Abstract
CBF transcription factors play central roles in the control of freezing tolerance in plants. The isolation of two additional CBF genes, EguCBF1c and EguCBF1d, from E. gunnii, one of the cold-hardiest Eucalyptus species, is described. While the EguCBF1D protein sequence is very similar to the previously characterized EguCBF1A and EguCBF1B sequences, EguCBF1C is more distinctive, in particular in the AP2-DBD (AP2-DNA binding domain). The expression analysis of the four genes by RT-qPCR reveals that none of them is specific to one stress but they are all preferentially induced by cold, except for the EguCBF1c gene which is more responsive to salt. The calculation of the transcript copy number enables the quantification of constitutive CBF gene expression. This basal level, significant for the four genes, greatly influences the final EguCBF1 transcript level in the cold. A cold shock at 4 °C, as well as a progressive freezing which mimics a natural frost episode, trigger a fast and strong response of the EguCBF1 genes, while growth at acclimating temperatures results in a lower but more durable induction. The differential expression of the four EguCBF1 genes under these cold regimes suggests that there is a complementary regulation. The high accumulation of the CBF transcript, observed in response to the different types of cold conditions, might be a key for the winter survival of this evergreen broad-leaved tree.
Keywords: CBF/DREB1 transcription factor, cold acclimation, Eucalyptus, freezing tolerance, gene expression, RT-qPCR, transcript copy number
Introduction
Freezing temperatures limit the geographical distribution of plants and reduce crop productivity and quality. Plant species differ greatly in their ability to acclimate and survive frost exposure. Eucalyptus, which is native to Australia, is the most widely planted hardwood tree in the world (FAO, 2000; Carbonnier et al., 2004). The expansion of Eucalyptus plantations is nonetheless limited to climatic regions without severe frosts. In fact, this genus is particularly vulnerable to freezing injury due to its opportunistic development which lacks both endodormancy and winterfall of the leaves. On the other hand, these characteristics make this woody plant an interesting model for studying cold tolerance without interference from the dormancy process. Indeed, the most frost-tolerant Eucalyptus species such as E. gunnii, which originated in the Tasmanian mountains, can only withstand freezing temperatures because of their capacity to tolerate the apoplastic occurrence of ice and subsequent cell dehydration. This ability to survive a rapid and severe temperature drop to below zero involves basal freezing tolerance together with the induced tolerance in response to low but non-freezing temperatures, a process known as cold acclimation. While the mechanism of basal tolerance is still poorly understood, most studies focus on the acclimation process and most often centre on herbaceous annuals such as Arabidopsis thaliana. Both transcript profiling analyses and traditional genetic studies indicate that it is a very complex response resulting from global changes in gene expression.
A significant part of these changes were shown to be under the control of the CBF (CRT/DRE-binding factor) pathway (Fowler and Thomashow, 2002; Cook et al., 2004; Gilmour et al., 2004). The CBF transcription factors bind specifically to the C-repeat (CRT)/dehydration responsive element (DRE)/low temperature responsive (LTR) motif present in a number of downstream genes that play important roles in cold acclimation and the development of freezing tolerance (Gilmour et al., 1998; Liu et al., 1998). The CBF pathway is conserved among distantly related species with various cold tolerance levels. In addition to Arabidopsis, CBF genes have been identified in 54 genera: 23 monocotyledons and 31 dicotyledons including 13 woody plants. The CBF expression level increasingly appears to be a crucial factor in the natural variability of freezing tolerance as has been suggested for wheat (Badawi et al., 2007), Arabidopsis (Hannah et al., 2006) or Citrus (Champ et al., 2007). However, the differential capacity of plants to tolerate freezing may also be related to the size of the CBF regulon, as suggested in tomato (Zhang et al., 2004).
The isolation and characterization of the first Eucalyptus CBF genes has previously been reported (El Kayal et al., 2006). The EguCBF1a/b genes have all the major structural characteristics of the Arabidopsis CBF1 gene. They also exhibit some of its standard behaviour such as very fast and strong cold regulation, and a poor response to other abiotic stresses. In contrast, some very specific features have also been observed in Eucalyptus genes, such as the positive effect of dark on EguCBF1 cold regulation and the duration of the cold induction during growth at chilling temperatures. So far, CBF orthologues have been described for three deciduous trees: Populus trichocarpa (Benedict et al., 2006), Prunus avium (Kitashiba et al., 2002), and Betula pendula (Welling and Palva, 2008), as well as for three evergreen species: Eucalyptus globulus (Gamboa et al., 2007), Poncirus trifoliata, and Citrus paradisi (Champ et al., 2007). The EgCBF1 gene isolated from the frost-sensitive E. globulus (Gamboa et al., 2007) is very similar to the EguCBF1b gene from E. gunnii (El Kayal et al., 2006), once again demonstrating the conservation of the CBF pathway between freezing-tolerant and freezing-sensitive species. Although the released coding sequence is very similar, the expression data presented here look very different from the EguCBF1 gene profiles. Does this contrast indicate a correlation between CBF gene regulation and freezing tolerance? The first evidence that the CBF expression pattern plays a role in the genetic variability of cold tolerance in trees was provided by comparing a sensitive Citrus species to the extremely frost-tolerant Poncirus (Champ et al., 2007). Altogether, these data highlight the importance of the CBF pathway in the cold response of woody perennials and demonstrate the interest of expanding CBF studies to these species to understand their unique overwintering capacity.
Among the numerous unanswered questions about CBF function in cold tolerance, is the involvement and role of the different members of the CBF families. For example, four CBF genes were characterized in Arabidopsis (Medina et al., 1999; Haake et al., 2002) and up to 25 are believed to be present in the wheat genome (Badawi et al., 2007). Are the different family members functionally redundant? Recent papers show either differences in transactivation of target genes as reported for two groups of rapeseed CBF genes (Zhao et al., 2006), or in cold expression profiles, as observed for perennial ryegrass (Tamura and Yamada, 2007), barley (Skinner et al., 2005), grape (Xiao et al., 2008) or Chinese cabbage (Zhang et al., 2006).
This paper presents a comparative study of the structure of the four CBF genes isolated from E. gunnii with particular attention to the AP2-DBD and the transactivating C-terminal domain. In addition to the quantitative analysis of cold induction, the global transcript level including the constitutive expression was calculated for each EguCBF1 gene. Stress response specificity and regulation in a range of cold conditions very close to natural cold occurrence are also analysed and the results are discussed in terms of complementarity/redundancy.
Materials and methods
Isolation and identification of new Eucalyptus gunnii CBF genes
In the screening of a 11 303 EST collection from E. gunnii cold-acclimated leaves (Keller et al., 2009), a partial new EguCBF1 gene was identified and designated EguCBF1c. To isolate additional CBF sequences from E. gunnii cell-suspension culture, genomic DNA, degenerated primers were designed in the conserved AP2/EREBP region of CBF/DREB1 sequences (see Supplementary Table S1 at JXB online). The resulting PCR product, cloned in pGEM-T Easy vector (Promega, France), was identified as a new CBF-like gene using BLAST analysis and designated EguCBF1d. As previously described (El Kayal et al., 2006), the extension of the upstream sequences of EguCBF1c/d genes was carried out using specific primers (see Supplementary Table S1 at JXB online).
Sequence analysis of EguCBF1 proteins
The comparison of 32 CBF proteins from 12 chosen dicotyledons was performed using CLUSTAL-W to align the AP2-DBD and flanking regions. A phylogenetic tree was generated using MEGA Version 3.1 Neighbor–Joining and Minimum Evolution methodologies on 1000 bootstrap replications.
Multiple amino acid sequence alignments of AP2-DBD of Arabidopsis, Populus, Vitis, and Eucalyptus were carried out with CLUSTAL-X (Thompson et al., 1997) and displayed with ESPript (Gouet et al., 1999).
The analysis of the Hydrophobic Cluster, based on the results from Arabidopsis (Wang et al., 2005; Badawi et al., 2007), was performed using CLUSTAL-W on aligned CBF sequences from Populus, Arabidopsis, and Eucalyptus. In addition, the pI of the acidic domain was calculated on the acid region starting after the DSAWR until the C-terminal end of the protein sequence with Biology Work Bench (http://workbench.sdx.edu/CGI/BW.cgi).
Three-dimensional structure modelling
The molecular modelling of the AP2-DBD of EguCBF1A/B/C/D was carried out on a Silicon Graphics O2 R10000 workstation, using the programs InsightII, Homology, and Discover3 (Accelrys, San Diego, CA, USA). The atomic coordinates of the AtERF1-DNA Binding Domain (RCSB Protein Data Bank code 1GCC) (Allen et al., 1998) were used as a template to build the three-dimensional model of the EguCBF1 AP2-DBD. Steric conflicts were corrected during the model-building procedure using the rotamer library (Ponder and Richards, 1987) and the search algorithm implemented in the Homology program (Mas et al., 1992) to maintain proper side-chain orientation. Energy minimization of the final models was carried out by 50 cycles of steepest descent using Discover3. PROCHECK (Laskowski et al., 1996) was used to assess the geometric quality of the three-dimensional models. All the residues of the EguCBF1 AP2-DBD were correctly assigned to the most suitable regions of the Ramachandran plot. Cartoons were drawn with PyMOL (WL DeLano, DeLano Scientific, Palo Alto, CA, USA. http://www.pymol.org).
Plant material
The Eucalyptus gunnii cell-suspension culture (line 665 from a plant breeding programme managed by AFOCEL) maintained as previously described by Teulières et al. (1989), was used to isolate the genes. Plantlets from the cold-tolerant hybrid Eucalyptus gunnii×Eucalyptus dalrympleana (‘E. gundal’, line 208) were provided by TEMBEC R&D KRAFT as material for gene expression studies. The 3-year-old plantlets were grown in controlled-environment chambers at 25/22 °C day/night, with a long-day photoperiod (16 h light=115 μmol m−2 s−1 supplied by Lumilux Daylight 58 W Osram). In addition, leaves from adult trees (about 10-years-old) of the same genotype, grown in an experimental field, were harvested in summer (at about 22 °C).
Abiotic stress treatments and ABA application on leaf discs
Ten leaf discs of fully expanded leaves from E. gundal line 208 were used for each experiment. Leaf discs were exposed for 2 h to cold (4 °C), heat (50 °C), NaCl (200 mM) or ABA (100 μM). In order to subtract gene expression due to wounding, leaf discs were incubated for similar periods in distilled water and room temperature as a control for cold and salt treatments, or in dimethylsulphoxide (DMSO) as a control for the ABA application. For drought stress, leaf discs on Whatman paper were incubated at 22 °C. After 2 h, the loss of fresh weight was about 30%. As a control, leaf discs were put on Whatman paper imbibed with water. The wounding effect was evaluated by comparing the control leaf discs from both cold and salt treatment to control whole leaves. For gene expression studies, the total RNA was extracted from the pool of the tested discs or whole leaves. The experiments were repeated three times.
Cold treatments on plantlets
The time-course of the EguCBF1 transcript accumulation was studied during a 24 h cold shock (direct transfer from 22 °C to 4 °C) in the dark, and during a cold acclimation programme as previously described by El Kayal et al. (2006). Briefly, for cold acclimation, three plantlets cultivated at 25/22 °C day/night were used as a control and then transferred to short-day conditions (12 h of reduced light intensity) and chilling treatment for 4 d at 12/8 °C day/night, then for 6 d at 4 °C.
To imitate the natural occurrence of frost, a progressive and controlled decrease of temperature to frost level was applied to detached leaves. Four leaves isolated from two plantlets were transferred into 20 ml of water before exposure to a cooling programme consisting of two steps: first, from 22 °C to 4 °C at a speed of –1 °C h−1, followed by a cooling from 4 °C to –8 °C at a speed of –2 °C h−1 (at –1 °C ice was added). At 4 °C, 0 °C, –4 °C, –6 °C, and –8 °C the leaves were collected for RNA extraction before expression analysis. The experiments were repeated three times.
Transcript quantification using RT-qPCR (reverse transcription-quantitative PCR)
The total RNA was extracted from E. gundal leaves using the SV Total RNA Isolation System (Promega, France). Using SuperScript II and random primers (Invitrogen, France), cDNAs were produced according to the manufacturer's instructions. The EguCBF1 specific primers were designed (see Supplementary Table S2 at JXB online) using the Primer Express software (version 2.0, Applied Biosystems, France). The PCRs were performed in 10 μl of 2× SYBR Green power plus Master mix (Applied Biosystems), using 300 nM of each primer and 240 ng of cDNA for the EguCBF1 genes. Three replicates of each PCR were run through an ABI PRISM 7900HT Sequence Detection System (Applied Biosciences, France) as previously described (El Kayal et al., 2006). Specific primers for 18S RNA were used as the internal control for the normalization of the RNA steady-state level, and the relative changes in gene expression were quantified using the 2–ΔΔCt method (Livak and Schmittgen, 2001). The results of the EguCBF1 relative transcript abundance were presented as a mean value of the three assay replicates compared with the mean of the three control values (leaves from control plants).
To quantify the transcript level, a standard curve (copy number as a function of Ct) was generated using a 10× mass dilution series of each EguCBF1 cDNA fragment (10–108 copies). The absolute copy number was determined by extrapolation of the Ct value for each cDNA sample on the standard curve and was expressed as copy number ng−1 of cDNA.
Results
Isolation and structural characterization of EguCBF1c and EguCBF1d genes
Two new EguCBF1 genes were isolated, one from the cold-acclimated leaf cDNA library available in the laboratory and the other using a genomic PCR-based approach. The corresponding full-length open reading frame (ORF) sequences designated EguCBF1c and EguCBF1d (EU794855, EU794856) encode two putative proteins of 229 and 196 amino acids, respectively. The two genes exhibit all the typical features of CBF genes including the absence of introns, the two signatures bordering the AP2 domain, the putative nuclear localization signal, and a putative acidic activation domain.
The alignment of the CBF amino acid sequences (see Supplementary Fig. S1 at JXB online) showed that the four EguCBF1 genes differ significantly from each other, since EguCBF1C/EguCBF1D are, respectively, 82.5/79.7% similar to EguCBF1A and 79.9/77.4% to EguCBF1B. When compared to the model plants, the four EguCBF1 amino acid sequences are from 59.1% to 66.1% similar to AtCBFs from Arabidopsis and from 59.5% to 74% similar to the PtCBFs from Populus trichocarpa. The closest dicotyledonous protein sequences are CBF1 from Eucalyptus globulus (from 76.5% to 99.1% similarity) and CBF4 from another woody plant, Vitis riparia (from 70.4% to 72.9% similarity).
To analyse the phylogenetic relationships of 12 dicotyledons (including woody plants), the most conserved were considered. The resulting phylogenetic tree (see Supplementary Fig. S2 at JXB online), based on 32 partial CBF protein sequences defines different groups based on similarity. A majority of CBF sequences are grouped by genus or species clusters as for Arabidopsis, Vitis or Populus. However, within the largest multigenic families the phylogenetic tree shows a divergent member for the AP2-DBD (AtCBF4, VvCBF4 or VrCBF4). Similarly, the EguCBF1A, 1B, and 1D from E. gunnii are grouped together with the EgCBF1 from E. globulus, while EguCBF1C is located on a different branch.
To predict the function of these transcription factors, the sequence comparison between Arabidopsis, Populus, and Eucalyptus was focused initially on the AP2-DBD, and, secondly, on the acidic COOH-terminal domain known to be responsible for the target gene transactivation.
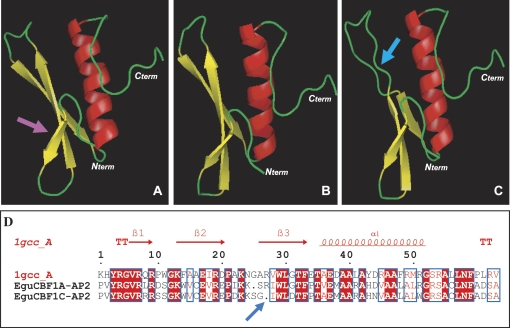
For the AP2 domain molecular modelling, EguCBF1 sequences were compared to AtERF, the only protein belonging to the AP2 family whose crystallographic structure is known (Allen et al., 1998). These data were used for computer modelling the tertiary structure of the Eucalyptus CBFs, since the AP2-DBD of EguCBF1A/B/C/D and AtERF are about 53% identical and 85% similar. Compared to the AtERF-DBD (Fig. 1A), the AP2-DBDs of EguCBF1A, B, and D exhibit the classic ERF fold consisting of three β-sheets connected by loops and a C-terminal α-helix (only the AP2-DBD of EguCBF1A is represented in Fig. 1B). This model predicts DNA binding for these three transcription factors. For EguCBF1C, the AP2-DBD conformation presents a small discrepancy in the third β-sheet (Fig. 1C), probably due to the deletion of arginine at position 27 (in the AP2 sequence) and the change of the residues on both sides (Fig. 1D). Nevertheless, since the greatest interactions for DNA binding occur at the second β-sheet (Sun et al., 2008), this small modification should not result in a loss of DNA binding activity. Only the DNA recognition and binding specificity may differ in EguCBF1C compared to the other EguCBF1s.
Fig. 1.
Three-dimensional structure of the AP2 DNA Binding Protein domain. (A) Ribbon diagram of the 1GCC-box binding domain of AtERF. The three β-sheets (yellow) and the α-helix (red) are connected by turns and loops (green). The pink arrow shows the area of the protein that is the most important for DNA interaction. (B) Ribbon diagram of the modelled AP2-DBD domain of EguCBF1A. (C) Ribbon diagram of the modelled AP2-DBD domain of EguCBF1C. The blue arrow indicates the main conformational modification. (D) AP2 sequence alignment of 1GCC (AtERF), EguCBF1A, and EguCBF1C. Identical residues are in red boxes and homologous residues are in colourless boxes. The deletion of the amino acid at position 27 of EguCBF1C is indicated by a blue arrow.
The pI values of the C-terminal acidic domain, of about 3.44 (EguCBF1C), 3.65 (EguCBF1D), 3.76 (EguCBF1B), and 3.91 (EguCBF1A), proved the presence of the characteristic acidic domain in these CBF sequences. The sequences resulting from the Hydrophobic Cluster Analysis which are known to be involved in α-helix formation in the COOH-terminal region were also fully conserved between Arabidopsis, Populus, and Eucalyptus, except for the clusters HC1 and HC5 (data not shown). According to the conserved number and length of the main hydrophobic clusters in the highly acidic C-terminal domain, this putative transactivating region is predicted to be functional in EguCBF1s.
In conclusion, the four genes differ from each other, with EguCBF1c being the most distinctive. Based on computer prediction, the four genes are supposed to be functional.
Regulation of EguCBF1 genes in response to various stimuli
Using the RT-qPCR method, the specificity of the EguCBF1 gene response was first investigated on leaf discs exposed to cold, salt, heat, drought, ABA, or wounding. This simplified system, previously used to study the specificity of the EguCBF1a and EguCBF1b gene response to stress, was found to be representative of EguCBF1 gene expression in whole plants, although the induction levels are much lower. The conditions used to apply the abiotic stresses are the standard treatments usually reported in the literature, such as the 4 °C shock for cold.
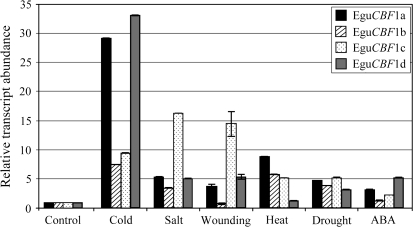
As shown in Fig. 2, the EguCBF1d and EguCBF1a genes were mainly cold responsive (with 33-fold and 29-fold induction, respectively) while EguCBF1b and EguCBF1c genes were less specific to one stress. In fact, the EguCBF1c gene appeared to be more induced by salt (16.5-fold) or wounding (14.5-fold) than by cold (9.3-fold). On the whole, the EguCBF1 genes were moderately regulated by drought or heat and poorly induced by ABA, which suggests that this hormone has a very limited influence on EguCBF1 gene regulation. In conclusion, the EguCBF1 genes are all induced by cold, but not exclusively.
Fig. 2.
Response of EguCBF1 genes to different environmental stimuli. Total RNA was extracted from leaf discs of E. gundal exposed for 2 h to cold (4 °C), NaCl (200 mM), heat (50 °C), drought (22 °C dessication) or exogenous ABA (100 μM). For evaluating the wounding effect, the leaf discs (distilled water, 22 °C) were compared to control whole leaves. Relative abundance of EguCBF1 transcripts was quantified in comparison to the 18S RNA transcript level using RT-qPCR with gene-specific primers (see Supplementary Table S2 at JXB online). The results correspond to the mean value of three assay replicates compared to the mean of the three control values. After normalization of the RNA steady-state level using 18S as an internal control, the EguCBF1 transcript levels of control tissues was used as a calibrator to determine the fold change of transcript abundance during treatment. A representative histogram with standard deviation from three replicates performed for each time point has been represented.
Quantification of EguCBF1 non-induced transcript level (constitutive expression) on plantlets and trees
RT-qPCR quantification has become the best method for the sensitive detection and precise quantification of mRNA. However, the resulting data are most often relative and expressed as induction levels between two situations (here stressed versus unstressed). The relative transcript abundance provides information about gene regulation but cannot indicate an absolute quantity of transcript for a given situation. To obtain these absolute data, the Ct values were converted into a EguCBF1 transcript copy number ng−1 of cDNA by using a standard curve (copy number as a function of Ct value, see the Materials and methods for details).
This calculation method makes it possible to estimate the constitutive expression of the EguCBF1 genes for plants in standard conditions. It also provides the EguCBF1 global transcript level corresponding to the constitutive+induced expression for plantlets after cold exposure. The resulting global copy number ng−1 of cDNA for each EguCBF1 gene predicts their involvement in the cold response. Finally, this calculation method allows the four individual EguCBF1 expression levels to be added and, therefore, provides an assessment of the total amount of EguCBF1 transcripts in a given situation.
The mean constitutive EguCBF1 transcript level, based on 14 independent experiments on a set of plantlets, ranged from 0.2 to 2.8 copies ng−1 of cDNA depending on the gene (Table 1). The EguCBF1c and EguCBF1b genes showed higher constitutive expression than the two other members. While the standard error (SE) was quite low for three of the genes, indicating good technical reproducibility, the highest SE value for the EguCBF1c gene was probably related to its high responsiveness to various stresses and, in particular, to wounding. The variation in expression between independent experiments could be due to slight differences in the physiological or phytosanitary state of the tested plantlets, despite their absolutely normal appearance.
Table 1.
Quantification of EguCBF1 constitutive transcript level in plantlets and trees
| Copy number ng−1 of cDNA |
||||
| EguCBF1a | EguCBF1b | EguCBF1c | EguCBF1d | |
| Plantlets | 0.2±0.002 | 1.1±0.003 | 2.8±0.008 | 0.3±0.002 |
| Trees | 0.06 | 0.23 | 0.21 | 0.05 |
In addition, the transcript level of the EguCBF1 genes was measured in summer conditions on adult trees (same genotype as the plantlets) from field trials. The EguCBF1 genes also appeared to be constitutively expressed in the trees (Table 1), but the corresponding transcripts were up to 13.3 times less abundant in the trees compared to the plantlets. Interestingly, the differential behaviour of the genes was similar for the two plant materials, with the EguCBF1c and EguCBF1b genes having the highest constitutive expression.
In conclusion, the EguCBF1 genes are constitutively but differentially expressed in standard conditions in both young and adult plants.
Effect of a cold shock on EguCBF1 gene expression
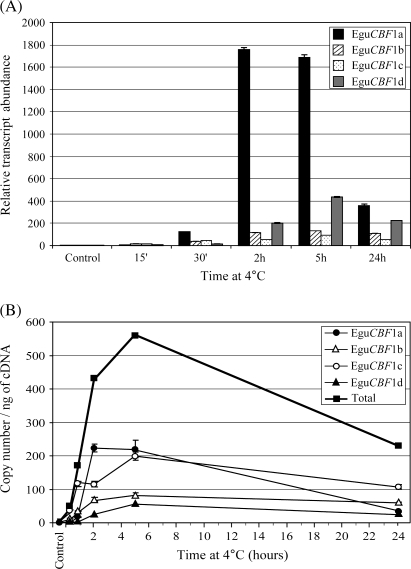
The expression of the four EguCBF1 genes was then quantified in plantlets during a 24 h cold exposure in the dark, after a direct transfer from 22 °C to 4 °C. The four EguCBF1 genes were strongly induced by cold (Fig. 3A) and exhibited quite similar expression profiles with contrasting relative transcript abundances. While the maximum induction was observed between 2 h and 5 h for the EguCBF1a gene (1760-fold and 1690-fold, respectively), there was a less impressive induction peak at 5 h for the EguCBF1d/b/c genes (436-, 131- and 91-fold, respectively). In addition, Fig. 3A shows that the EguCBF1 gene cold response was early and durable: the induction was already detectable at 15 min and remained high at 24 h, but much lower than at 5 h. This profile is in agreement with the data previously published for EguCBF1a/b genes (El Kayal et al., 2006), except for slight differences in induction levels due to the use here of mean control values that were more representative (basal level in Table 1) instead of the specific control value.
Fig. 3.
Time-course of EguCBF1 gene expression at 4 °C in the dark. Total RNA was extracted from a pool of leaves harvested before cold exposure (control), after 15 min, 30 min, 2 h, 5 h, and 24 h. (A) The relative EguCBF1 transcript abundance was determined as described in the legend of Fig. 2. (B) Copy number ng−1 of cDNA of EguCBF1 genes was calculated using a standard curve (see the Materials and methods for details). For transcript copy number ng−1 of cDNA values, closed circles, open triangles, open circles, closed triangles, and closed squares correspond to EguCBF1a, EguCBF1b, EguCBF1c, EguCBF1d genes, and the total amount of the EguCBF1 genes, respectively.
When the raw data were converted into the transcript copy number ng−1 of cDNA (Fig. 3B), the resulting profiles were quite similar to the induction ones (Fig. 3A), with a spectacular increase during the first hours of treatment. At 5 h, the total amount of EguCBF1 transcripts ng−1 of cDNA was mainly made up of EguCBF1a (39%) and EguCBF1c (36%). It is interesting to note that a similar number of EguCBF1 transcripts ng−1 of cDNA was generated after 5 h at 4 °C from both the EguCBF1a gene, which exhibited the lowest constitutive level and the strongest induction, and from the EguCBF1c gene, which presented the strongest basal level and the lowest induction. EguCBF1b/d transcripts accounted for 25% of the total EguCBF1 at 5 h, thus representing a minority of these transcripts.
In conclusion, these data strongly stress the importance of the constitutive expression of EguCBF1 genes and the specific impact of each one on the total EguCBF1 final expression in the cold.
Response of EguCBF1 genes to a progressive freezing exposure
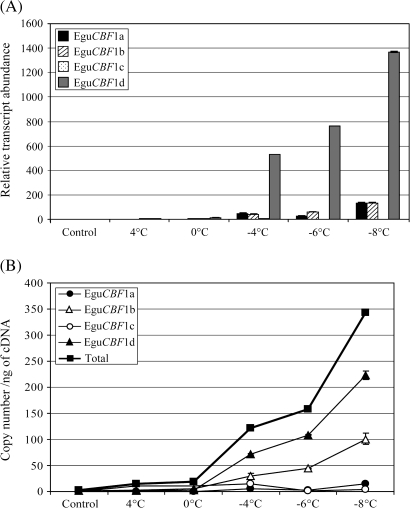
In order to study the influence of the occurrence of frost on EguCBF1 gene regulation, a new cold programme, intended to mimic a natural frost episode, was applied. Whole leaves from plantlets (in demineralized water) were exposed to a controlled cooling programme, consisting of a first step from 22 °C to 4 °C with a speed of –1 °C h−1 followed by freezing at –2 °C h−1 from 4 °C to –8 °C. The EguCBF1 transcript levels were measured at 4 °C, 0 °C, –4 °C, –6 °C, and –8 °C.
During the temperature decrease to freezing, the EguCBF1d gene was found to exhibit a strong, progressive increase in induction up to 1367-fold at –8 °C (Fig. 4A), which was in the same range as the maximum induction measured for the EguCBF1a gene (1760-fold), after a cold shock at 4 °C (Fig. 3A). The induction at 4 °C was found to be much lower following progressive freezing (up to 9-fold, Fig. 4A) than following a cold shock (up to 1760-fold, Fig. 3A). It is difficult to compare the two experiments for a given temperature since harvesting occurred immediately after the transition to 4 °C for the progressive cooling experiment, whereas it occurred after 2 h for the cold shock experiment. Moreover, cold treatment was directly applied to isolated leaves in water for the progressive cooling whereas whole plants were used for the cold shock, preventing a comparison of absolute induction values. In any case, the results presented in Fig. 4A reveal that the four EguCBF1 genes are differentially regulated by this cold treatment as well, with the EguCBF1d gene being the most strongly induced compared to the moderate induction (up to 135-fold at –8 °C) of the EguCBF1a/b genes and the insignificant response of the EguCBF1c gene. The data presented show that EguCBF1 transcripts can be generated at freezing temperatures after a progressive freezing programme, whereas after a cold shock at negative temperatures (El Kayal et al., 2006), the EguCBF1a/b genes were not significantly induced.
Fig. 4.
Response of EguCBF1 genes to exposure to progressive freezing. Four leaves picked from two plantlets were exposed to a cooling programme in two steps: first from 22 °C to 4 °C with a speed of –1 °C h−1, followed by freezing from 4 °C to –8 °C with a speed of –2 °C h−1 (at –1 °C ice was added). During the cooling programme, leaves were collected at 4 °C, 0 °C, –4 °C, –6 °C, and –8 °C for total RNA extraction. Relative transcript abundance (A) and copy number ng−1 of cDNA of EguCBF1 genes (B) were determined using RT-qPCR as described in the legend of Fig. 2 and in the Materials and methods. For transcript copy number ng−1 of cDNA values, closed circles, open triangles, open circles, closed triangles, and closed squares correspond to EguCBF1a, EguCBF1b, EguCBF1c, EguCBF1d genes, and the total amount of the EguCBF1 genes, respectively.
When the total EguCBF1 transcript level was considered during the progressive cold exposure, a very strong and progressive increase of the global transcript copy number ng−1 of cDNA was observed (Fig. 4B), with a maximum of 343 at –8 °C, made up of EguCBF1d (66%), EguCBF1b (29%), EguCBF1a (4%), and EguCBF1c (1%). Despite its low constitutive level, the quantitative significance of the EguCBF1d gene in this cold response was due to its strong induction. EguCBF1b exhibited both an intermediate induction rate and basal transcription level, and thus proved to be more responsive to this progressive freezing than to a shock. By contrast, The EguCBF1a/c genes have a slight impact on the global transcript level because they are poorly regulated in these conditions.
In conclusion, these results suggest that the way frost occurs, through differences in temperatures and the speed of the temperature drop, have a great influence on EguCBF1 gene regulation in a differential way depending on the gene. Conditions of cold treatment affect not only the kinetics and amplitude of the CBF response but also the differential response of the genes. In experimental conditions more representative of environmental cold events, the EguCBF1 genes are strongly and progressively induced, even at freezing temperatures.
Expression of EguCBF1 genes during cold acclimation
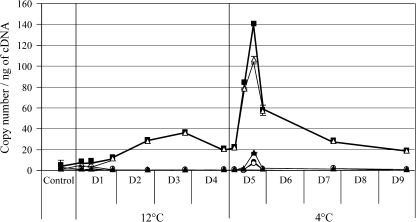
As previously described by El Kayal et al. (2006), plantlets were cold-acclimated with a short-day photoperiod after transfer for 4 d at 12/8 °C day/night and then for 12 d at 4 °C night and day. To avoid any overlap with the data presented in the previous paper (El Kayal et al., 2006), detailed induction results are not provided here. Briefly, the robust induction of the EguCBF1a/b genes (up to 118-fold and 190-fold, respectively) already reported, contrasted with the slight effect of the acclimation programme on EguCBF1c/d regulation, 5-fold and 22-fold, respectively (data not shown). The EguCBF1 total transcription level and the specific amount for each gene all along the acclimation programme are presented in Fig. 5. Interestingly, a fast induction was observed for the EguCBF1b gene after each temperature transition that was much more pronounced at 4 °C than at 12 °C. The corresponding total transcript number ng−1 of cDNA progressively increased over 3 d at 12 °C, in contrast to the stronger and faster accumulation that was observed during the first day at 4 °C. Then, for both temperatures, the total transcript copy number ng−1 of cDNA came back after 4 d to an intermediate level which was still four times higher than the basal level. Throughout the acclimation programme, the EguCBF1 global transcript amount coincided with the EguCBF1b gene expression profile (Fig. 5). In contrast to the EguCBF1a/c genes which were more responsive to cold shock, the EguCBF1b gene appeared to be the ‘acclimation candidate’ since, during this hardening treatment, it was by far the most transcribed.
Fig. 5.
EguCBF1 transcript accumulation during an acclimation programme of plantlets in short-day conditions (SD). Total RNA was extracted from Eucalyptus leaves randomly harvested at different times during the cold programme: 30 min, 2 h after transfer to 12 °C (D1), and 30 min, 2 h, and 5 h after transfer to 4 °C (D5), and at the end of each day (except D6 and D8). The copy number ng−1 of cDNA of EguCBF1 during the plantlet acclimation programme was quantified using RT-qPCR as described in the legend of Fig. 2 and in the Materials and methods. For transcript copy number ng−1 of cDNA values, closed circles, open triangles, open circles, closed triangles, and closed squares correspond to EguCBF1a, EguCBF1b, EguCBF1c, EguCBF1d, and the total amount of the EguCBF1 genes, respectively.
In conclusion, chilling exposure leads to a moderate and durable EguCBF1 gene expression depending on the temperature. The EguCBF1b gene was the most strongly induced in this condition.
Discussion
This paper presents the identification and characterization of two new Eucalyptus CBF genes in addition to the previously described EguCBF1a/b genes (El Kayal et al., 2006). The four genes, all closest to the AtCBF1 gene in the reference Arabidopsis family, are very distinct from each other, with the EguCBF1c gene encoding the most distant protein on the AP2-based phylogenetic tree.
This paper mainly describes the expression of the four Eucalyptus CBF genes, analysed through a series of transcript quantifications during different controlled environmental treatments. The Eucalyptus CBF genes are preferentially cold-regulated with a strong, quick, and durable response, depending on the gene and on the treatment. When the specificity of the EguCBF1 gene response was investigated, none of the four genes described was found to be cold specific. Moreover, the EguCBF1c gene is more induced by salt or wounding than by cold. Overall, the regulation appeared to be ABA-independent. The CBF genes are described in the recent literature to be more or less specific to cold stress depending on the plant species (Gao et al., 2002; Tang et al., 2005; Huang et al., 2007). Sometimes, different profiles are also reported within the same CBF family. For example, in Vitis sp., the CBF4 gene was mostly induced by cold while the CBF1–3 genes showed a better response to drought compared to cold (Xiao et al., 2008).
To date, quantitative data on CBF gene expression are still limited (Badawi et al., 2007; Miura et al., 2007; McKhann et al, 2008; Welling and Palva, 2008). Although unnatural, the cold exposure consisting of a direct shock at 4 °C provides a comparison with commonly reported data on other species. With an induction up to 1760-fold, Eucalyptus CBF1 genes are among the most strongly up-regulated, compared to, for example, birch, which exhibits a maximum CBF induction of 300-fold. Within the Eucalyptus genus, through a semi-quantitative RT-PCR, the EgCBF1 gene from E. globulus was reported to be weakly cold-induced (Gamboa et al., 2007).
As a very common feature in transcription factor genes and, in particular, the CBF genes described to date, the EguCBF1 cold induction was very fast but also particularly durable. At 4 °C, the Eucalyptus CBF response was found to be fast and durable (in particular for EguCBF1a) since it remained significant at 24 h. By contrast, for the cold-sensitive E. globulus, EgCBF1 induction after a shock at 4 °C was reported to be very transient (Gamboa et al., 2007). For other woody plants, CBF gene induction was found to last for long periods, up to 4 d for the Citrus CBF1 gene (Champ et al., 2007), and 5 d for the Vitis CBF3 gene (Xiao et al., 2006). In these cases, as suggested for Eucalyptus species, when the cold-tolerant and cold-sensitive species were compared, the most durable induction of CBF genes was observed in the cold-tolerant species.
In addition to the direct shock from 22 °C to 4 °C, two more natural cold treatments were applied in this study. These treatments roughly represent the most common cold conditions that Eucalyptus has to cope with, not only in temperate winters when progressive freezing may occur, either in midwinter when the trees are cold-acclimated but also in early spring or late autumn without acclimation. The EguCBF1 genes (in particular, EguCBF1b) were durably induced during chilling exposure, during which a succession of expression peaks render this induction significant for at least a 9 d period (El Kayal et al., 2006).
The controlled freezing down to –8 °C led to a specific expression profile with a very strong increase in the induction up to a maximum very close to the maximum induction after a shock at 4 °C. This progressive freezing, much more natural than cold shock, also proved to be an efficient way to induce the EguCBF1 genes, in particular, EguCBF1d. In addition, the strong response to sub-zero temperatures observed for the Eucalyptus CBF1 genes was also reported for Arabidopsis (Zarka et al., 2003) and birch (Welling and Palva, 2008).
These results show that differences in temperature, the temperature itself, and cooling or freezing rates are the three critical factors which control EguCBF1 gene expression. This set of expression data confirms the very complex fine-tuning of the EguCBF1 genes in response to various cold regimes.
Beyond these regulation aspects, this paper reports a quantification of basal CBF transcript levels in the absence of any stress. The measurement of CBF constitutive gene expression in Eucalyptus was achieved through the combination of the sensitive RT-qPCR analysis and the calculation of transcript copy number ng−1 of cDNA, which has, to date, mainly been used in the medical field (Nunan et al., 2004; Bustin et al., 2005) and, recently, for plants (Peng et al., 2008). This calculation also provides the total CBF transcript quantity in a given situation, both for one individual gene and for the pool of isolated members. Constitutive expression does actually exist for the EguCBF1 genes, at different levels according to the gene. It reaches nearly 2.8 copies ng−1 of cDNA for the EguCBF1c gene and was observed in independent reproducible experiments conducted in a phytotron. This expression was also confirmed on adult trees in the field in the absence of cold. According to the recent literature, CBF gene expression in unstressed conditions is observed for several plant species (Tang et al., 2005; Xiao and Nassuth, 2006; Xiao et al., 2008). Morever, the involvement of a CBF basal gene expression in cold tolerance was suggested in Hordeum vulgare when Stockinger and co-workers (Stockinger et al., 2007) compared two recombinant populations differing by the allele at FRH2 (Frost Resistant 2 locus). In Eucalyptus, one may hypothesize that the permanent basal level of CBF gene expression, if the corresponding transcription factors are functional, could help the plants to face constant diverse moderate stresses like drought or low temperatures occurring before or after winter.
Interestingly, the EguCBF1 transcript quantity is globally ten times higher in young plants grown in a phytotron (25/22 °C) than in trees in summer conditions, as was also observed on a set of E. gunnii genotypes cultured in controlled conditions and tested at different ages (Marque, 2008). In addition to a diminution in constitutive EguCBF1 expression, the older plantlets exhibit a lower level of EguCBF1 induction after cold treatment. This attenuation of EguCBF1 response with age would explain the moderate seasonal changes in CBF expression observed when comparing adult trees in the field in summer and winter conditions. The particular importance of the CBF genes in the early stages of plant development was also suggested for Arabidopsis, which exhibits basal CBF gene expression only during the 3 weeks after germination (Novillo et al., 2007). One may hypothesize that cell protection against cold-induced injuries via the CBF response would be essential for young Eucalyptus tissues, more fragile than older ones. For protecting adult plants, additional adaptive mechanisms at the morphological level, such as cuticle thickening, may take over.
The basal expression level observed for EguCBF1 genes in unstressed conditions is apparently essential in determining the extent of the CBF gene response to chilling or freezing temperatures. In fact, this basal expression could be quickly and transiently increased by a more brutal frost episode or more durably in autumn during cold acclimation. Indeed, this permanent, changing EguCBF1 gene expression is likely to be an important adaptive feature of Eucalyptus for coping with long-term cold exposure.
Among the main features of the EguCBF1 gene expression, the differential behaviour of the four genes was obvious in all the analyses presented. The four genes differ in their constitutive expression, stress specificity, type of cold specificity, and/or kinetics of response. Based on the described gene regulation profiles, a predicted specific role for each EguCBF1 factor, if functional, is proposed.
(i) The EguCBF1c gene is the ‘constitutive and non-specific member’. It could be involved in the permanent protection of the cell without stress or in response to diverse stimuli.
(ii) The EguCBF1b gene is the ‘cold acclimation member’. It may participate in the winter basal protection over winter and, secondarily, in the response to a progressive frost.
(iii) The EguCBF1d gene is the ‘natural frost member’. It could help the cell to tolerate frost episodes with or without cold acclimation. It might be involved in protection against diurnal changes of temperature as suggested by expression measurements throughout the day in the field (data not shown).
(iv) The EguCBF1a gene is the ‘cold shock member’, likely to be involved in the reaction to exceptional, very brutal temperature drops.
Of the four profiles, the EguCBF1c gene regulation is the most distinctive, which is not surprising given its phylogenetic distance from the others. Although not always so obvious, structural and/or functional differences within the same family are quite common for CBF genes. The CBF1–3 genes from Vitis sp. exhibit a similar regulation while CBF4 gene expression contrasts in its tissue specificity and kinetics (Xiao et al., 2008). In addition, very strong differences in the level or kinetics of induction have been observed within the CBF families of Triticum sp. (Badawi et al., 2007), Lolium perenne (Tamura and Yamada, 2007), and Brassica napus (Gao et al., 2002). For Arabidopsis, the issue of the functional redundancy of the AtCBF1–3 genes had long been assumed until recent evidence showed that AtCBF2 negatively regulates AtCBF1/3 genes without reciprocity (Novillo et al., 2004). Further, AtCBF1/3 genes seem to be involved only in cold acclimation whereas the AtCBF2 gene could act in both cold acclimation and basal tolerance (Novillo et al., 2007).
The sequence analyses (AP2-DBD and C-terminal domains) suggest functionality of the four EguCBF1 factors. If these proteins accumulate proportionally to transcript abundance, a significant number of transcription factors would be present in any stressful conditions. Nonetheless, differences in binding and/or transactivation efficiency between the four proteins can not be excluded. According to the molecular model, EguCBF1C, which exhibits a modified β-sheet conformation, may, in particular, differ from the other members in its DNA binding specificity. Differential binding preferences have already been demonstrated in vitro for CBF transcription factors from Brassica napus (Gao et al., 2002). Although evidence of EguCBF1 differential gene regulation is provided in this paper, further experiments will be needed to compare the binding and activation ability of the corresponding proteins.
If these transcription factors prove to be efficient, the complementary expression profiles of Eucalyptus CBF1 genes may provide a finely tuned balance of CBF gene responses enabling fitness in an environment subject to various simultaneous or successive stress episodes. The physiology of this tree is mostly directed towards growth and productivity at the expense of adaptive traits such as winter leaf drop, anatomical organ protection or endodormancy, making this genus a special plant system with regard to cold tolerance studies. Herbaceous annual and woody perennial plants share molecular responses to cold, including the CBF pathway (Welling and Palva, 2006). Similarities in CBF regulation mainly concern acclimation to short frost episodes during growth and, to a lesser extent, seasonal cold acclimation. CBF genes appear to be differentially regulated during the endodormancy period associated with overwintering as shown in birch (Welling and Palva, 2008) and poplar (Benedict et al., 2006). That suggests that the perennial-driven evolution of winter dormancy has given rise to specificities in the CBF response. Moreover, when an evergreen Prunus genotype lacking dormancy was compared to the common deciduous and dormant one (Artlip et al., 1997), the higher cold sensitivity observed was associated with a more transient dehydrin transcript accumulation throughout the year, suggesting a distinct CBF control. In the absence of true dormancy, the CBF response of Eucalyptus might exhibit some intermediate features between annual and winter-dormant perennial plants. In particular, E. gunnii is able to tolerate temperatures as low as –18 °C for adult trees, despite its lack of bud insulation and dormancy. The richness and complexity of the CBF response suggested by the present study on the EguCBF1 genes could be a key for reaching this relatively high tolerance. Thanks to its magnitude and flexibility, this response could be more efficient during episodic frosts than in annuals like Arabidopsis. Thanks to the basal expression and the specialization of one paralogue (EguCBF1b), this response could also trigger a longer term seasonal acclimation, quite as efficiently as in dormant trees. The first data on the much more sensitive species, E. globulus, tend to support this hypothesis, as the isolated CBF gene shows weaker and more transient cold induction.
As a general conclusion, this paper presents a set of structure and expression analyses of the EguCBF1 genes. The results suggest a possible involvement of these genes in the permanent protection of Eucalyptus cells against abiotic stresses and particularly frost, participating in basic tolerance, cold acclimation, and a fast adaptive response. This permanent high and versatile CBF gene expression fits in quite well with the opportunistic growth characteristics of Eucalyptus. Is this complexity and efficiency of the Eucalyptus CBF family at a regulatory level also true for the binding and transactivation activities of the corresponding factors? The next experiments investigating the impact of EguCBF1-overexpression on transgenic plants should answer this question.
Supplementary data
The following supplementary data can be found on JXB online.
Supplementary Table S1. List of oligonucleotide PCR primer sequences used for isolation of the EguCBF1 genes.
Supplementary Table S2. List of oligonucleotide sequences used in RT-qPCR.
Supplementary Fig. S1. Multiple alignment of the amino acid sequences of CBF/DREB proteins.
Supplementary Fig. S2. Phylogenetic relationships between partial protein CBF sequences.
Acknowledgments
We thank AFOCEL (FCBA) for the supply of the Eucalyptus cuttings and Toulouse Genotoul for genomics facilities. Research support, salaries, and grants were provided by the French Ministry of Research and Technology (MNRT), ANR-06-ERAPG-0008, the ‘Midi Pyrénées French Council’, and TEMBEC SA R&D KRAFT (St Gaudens, France). We thank Victoria McBride and Heather McKhann for language proofreading.
Glossary
Abbreviations
- ABA
abscisic acid
- AFOCEL
Association Forêt Cellulose
- AP2-DBD
AP2-DNA binding domain
- CBF/DREB1
C-repeat binding factor/dehydration-responsive element binding1
- DBD
DNA binding domain
- FCBA
Forêt Cellulose Bois Ameublement
- ORF
open reading frame
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
References
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO Journal. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artlip TS, Callahan AN, Basett CL, Wisniewski ME. Seasonal expression of a dehydrin gene in sibling deciduous and evergreen genotypes of peach (Prunus persica [L.] Batsch) Plant Molecular Biology. 1997;33:61–70. doi: 10.1023/a:1005787909506. [DOI] [PubMed] [Google Scholar]
- Badawi M, Daniluk J, Boucho BMH, Sarhan F. The CBF gene family in hexaploïd wheat and its relationship to the phylogenetic complexity of cereals CBFs. Molecular Genetics and Genomics. 2007;277:533–554. doi: 10.1007/s00438-006-0206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict C, Skinner JS, Meng R, Chang Y, Bhalerao R, Huner NPA, Finn CE, Chen THH, Hurry V. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant, Cell and Environment. 2006;29:1259–1272. doi: 10.1111/j.1365-3040.2006.01505.x. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR: a perspective. Journal of Molecular Endocrinology. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- Carbonnier L, Marques C, Coutinho J, Madeira M, Tomé M. Borralho N.M.G. PJS, IUFRO on silviculture and improvement of Eucalypts: ‘Eucalyptus in a changing world’. Portugal: Aveiro; 2004. The future of Eucalyptus plantations. [Google Scholar]
- Champ KI, Febres VJ, Moore BD. The role of CBF transcriptional activators in two Citrus species (Poncirus and Citrus) with contrasting levels of freezing tolerance. Physiologia Plantarum. 2007;129:529–541. [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kayal W, Navarro M, Marque G, Keller G, Marque C, Teulieres C. Expression profile of CBF-like transcriptional factor genes from Eucalyptus in response to cold. Journal of Experimental Botany. 2006;57:2455–2469. doi: 10.1093/jxb/erl019. [DOI] [PubMed] [Google Scholar]
- FAO. Global forest resource assessment. Main report. 2000 [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa MC, Rasmussen-Poblete S, Valenzuela PD, Krauskopf E. Isolation and characterization of a cDNA encoding a CBF transcription factor from E. globulus. Plant Physiology and Biochemistry. 2007;45:1–5. doi: 10.1016/j.plaphy.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Gao MJ, Allard G, Byass L, Flanagan AM, Singh J. Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Molecular Biology. 2002;49:459–471. doi: 10.1023/a:1015570308704. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Molecular Biology. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. The Plant Journal. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiology. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiology. 2006;142:98–112. doi: 10.1104/pp.106.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang BO, Jin LG, Liu JY. Molecular cloning and functional characterization of a DREB1/CBF-like gene (GhDREB1L) from cotton. Science China, Series C, Life Sciences. 2007;50:7–14. doi: 10.1007/s11427-007-0010-8. [DOI] [PubMed] [Google Scholar]
- I McKhann H, Gery C, Bérard A, Lévêque S, Zuther E, Hincha KD, De Mita S, Brunel D, Téoulé E. Natural variation in CBF gene sequence, gene expression and freezing tolerance in the Versailles core collection of Arabidopsis thaliana. BMC Plant Biology. 2008;8:105. doi: 10.1186/1471-2229-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G, Marchal T, SanClemente H, Navarro M, Ladouce N, Winckler P, Couloux A, Teulieres C, Marque C. Development and functional annotation of a 11,303 EST collection from Eucalyptus for studies of cold tolerance. Tree Genetics and Genomes. 2009;5:317–327. [Google Scholar]
- Kitashiba H, Matsuda N, Ishizaka T, Nakano H, Suzuki T. Isolation of two genes similar to DREB1/CBF from the sweet cherry (P. avium) Journal of the Japanese Society for Horticultural Science. 2002;71:651–657. [Google Scholar]
- Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marque G. 2008. Isolement et caractérisation chez l’Eucalyptus de gènes codant les facteurs de transcription CBF impliqués dans la tolérance au froid. PhD thesis, France, Toulouse University. [Google Scholar]
- Mas MT, Smith KC, Yarmush DL, Aisaka K, Fine RM. Modelling the anti-CEA antibody combining site by homology and conformational search. Proteins. 1992;14:483–498. doi: 10.1002/prot.340140409. [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Pérez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiology. 1999;119:463–469. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. The Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Medina J, Salinas J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proceedings of the National Academy of Sciences, USA. 2007;104:21002–21007. doi: 10.1073/pnas.0705639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunan LM, Tang-Nelson K, Lighter DV. Real time RT-PCR determination of viral copy number in Penaeus vannamei experimentally infected with Taura syndrome virus. Aquaculture. 2004;229:1–10. [Google Scholar]
- Peng Y, Lin W, Wei H, Krebs SL, Arora R. Phylogenetic analysis and seasonal cold acclimation-associated expression of early light-induced protein genes of Rhododendron catawbiense. Physiologia Plantarum. 2008;132:44–52. doi: 10.1111/j.1399-3054.2007.00988.x. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Richards FM. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. Journal of Molecular Biology. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen TH, Hayes PM. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant and Molecular Biology. 2005;59:533–551. doi: 10.1007/s11103-005-2498-2. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N. Expression levels of barley Cbf genes at the frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. The Plant Journal. 2007;51:308–321. doi: 10.1111/j.1365-313X.2007.0141.x. [DOI] [PubMed] [Google Scholar]
- Sun S, Yu JP, Chen F, Zhao TJ, Fang XH, Li YQ, Sui SF. TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. Journal of Biological Chemistry. 2008;283:6261–6271. doi: 10.1074/jbc.M706800200. [DOI] [PubMed] [Google Scholar]
- Tamura K, Yamada T. A perennial ryegrass CBF gene cluster is located in a region predicted by conserved synteny between Poaceae species. Theoretical and Applied Genetics. 2007;114:273–283. doi: 10.1007/s00122-006-0430-z. [DOI] [PubMed] [Google Scholar]
- Tang M, Lu S, Jing Y, Zhou X, Sun J, Shen S. Isolation and identification of a cold-inducible gene encoding a putative DRE-binding transcription factor from Festuca arundinacea. Plant Physiology and Biochemistry. 2005;43:233–239. doi: 10.1016/j.plaphy.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Teulières C, Feuillet C, Boudet AM. Differential characteristics of cell suspension cultures initiated from Eucalyptus gunnii clones differing by their frost tolerance. Plant Cell Reports. 1989;8:407–410. doi: 10.1007/BF00270080. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Triezenberg SJ, Thomashow MF, Stockinger EJ. Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundancy in trans-activation. Plant Molecular Biology. 2005;58:543–559. doi: 10.1007/s11103-005-6760-4. [DOI] [PubMed] [Google Scholar]
- Welling A, Palva ET. Molecular control of cold acclimation in trees. Physiologia Plantarum. 2006;127:167–181. [Google Scholar]
- Welling A, Palva ET. Involvement of CBF transcription factors in winter hardiness in Birch. Plant Physiology. 2008;147:1199–1211. doi: 10.1104/pp.108.117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Nassuth A. Stress- and development-induced expression of spliced and unspliced transcripts from two highly similar dehydrin 1 genes in V. riparia and V. vinifera. Plant Cell Reports. 2006;25:968–977. doi: 10.1007/s00299-006-0151-4. [DOI] [PubMed] [Google Scholar]
- Xiao H, Siddiqua M, Braybrook S, Nassuth A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant, Cell and Environment. 2006;29:1410–1421. doi: 10.1111/j.1365-3040.2006.01524.x. [DOI] [PubMed] [Google Scholar]
- Xiao H, Tattersall EA, Siddiqua MK, Cramer GR, Nassuth A. CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant, Cell and Environment. 2008;31:1–10. doi: 10.1111/j.1365-3040.2007.01741.x. [DOI] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiology. 2003;133:910–918. doi: 10.1104/pp.103.027169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fowler SG, Cheng H, Lou Y, Rhee S, Y Stockinger EJ, Thomashow MF. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. The Plant Journal. 2004;39:905–919. doi: 10.1111/j.1365-313X.2004.02176.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang TW, Zhang LJ, Di CX, Xu SJ, An LZ. Isolation and expression analysis of two cold-inducible genes encoding putative CBF transcription factors from Chinese cabbage (Brassica pekinenesis Rupr.) Journal of Integrative Plant Biology. 2006;48:848–856. [Google Scholar]
- Zhao TJ, Sun S, Liu Y, Liu JM, Liu Q, Yan YB, Zhou HM. Regulating the DRE-mediated signaling pathway by synergic functions of trans-active and trans-inactive DREBs in Brassica napus. Journal of Biological Chemistry. 2006;281:10752–10759. doi: 10.1074/jbc.M510535200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.