two latter genes are expressed to a higher level in root than in shoot of Arabidopsis (Tsay et al., 2007), but substrate selectivity has not been determined. Functionally characterized di-/tripeptide transporters of subfamily II and subfamily III are less similar (36–43% identity) to HaPTR5 and HaPTR12 (Miranda et al., 2003; Dietrich et al., 2004; Karim et al., 2007; Tsay et al., 2007).
Fig. 3.
Phylogenetic relationship between HaPTRs and related proteins. The analysis was performed using the aligned protein sequences of the Arabidopsis NRT1/PTR gene family, including proteins from other plant species for proteins of subfamily II (PTR1-like proteins, Rentsch et al., 2007; Tsay et al., 2007; AtPTR3, Karim et al., 2007; VfPTR1, Miranda et al., 2003; NaNTR1, Schulze et al., 1999; PdPTR2, Campalans et al., 2001; LeNTR1, AF016713; HvPTR1, West et al., 1998). Maximum parsimony analysis was performed using PAUP 4.0b10 with all characters unweighted and gaps scored as missing characters (Swofford, 2003). The complete alignment was based on 1409 amino acids; 633 characters were parsimony informative. The yeast peptide transporter ScPTR2 was used as the outgroup.
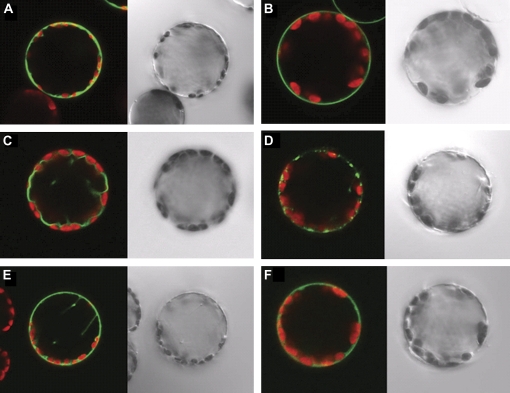
Expression in protoplasts indicates localization of HaPTRs in different membranes
To demonstrate intracellular localization, GFP fusion proteins were transiently expressed in tobacco protoplasts. Free GFP showed characteristic fluorescence in the cytosol (Fig. 4A). AtProT2–GFP fusion protein was the positive control for proteins localized at the plasma membrane (Fig. 4B; Grallath et al., 2005). Protoplasts expressing HaPTR4–GFP or GFP–HaPTR4 showed localization at the tonoplast (Fig. 4C). Protoplasts expressing HaPTR5–GFP or GFP–HaPTR5 showed fluorescence in small vesicular structures (Fig. 4D). To investigate the possibility that HaPTR5 localizes to the mitochondrial membrane, co-localization with a mitochondrion-specific dye, Mitotracker (Molecular Probes™, Eugene, OR, USA), was performed; however, no clear co-localization was observed (data not shown). Protoplasts transformed with the HaPTR12–GFP construct showed localization at the plasma membrane (Fig. 4F) while the GFP–HaPTR12 construct showed fluorescence at the plasma membrane as well as some endomembranes (Fig. 4E), possibly due to incomplete targeting.
Fig. 4.
Localization of fusion proteins of HaPTR with GFP in tobacco (Nicotiana tabacum) protoplasts. Confocal laser scanning microscope pictures (left) and corresponding bright field images (right) of tobacco protoplasts transiently expressing fusion proteins of HaPTRs with GFP or free GFP. Free GFP (A), ProT2–GFP (B), HaPTR4–GFP (C), HaPTR5–GFP (D), GFP–HaPTR12 (E), and HaPTR12–GFP (F). Merged images show GFP fluorescence (green) and chlorophyll fluorescence (red). Protoplast diameter ∼40 μm.
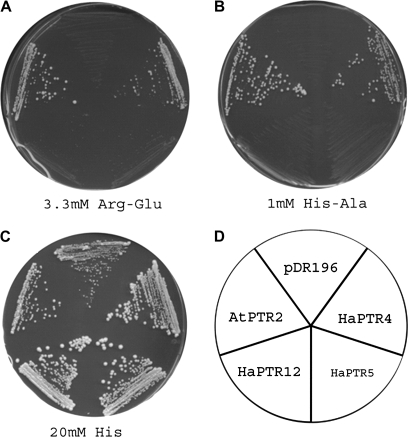
HaPTR4 mediates transport of di- and tripeptides
To determine gene function, cDNAs were expressed in the peptide transport-deficient yeast strain LR2 (Rentsch et al., 1995). This yeast strain is unable to grow on media containing peptides as the sole N source or on media containing histidine in the form of di- or tripeptides. In contrast to HaPTR5 and HaPTR12, which did not support growth, HaPTR4 mediated growth on 1 mM His-Ala as the sole source of histidine, similar to cells expressing the Arabidopsis di- and tripeptide transporter AtPTR2 (Fig. 5). HaPTR4 also mediated growth on the di- and tripeptides Arg-Glu, Ala-Asp, His-Lys, Leu-Leu, Phe-Ala, Val-Ala, Ala-Ala-Lys, and Leu-Leu-Leu as the sole N source (data not shown). Thus, HaPTR4 appears to have a low selectivity towards the amino acid side chain of the tested di- and tripeptides.
Fig. 5.
Complementation of Saccharomyces cerevisiae strain LR2 deficient in the uptake of di- and tripeptides. Growth of LR2 cells expressing the peptide transporters AtPTR2 (positive control), HaPTR4, HaPTR5, and HaPTR12, and the strain transformed with the vector pDR196 (negative control) is shown. (A) MM medium containing 3.3 mM Arg-Glu, (B) SC medium containing 1 mM His-Ala, (C) SC medium containing 20 mM histidine. (D) Schematic representation of the strains used in (A–C).
To determine if HaPTRs transport larger peptides, yeast strain BY4730 was used which is auxotrophic for leucine and methionine and can use peptides as a source for these amino acids provided that the mutation in the yeast oligopeptide transporter gene is complemented by a homologue. None of the HaPTRs mediated growth of the yeast mutant BY4730 when supplied with leucine-containing tetra-, penta-, or hexapeptides in the growth medium. When expressed in Xenopus oocytes, no transport functions of HaPTR5 and 12 were demonstrated, despite testing diverse compounds (data not shown).
Tissue-specific expression of HaPTRs
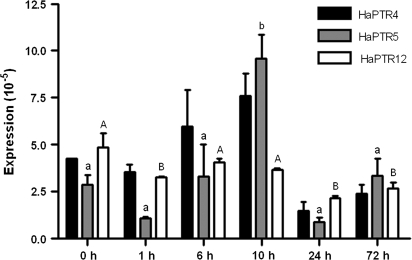
Experiment 3 was designed to examine if expression of HaPTR genes was altered in the short term in response to protein supply. Over 72 h, expression of HaPTR4 was unaltered by protein re-supply, while expression of HaPTR12 decreased after 24 h and 72 h (Fig. 6). Expression of HaPTR5 was significantly higher at 10 h than at all other times (Fig. 6).
Fig. 6.
Time course of HaPTR expression in cluster roots. Expression levels were determined with quantitative real-time PCR and standardized using 18S rRNA. Plants were grown hydroponically in the glasshouse under nutrient starvation for 7 months and resupplied for 1, 6, 19, 24, and 72 h with protein as the N source. Data represent averages and the SD of three independent samples (two replicates where no SD is shown). Significant differences (P <0.05, one-way ANOVA, Tukey's post hoc test) are indicated with lower case (HaPTR5) and upper case letters (HaPTR12).
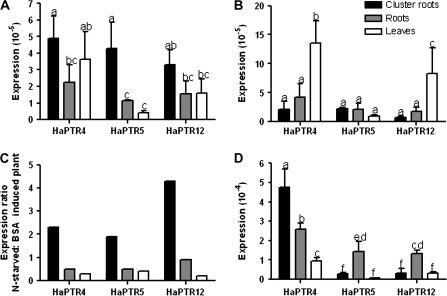
A longer term response of HaPTR expression to protein resupply was determined in plants grown under nutrient starvation condition for 7 months and resupplied for 3 d with protein as the N source (experiment 3). Expression of HaPTR genes in cluster roots under conditions of nutrient starvation was up to 2- to 4-fold higher than after re-supply with protein (Fig. 7A–C). In contrast, expression of HaPTR genes was lower in leaves of nutrient-starved than in N-resupplied plants (Fig. 7A–C). The lower expression of HaPTR genes in plants with higher N supply was in agreement with expression of hydroponically grown plants (experiment 1). Overall, expression of HaPTR genes was higher in plants grown without N or with 15 μM N than in plants grown with 150 μM N, except in cluster roots in the 15 μM glycine treatment (Table 2). Expression of HaPTRs was comparatively low because, following standard practice, expression of target genes is compared with the expression of highly abundant 18S rRNA.
Fig. 7.
Expression of HaPTR genes of Hakea actites in hydroponically grown (A, B) and field-grown plants (D). (C) Expression ratio of HaPTR genes in nutrient-starved (A) versus N-resupplied plants with protein as the N source (B). Expression levels of mRNA were determined with quantitative real-time PCR and standardized against 18 rRNA. Data are averages ±SD of 5–11 replicate samples in field-grown plants and three replicates in hydroponically grown plants. Field-grown plants were collected in heathland habitat. Hydroponically grown plants were cultivated in the glasshouse under nutrient starvation for 7 months and resupplied for 3 d with protein.
Table 2.
Expression ratio of peptide transporter genes HaPTR4, HaPTR5, and HaPTR12 in hydroponically grown Hakea actites
| Tissue | Transporter | Values | No N | 15 μM N concentration |
150 μM N concentration |
||||
| BSA | Glycine | NH4NO3 | BSA | Glycine | NH4NO3 | ||||
| Cluster root | HaPTR4 | Expression (×10−5) | 6.67±4.28 | 4.77±4.05 | 11.75±7.49 | n/a | n/a | n/a | n/a |
| Comparative | 1 | 1.4 | 0.6 | n/a | n/a | n/a | n/a | ||
| HaPTR5 | Expression (×10−5) | 6.36±3.82 | 5.45±4.05 | 12.17±8.13 | n/a | n/a | n/a | n/a | |
| Comparative | 1 | 1.2 | 0.5 | n/a | n/a | n/a | n/a | ||
| HaPTR12 | Expression (x10-5) | 4.65±3.64 | 2.16±0.50 | 14.57±1.02 | n/a | n/a | n/a | n/a | |
| Comparative | 1 | 2.2 | 0.3 | n/a | n/a | n/a | n/a | ||
| Non-cluster root | HaPTR4 | Expression (×10−5) | 15.78±15.43 | 23.10±24.96 | 8.82±9.63 | 6.13±5.01 | 5.36±0.92 | 4.77±5.06 | 2.42±1.14 |
| Comparative | 1 | 0.7 | 1.8 | 2.6 | 2.9 | 3.3 | 6.5 | ||
| HaPTR5 | Expression (×10−5) | 10.03±9.29 | 14.33±15.33 | 4.00±4.02 | 4.44±4.71 | 2.74±0.78 | 0.98±0.15 | 1.28±0.59 | |
| Comparative | 1 | 0.7 | 2.5 | 2.3 | 3.7 | 10.2 | 7.8 | ||
| HaPTR12 | Expression (×10−5) | 7.14±7.28 | 7.26±7.47 | 4.15±5.21 | 2.23±1.99 | 1.58±0.38 | 3.75±5.83 | 0.65±0.27 | |
| Comparative | 1 | 1.0 | 1.7 | 3.2 | 4.5 | 1.9 | 11.0 | ||
| Leaves | HaPTR4 | Expression (×10−5) | 28.32±21.62 | 20.12±15.18 | 24.75±5.43 | 15.07±9.19 | 12.69±9.80 | 4.91±2.45 | 14.72±11.1 |
| Comparative | 1 | 1.4 | 1.1 | 1.9 | 2.2 | 5.8 | 1.9 | ||
| HaPTR5 | Expression (×10−5) | 4.23±4.09 | 1.39±0.95 | 1.66±0.17 | 1.29±0.59 | 0.63±0.42 | 1.03±0.99 | 1.12±0.75 | |
| Comparative | 1 | 3.0 | 2.5 | 3.3 | 6.7 | 4.1 | 3.8 | ||
| HaPTR12 | Expression (×10−5) | 11.03±8.79 | 7.40±5.54 | 11.79±4.53 | 5.11±3.81 | 6.46±5.39 | 2.84±2.16 | 5.75±4.82 | |
| Comparative | 1 | 1.5 | 0.9 | 2.2 | 1.7 | 3.9 | 1.9 | ||
Plants were cultivated without N for 3 months and then grown for 7 months with different N treatments. Data represent averages ±SDs of three independent replicates. Expression values were normalized by comparing them with 18S rRNA, and comparative values are the expression results of plants grown on different N compared with expression of plants grown without N supply. See Table 1 for biomass and number of cluster roots.
n/a, plants produced no or insufficient (15 μM NH4NO3) clusters.
Expression levels of HaPTR genes in field-grown plants in all tissues were considerably higher than in plants from the controlled condition (Fig. 7). HaPTR genes were expressed more strongly in root tissues than in leaves (Fig. 7D). Expression of HaPTR4 was significantly higher in cluster roots than in other tissues, while expression of HaPTR5 and HaPTR12 was highest in roots (Fig. 7D).
Discussion
N form had a strong effect on biomass production and cluster root formation of H. actites. The biomass of plants grown with limiting N supply was highest with glycine as the N source, followed by inorganic N and protein. To enhance cluster root formation, phosphorus was omitted from the nutrient solution in the last 4 weeks of growth. No cluster roots were produced by plants supplied with adequate N irrespective of the N source, while numerous large cluster roots were formed by plants grown without N, accounting for 11% of total root dry weight. Growth-limiting supply with inorganic N or amino acid N resulted in few and small cluster roots (∼1–2% of root dry weight), while plants supplied with protein as the N source produced many small cluster roots (∼3% of root dry weight). The results indicate that under N-limiting growth conditions, N form affects cluster root formation.
Similar observations were made in other species: Myrica gale produced cluster roots more readily when supplied with urea than when supplied with nitrate (Crocker and Schwintzer, 1993), and Gymnostoma papuanum formed cluster roots in the order nitrate>no added N and N2 fixation>ammonium (Racette et al., 1990), while N form did not affect cluster root formation in Myrica cerifera (Louis et al., 1990). This highlights that taxa differ in their responses to cluster root formation, although the plants’ nutritional status and supply of other nutrients, especially phosphorus and iron, also affect cluster root formation (Dinkelaker et al., 1995; Neumann and Martinoia, 2002; Shane and Lambers, 2005).
N supply determines cluster root initiation in several taxa. Formation of cluster roots in Hakea species increased with low N and low phosphorus supply and decreased with high N and low phosphorus supply (Lamont, 2003). Here, omission of phosphorus increased cluster root formation only under low N supply. In contrast, cluster root formation was greater in M. gale and Grevillea robusta supplied with adequate N than in those grown without N (Moore and Keraitis, 1966; Crocker and Schwintzer, 1993). However, these seemingly conflicting findings have to be viewed in an ecological context. Cluster-rooted Proteaceae have the highest abundance on highly weathered soils poor in phosphorus and N, while other cluster-rooted species such as N2-fixing species occur in habitats with higher phosphorus or N status (Lambers et al., 2008). In its heathland habitat, H. actites produces abundant cluster roots in the organic matter-rich upper soil layer, which may indicate that nutrients are accessed from organic compounds, but does not exclude the possibility that nutrients are derived from mineral sources.
Proteolytic enzymes are exuded from cluster roots and non-cluster roots (Paungfoo-Lonhienne et al., 2008). To assess the possible role of cluster roots for accessing organic N forms, membrane transporters were examined because transport of peptides across biomembranes is a likely prerequisite for uptake and metabolism of peptide-based N. While HaPTR4 was most similar to the functionally characterized di- and tripeptide transporters AtPTR1, AtPTR2, and AtPTR5 (60, 73, and 59% identity, respectively), HaPTR5 and HaPTR12 were less similar (41% and 43% identity, respectively) to HaPTR4. HaPTR4 mediated transport of di- and tripeptides, but no transport function could be assigned to HaPTR5 and HaPTR12. Similarly to AtPTR1, AtPTR2, and AtPTR5, HaPTR4 had a low selectivity for di- and tripeptides when expressed in yeast, agreeing with other reports and indicating that di- and tripeptide transporters have no strict side chain specificity (Becker and Naider, 1980; Payne and Smith, 1994). Similarly to AtPTR1 and AtPTR2 (Chiang et al., 2004; Dietrich et al., 2004), HaPTR4 did not facilitate transport of oligopeptides including tetra-, penta-, and hexapeptides. Therefore, the results provide further evidence that genes in the PTR subgroup II encode functional di- and tripeptide transporters in herbaceous and woody taxa.
Examining membrane localization, GFP fusion in tobacco protoplasts showed that HaPTR4 was localized at the tonoplast, similar to AtPTR2, which was identified as a tonoplast protein in proteome analysis (Shimaoka et al., 2004). Vacuoles perform multiple functions, including storage and degradation of proteins (Bassham and Raikhel, 2000; De, 2000), and localization of HaPTR4 at the tonoplast may point to a role for HaPTR4 for transport of protein degradation products into or out of the vacuole (Rentsch et al., 2007). In contrast, HaPTR12–GFP localized to the plasma membrane, whereas GFP–HaPTR12 was detected at the plasma membrane and some endomembranes, indicating that GFP at the N-terminus interferes with efficient targeting to the plasma membrane. HaPTR5 localized to small vesicles; however, the identity of these structures is unknown. It is interesting that these putative transporters localized to a range of membranes, including possibly vesicles. It was recently proposed that endocytosis could be a pathway for transporting protein into roots (Paungfoo-Lonhienne et al., 2008), and further research has to ascertain if the studied transporters participate in this process.
PTRs localized at the plasma membrane include AtPTR1, AtPTR5, and HvPTR1 (Waterworth et al., 2000; Dietrich et al., 2004; Komarova et al., 2008). Using atptr1 knockout and AtPTR5-overexpressing Arabidopsis, Komarova et al. (2008) showed, for the first time, that AtPTRs facilitate transport of di- and tripeptides into roots, confirming that peptides are an N source for non-mycorrhizal plants. It is demonstrated here that intact Gly-Gly was taken up by roots. Although the experiment does not allow differentiation between glycine originating from Gly-Gly cleaved prior to uptake into the root and glycine generated from Gly-Gly in planta, H. actites rapidly metabolizes glycine to serine when supplied with glycine as the sole N source (Schmidt and Stewart, 1999). There is now good evidence that dipeptides are taken up into roots intact, and the ecological relevance of peptides as N sources has now to be established.
To investigate a possible function of the HaPTRs, tissue-specific expression was studied in H. actites seedlings grown in heathland and controlled conditions. Expression of HaPTR genes was generally higher in cluster roots and non-cluster roots than in leaves, indicating a role for HaPTRs for nutrient acquisition and/or root function. In cluster roots and non-cluster roots of field-grown plants, expression of HaPTR4 was 5- and 3-fold higher than in leaves, respectively, while expression of the two uncharacterized transporters HaPTR5 and HaPTR12 was 25- and 4-fold higher in non-cluster roots than in leaves. Starved of nutrients, cluster roots had mostly higher expression of HaPTR4, HaPTR12, and HaPTR5 relative to leaves. Expression of HaPTR4 and HaPTR12 decreased after 6 h incubation with protein as the N source, similar to the expression pattern of the di- and tripeptide transporter HcPTR2A of Hebeloma cylindrosporum (Benjdia et al., 2006). In contrast to HcPTR2A which had higher expression with inorganic N sources (Benjdia et al., 2006), HaPTR genes were expressed more strongly in the presence of ON sources. Considering the expression of HaPTR genes at different stages of cluster root development, there is evidence that expression of HaPTR4 increases throughout cluster root development (Schmidt et al., 2003). This observation was supported by initial results of quantitative real-time PCR, and transcript levels and tissue-specific localization of HaPTRs have to be ascertained, for example with mRNA in situ localization in cluster roots throughout development.
Overall, the studied HaPTR genes were expressed to a greater extent in cluster roots than in other tissues under conditions of nutrient or N starvation. Similarly, expression of the di- and tripeptide transporter HcPTR2A of H. cylindrosporum (Benjdia et al., 2006), and ammonium transporters AtAMT1;1, AtAMT1;3, and LeAMT1.1 (Lauter et al., 1996; Gazzarrini et al., 1999; von Wirén et al., 2000) increased in response to N deficiency. It was argued that up-regulation of N transporters increases N uptake from the soil under conditions of low N availability. In contrast, the PTR transporter VfPTR1 had lower transcript levels in roots when Leu-Leu or glutamine was present (Miranda et al., 2003), while amino acids down-regulated HvPTR1 protein activity at the post-translational level (Waterworth et al., 2005).
In summary, it is proposed that H. actites is a useful model for studying ON acquisition. While only HaPTR4 could be fully characterized, the higher expression of the studied HaPTRs in cluster roots and non-cluster roots compared with leaves, and the strong effect of N levels, form, and concentration on expression levels indicate that the studied HaPTRs are involved in root processes.
Acknowledgments
We are grateful to the following people: Bob Simpson (quantitative real-time-PCR), Marianne Suter-Grotemeyer (yeast complementation), Andreas Meyer and Christophe Gumy (electrophysiology), Nataliya Komarova (phylogenetic tree construction), and Nicole Robinson (comments on the manuscript). Thanks to the ARC Centre of Excellence for Integrative Legume Research access to research facilities. This research was supported by an Australian Research Council Discovery Grant DP0452559 (to SS), the Swiss National Science Foundation SNF3100A0-107507 (to DR), and the Centre of Excellence for Integrative Legume Research (to TGAL).
Glossary
Abbreviations
- N
nitrogen
- ON
organic nitrogen
- PTR
peptide transporter
References
- Arai M, Mitsuke H, Ikeda M, Xia J-X, Kikuchi T, Satake M, Shimizu T. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Research. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV. Unique features of the plant vacuolar sorting machinery. Current Opinion in Cell Biology. 2000;12:491–495. doi: 10.1016/s0955-0674(00)00121-6. [DOI] [PubMed] [Google Scholar]
- Baukrowitz T, Tucker SJ, Schulte U, Benndorf K, Ruppersberg JP, Fakler B. Inward rectification in K-ATP channels: a pH switch in the pore. EMBO Journal. 1999;18:847–853. doi: 10.1093/emboj/18.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JM, Naider F. Transport and utilization of peptides by yeast. In: Payne JW, editor. Microorganisms and nitrogen sources. New York: John Wiley and Sons; 1980. pp. 257–279. [Google Scholar]
- Benjdia M, Rikirsch E, Muller T, Morel M, Corratge C, Zimmermann S, Chalot M, Frommer WB, Wipf D. Peptide uptake in the ectomycorrhizal fungus Hebeloma cylindrosporum: characterization of two di- and tripeptide transporters (HcPTR2A and B) New Phytologist. 2006;170:401–410. doi: 10.1111/j.1469-8137.2006.01672.x. [DOI] [PubMed] [Google Scholar]
- Campalans A, Pages M, Messeguer R. Identification of differentially expressed genes by the cDNA-AFLP technique during dehydration of almond (Prunus amygdalus) Tree Physiology. 2001;21:633–643. doi: 10.1093/treephys/21.10.633. [DOI] [PubMed] [Google Scholar]
- Chiang C-S, Stacey G, Tsay Y-F. Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. Journal of Biological Chemistry. 2004;279:30150–30157. doi: 10.1074/jbc.M405192200. [DOI] [PubMed] [Google Scholar]
- Crocker LJ, Schwintzer CR. Factors affecting formation of cluster roots in Myrica gale seedlings in water culture. Plant and Soil. 1993;152:287–298. [Google Scholar]
- De DN. Plant cell vacuoles: an introduction. Collingwood, Victoria, Australia: CSIRO Publishing; 2000. [Google Scholar]
- Dietrich D, Hammes U, Thor K, Suter-Grotemeyer M, Fluckiger R, Slusarenko AJ, Ward JM, Rentsch D. AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. The Plant Journal. 2004;40:488–499. doi: 10.1111/j.1365-313X.2004.02224.x. [DOI] [PubMed] [Google Scholar]
- Dinkelaker B, Hengeler C, Marschner H. Distribution and function of proteoid roots and other root clusters. Botanica Acta. 1995;108:183–200. [Google Scholar]
- Di Sansebastiano GP, Paris N, Marc-Martin S, Neuhaus JM. Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. The Plant Journal. 1998;15:449–457. doi: 10.1046/j.1365-313x.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AW, Honer CB, Hollenberg CP. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Rentsch D. Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Letters. 1994;347:185–189. doi: 10.1016/0014-5793(94)00533-8. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay T, Gojon A, Ninnemann O, Frommer WB, von Wirén N. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into arabidopsis roots. The Plant Cell. 1999;11:937–947. doi: 10.1105/tpc.11.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallath S, Weimar T, Meyer A, Gumy C, Suter-Grotemeyer M, Neuhaus J-M, Rentsch D. The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiology. 2005;137:117–126. doi: 10.1104/pp.104.055079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M, Donhardt AM, Barnes D, Naider F, Becker JM. Enkephalins are transported by a novel eukaryotic peptide uptake system. Journal of Biological Chemistry. 2000;275:3037–3041. doi: 10.1074/jbc.275.5.3037. [DOI] [PubMed] [Google Scholar]
- Karim S, Holmström K-O, Mandal A, Dahl P, Hohmann S, Brader GN, Palva E, Pirhonen M. AtPTR3, a wound-induced peptide transporter needed for defence against virulent bacterial pathogens in Arabidopsis. Planta. 2007;225:1431–1445. doi: 10.1007/s00425-006-0451-5. [DOI] [PubMed] [Google Scholar]
- Komarova NY, Thor K, Gubler A, Meier S, Dietrich D, Weichert A, Suter Grotemeyer M, Tegeder M, Rentsch D. AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiology. 2008;148:856–869. doi: 10.1104/pp.108.123844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Raven JA, Shaver GR, Smith SE. Plant nutrient-acquisition strategies change with soil age. Trends in Ecology and Evolution. 2008;23:95–103. doi: 10.1016/j.tree.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Lamont BB. Structure, ecology and physiology of root clusters—a review. Plant and Soil. 2003;248:1–19. [Google Scholar]
- Lauter FR, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB. Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proceedings of the National Academy of Sciences, USA. 1996;93:8139–8144. doi: 10.1073/pnas.93.15.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis I, Racette S, Torrey J. Occurrence of cluster roots on Myrica cerifera L. (Myricaceae) in water culture in relation to phosphorus nutrition. New Phytologist. 1990;115:311–317. doi: 10.1111/j.1469-8137.1990.tb00457.x. [DOI] [PubMed] [Google Scholar]
- Mason MG, Schmidt S. Rapid isolation of total RNA and genomic DNA from Hakea actities. Functional Plant Biology. 2002;29:1013–1016. doi: 10.1071/PP01151. [DOI] [PubMed] [Google Scholar]
- Matkin OA, Chandler PA. The U.C.-type soil mixes. In: Baker K, editor. The UC system for producing healthy container-grown plants through the use of clean soil, clean stock and sanitation, manual 23. Berkeley, CA: University of California, Division of Agricultural Sciences, Agricultural Experiment Station, Extension Services; 1957. pp. 68–85. [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Dietrich D, Rentsch D, Weber H, Wobus U. Peptide and amino acid transporters are differentially regulated during seed development and germination in Faba bean. Plant Physiology. 2003;132:1950–1960. doi: 10.1104/pp.103.024422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C, Keraitis K. Nutrition of Grevillea robusta. Australian Journal of Botany. 1966;14:151–163. [Google Scholar]
- Näsholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. New Phytologist. 2009;182:31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- Neumann G, Martinoia E. Cluster roots—an underground adaptation for survival in extreme environments. Trends in Plant Science. 2002;7:162–167. doi: 10.1016/s1360-1385(02)02241-0. [DOI] [PubMed] [Google Scholar]
- Pate JS, Jeschke WD. Mineral uptake and transport in xylem and phloem of the proteaceous tree Banksia prionotes. In: Barrow NJ, editor. Plant nutrition: from genetic engineering to field practice. Dordrecht: Kluwer Academic Publishers; 1993. pp. 313–316. [Google Scholar]
- Paungfoo-Lonhienne C, Lonhienne TGA, Rentsch D, Robinson N, Christie M, Webb RI, Gamage HK, Carroll BJ, Schenk PM, Schmidt S. Plants can use protein as a nitrogen source without assistance from other organisms. Proceedings of the National Academy of Sciences, USA. 2008;11:4524–4529. doi: 10.1073/pnas.0712078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne JW, Smith MW. Peptide transport by microorganisms. Advances in Microbial Physiology. 1994;36:52–69. doi: 10.1016/s0065-2911(08)60176-9. [DOI] [PubMed] [Google Scholar]
- Racette S, Louis I, Torrey JG. Cluster root formation by Gymnostoma papuanum (Casuarinaceae) in relation to aeration and mineral nutrient availability in water culture. Canadian Journal of Botany-Revue Canadienne De Botanique. 1990;68:2564–2570. [Google Scholar]
- Read DJ. Mycorrhizas in ecosystems. Experientia. 1991;47:376–391. [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Letters. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Letters. 2007;581:2281–2289. doi: 10.1016/j.febslet.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Mason M, Sangtiean T, Stewart GR. Do cluster roots of Hakea actities (Proteaceae) acquire complex organic nitrogen? Plant and Soil. 2003;248:157–165. [Google Scholar]
- Schmidt S, Stewart GR. Waterlogging and fire impacts on nitrogen availability and utilization in a subtropical wet heathland (wallum) Plant, Cell and Environment. 1997;20:1231–1241. [Google Scholar]
- Schmidt S, Stewart GR. Glycine metabolism by plant roots and its occurrence in Australian plant communities. Australian Journal of Plant Physiology. 1999;26:253–264. [Google Scholar]
- Schulze W, Frommer WB, Ward JM. Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. The Plant Journal. 1999;17:637–646. doi: 10.1046/j.1365-313x.1999.00414.x. [DOI] [PubMed] [Google Scholar]
- Shane MW, Lambers H. Cluster roots: a curiosity in context. Plant and Soil. 2005;274:101–125. [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki K-I, Maeshima M, Yokota A, Tomizawa K-I, Mimura T. Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant and Cell Physiology. 2004;45:672–683. doi: 10.1093/pcp/pch099. [DOI] [PubMed] [Google Scholar]
- Steiner HY, Naider F, Becker JM. The PTR family—a new group of peptide transporters. Molecular Microbiology. 1995;16:825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony and other methods, Version 4.0b10. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. Nitrate transporters and peptide transporters. FEBS Letters. 2007;581:2290–2300. doi: 10.1016/j.febslet.2007.04.047. [DOI] [PubMed] [Google Scholar]
- Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- von Wirén N, Lauter FR, Ninnemann O, Gillissen B, Walch-Liu P, Engels C, Jost W, Frommer WB. Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. The Plant Journal. 2000;21:167–175. doi: 10.1046/j.1365-313x.2000.00665.x. [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Ashley MK, West CE, Sunderland PA, Bray CM. A role for phosphorylation in the regulation of the barley scutellar peptide transporter HvPTR1 by amino acids. Journal of Experimental Botany. 2005;56:1545–1552. doi: 10.1093/jxb/eri149. [DOI] [PubMed] [Google Scholar]
- Waterworth WM, West CE, Daws MI, Bray CM. The barley scutellarpeptide transporter: relationship to germination and lost of seed viability. In: Black M, Bradford KJ, Vázquez-Ramos J, editors. Seed biology: advances and applications. Wallingford, UK: CAB International; 2000. pp. 297–308. [Google Scholar]
- West CE, Waterworth WM, Stephens SM, Smith CP, Bray CM. Cloning and functional characterisation of a peptide transporter expressed in the scutellum of barley grain during the early stages of germination. The Plant Journal. 1998;15:221–229. doi: 10.1046/j.1365-313x.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Mattick JS, Teasdale RD. SVMtm: support vector machines to predict transmembrane segments. Journal of Computer Chemistry. 2004;25:632–636. doi: 10.1002/jcc.10411. [DOI] [PubMed] [Google Scholar]