Abstract
Stem lodging-resistance is an important phenotype in crop production. In the present study, the expression of the wheat COMT gene (TaCM) was determined in basal second internodes of lodging-resistant (H4564) and lodging-susceptible (C6001) cultivars at stem elongation, heading, and milky endosperm corresponding to Zadoks stages Z37, Z60, and Z75, respectively. The TaCM protein levels were analysed by protein gel blot and COMT enzyme activity was determined during the same stem developmental stages. TaCM mRNA levels were higher in H4546 from elongation to the milky stages and in C6001 the TaCM mRNA levels decreased markedly at the heading and milky stages. The TaCM protein levels and COMT activity were also higher in H4564 than that in C6001 at the heading and milky stages. These results corresponded to a higher lignin content measured by the Klason method and stem strength and a lower lodging index in H4564 than in C6001 at the heading and milky stages. Therefore, the TaCM mRNA levels, protein levels, and enzyme activity in developing wheat stems were associated with stem strength and lodging index in these two wheat cultivars. Southern analysis in a different population suggested that a TaCM locus was located in the distal region of chromosome 3BL, which has less investigated by QTL for lodging-resistant phenotype.
Keywords: Caffeic acid 3-O-methyltransferase, lignin biosynthesis, lodging resistance, Triticum aestivum L
Introduction
Crop lodging is a complicated phenomenon that is influenced by many factors from physiology and genetics, to husbandry and environment. Lodging is a major problem in crop production which causes poor grain filling and yield loss, reduces grain quality and mechanical harvesting efficiency (see review by Berry et al., 2004, and references therein). The dwarfness of the crop using dwarfing genes and the application of plant growth regulators have been very effective tools for reducing the risk of lodging and maintaining a steady yield. However, they have not completely eradicated lodging risk. Furthermore, a reduction in plant height for lodging resistance may reduce the photosynthetic capacity of a canopy which would then limit the final yield (Wu et al., 2002). An analysis of the dwarfing genes on crop yield has shown that the optimum plant height for maximum photosynthetic capacity is between 70 cm and 100 cm in wheat (Flintham et al., 1997). Therefore, alternative strategies for further improvement in lodging resistance of crops must be developed.
According to Sterling et al. (2003), two types of lodging occur in wheat. Stem lodging results from the bending or breaking of the lower culm internodes, while root lodging refers to intact and unbroken culms leaning from the crown due to failure of root anchorage on ground. Stem strength, the bending or breaking strength of the culm, is important for stem lodging resistance, particularly for the basal internodes of crops (Zuber et al., 1999). Increasing stem strength will be a very promising strategy to breed crops with high lodging resistance and yield. Except for stem weight and diameter which are closely related to physical strength, stem strength is also the product of its chemical and biochemical components. Stem tissues of crops mainly contain the storage and structural carbohydrates. Recent reports have shown that higher starch accumulation in the stem tissues is responsible for the higher lodging resistance in rice during typhoons (Kashiwagi and Ishimaru, 2004; Kashiwagi et al., 2006; Ishimaru et al., 2008). In theory, the structural carbohydrates (cellulose and lignin) should also play the (even more) important roles for material strength in stems.
Lignin is a phenolic cell wall polymer closely linked to cellulose and hemicelluloses, and is, second to cellulose, the most abundant biopolymer on earth. In plants, lignin mainly deposits in the walls of certain specialized cells such as tracheary elements, sclerenchyma, and phloem fibres. This leads to a dramatic change in cell wall properties, which impart strength and structural support to the wall and assists in the transport of water and nutrients within xylem tissue by decreasing the permeability of the cell wall (Boerjan et al., 2003). It has long been proposed that lignin synthesis might be related to stem strength. This issue is especially important in monocotyledonous crop plants, where weak stem strength will lead to a lodging phenotype (Hai et al., 2005). To date, however, there is little information at the molecular level as to how the regulation of lignin synthesis affects crop lodging. Reduced lignin levels have been observed in maize brown-midrib (bm) mutants and this has been associated with reduced stem strength. Recent work has confirmed that the bm3 mutant harboured a mutation in the gene for caffeic acid 3-O-methyltransferase (EC 2.1.1.68, COMT) (Vignols et al., 1995), while bm1 was shown to affect directly expression of the cinnamyl alcohol dehydrogenase (EC 1.1.1.195, CAD) (Halpin et al., 1998). Some brittleness mutants have been reported in barley (Kokubo et al., 1991) and rice (Kinoshita, 1998). A COBRA-like regulation protein has been reported to be involved in brittle stalk mutants in rice (Li et al., 2003) and maize (Sindhu et al., 2007). The actions of these mutants on gene levels, however, are largely unknown.
A cDNA fragment for COMT from wheat has been isolated and RNA gel blot analysis showed that this gene was differentially expressed in lodging-sensitive and -resistant cultivars (Ma et al., 2002). The full-length cDNA for this COMT, TaCM, was obtained by screening wheat stem cDNA library and the functional expression and transgenic approach have demonstrated that TaCM is an authentic COMT involved in lignin biosynthesis (Ma and Xu, 2008). The analyses of TaCM gene expression, enzyme activity, and protein content in relation to the lodging-resistant phenotype in wheat is reported here. The TaCM locus was mapped in the chromosome.
Materials and methods
Plant materials
Wheat (Triticum aestivum L.) plants were grown in a naturally lit greenhouse with normal irrigation and fertilization. Two cultivars, C6001 and H4564, were chosen for our analyses. C6001 and H4564 developed from the same pedigree of Jimai from the Hebei province of China. Both cultivars have the same growth period (240 d) and vernalization type (winter), similar morphological and agricultural phenotypes (with plant height of 75–80 cm, diameter of the mature stem of 2.97–3.27 mm, kernel number per spike of 18–20, and grain number per kernel of 28–36 with mature seed weight 38–41 g per 1000 seeds), but differ in stem lodging phenotype, in which C6001 is lodging-sensitive while H4564 is lodging-resistant. The stem lodging level (as judged by the angle of the stem to the vertical position of more than 30o) just before harvest was 25–30% for C6001 and 2–4% for H4564 in the flood irrigation field with 10–15% and 0%, respectively, in dry farmland. The majority of lodgings observed are due to stem breakage so they belong to stem lodging predominates. Basal second internodes were collected from the wheat plants with three internodes, and then collected at 3 week intervals at the stem elongation, heading, and milky endosperm stages, corresponding to Zadoks stages (Zadoks et al., 1974) Z37, Z60, and Z75, respectively. Previous investigations have shown that the second internodes play the more important role in wheat lodging resistance (Zuber et al., 1999; Dong et al., 2003).
RNA isolation, Northern blot, and RT-PCR analysis
Total RNA was isolated from wheat internode tissue by TRI reagent (Molecular Research Center, Inc, Cincinnati, USA) following the manufacturer's instructions. Ten μg of each total RNA sample was electrophoresed on formaldehyde 1.4% (w/v) agarose gels. RNA was blotted onto Hybond-N+ membrane (Amersham, GE Healthcare Bio-Sciences Corp., Piscataway, NJ 08855-1327, USA) using the standard transfer procedure of Sambrook et al. (1989). The blots were hybridized at 42 oC with 6× SSC, 5× Denhardt, 0.5% SDS, 100 μg ml−1 salmon sperm DNA with 50% formamide, and washed with 0.1× SSC plus 0.1% SDS at 65 °C. Probes were 32P-labelled using a Ready-to-Go DNA Labeling Kit (Amersham). RNA hybridization signals were normalized by a soybean 18S ribosomal RNA.
The first-strand cDNA was synthesized using the RT-PCR kit (TakaRa, Japan) with total RNA. Reverse transcription reactions were carried out at 42 °C for 60 min and terminated by heating to 95 °C for 10 min. One microlitre of the reaction mixture was used as a template in the PCR reaction. The parameters for 30 cycles were 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 1.5 min, with a final extension at 72 °C for 10 min. The PCR products were loaded onto 1% agarose gel to visualize the amplified cDNAs. Primers for wheat TaCM were: sense primer 5′-AAGGTCCTCATGGAGAGCTG-3′ and antisense primer 5′-CGGCAGGATGCATCCACGGA-3′. Primers for α-tubulin were: sense primer 5′-CTCTACTGCCTCGAGCAT-3′ and antisense primer 5′-GAGGAAAGCATGAAGTGG-3′.
Enzyme extraction and assay
Enzyme extraction followed Parvathi et al. (2001). In brief, wheat internode tissue was ground in liquid nitrogen and then the tissue powder was mixed with the extraction buffer (100 mM TRIS-HCl pH 7.5, 0.2 mM MgCl2, 2 mM DTT, 10% glycerol) and incubated at 4 °C for 1 h. The samples were spun at 12 000 g for 10 min at 4 °C. The extracts were desalted on PD-10 columns (Pharmacia, Piscataway, NJ08855-1327, USA). The soluble protein fractions were used for the COMT enzyme assay. COMT enzyme activities were measured according to the method of Ni et al. (1996). Protein concentrations were determined by the Bradford assay (Bradford, 1976) with bovine serum albumin as the standard.
Protein gel blot analysis
Protein samples were resolved by 12% SDS-PAGE, then electro-transferred onto nitrocellulose membrane. The membrane was incubated in blocking buffer (PBS containing 0.05% Tween 20 and 5% skim milk) at room temperature overnight, and then incubated with rabbit anti-alfalfa COMT serum at 1:10 000 dilution (Kersey et al., 1999) in blocking buffer for 2 h. The membranes were washed with PBST (PBS with 0.05% Tween 20) and incubated with peroxidase-conjugated goat anti-rabbit polyclonal antibodies (Bio-Rad Laboratories Beijing Ltd., Beijing 100086, China) at 1:10 000 dilution in PBST for 1 h. The signals were detected with a chemiluminescent reaction reagent (ECL, Amersham) according to the manufacturer's protocol.
Lignin content analysis
Lignin content in air-dried basal second internodes was quantitatively measured by using the Klason method (Kirk and Obst, 1988) and expressed as a percentage of the cell wall residue (% CWR).
Mapping analysis
The mapping population consists of 114 recombinant inbred lines (RILs) and has been the subject of an extensive genome mapping effort by investigators of the International Triticeae Mapping Initiative (ITMI) (Marino et al., 1996). Genomic DNA isolation from Chinese Spring NT lines, W-7984, Opata 85, and the RILs, restriction enzyme digestion, gel electrophoresis, Southern blotting, and hybridization were performed as described in Faris et al. (2000). TaCM cDNA (GenBank accession no. EF413031) was 32P-labelled using a Ready-to-Go DNA Labelling Kit (Amersham) to be used as the probe. The genetic mapping and linkage relationships were analysed as reported previously (Ma, 2007).
Stem strength and lodging index determination
Wheat basal second internodes were collected from the field. The stem strength and lodging index (LI) was determined according to the method of Wang and Li (1996). In brief, stem strength was measured using a three point bending method where the freshly collected 5 cm stem segments (span more than 10× the diameter) were placed horizontally and mounted on two supports in the rack of the apparatus. The force exerted to break the stem was recorded with a universal force testing device (model DC-KZ300, Sichuan, China) to determine the stem strength (S). The fresh weight (G) of the aerial part, and height (H) to the centre of gravity (measured by balancing the whole plant in the centre position) of each single wheat plant were determined. LI was calculated by following formula: LI=G×H/S. Fifteen stem samples were determined.
Student's t test for independent samples (Geng and Hills, 1989) was applied to determine the difference between C6001 and H4564 cultivars and the probability value was estimated at the P0.05 and P0.01 levels. Data were shown as mean ±standard deviation. All statistical analyses were conducted using the SPSS 11.0 software package.
Results
Stem strength and lodging index at different developmental stages in wheat stem
As the basal internodes play a crucial role in stem lodging resistance in wheat, the basal second internodes were used to determine the stem strength and lodging index from three different developing stages (Table 1). The stem strength of H4564 was always higher than that of C6001, and this difference was particularly remarkable at the heading and milky stages, which was more than 95.1% and 78.4%, respectively. Consistent with the high stem strength, the lodging index of H4564 was significant lower than that of C6001, consistent with its lodging-resistant phenotype.
Table 1.
Comparison of stem bending strength (g), lodging index, Klason lignin content (as % CWR), and COMT activity (pkat mg-1 protein) of basal second internodes in wheat cultivars H4564 and C6001 at different developmental stages
| Developmental stages | Cultivars | Stem bending strength | Lodging index | Lignin content | COMT activity |
| Elongation | C6001 | 167±12 | n.d. | 2.85±0.37 | 12.6±1.33 |
| H4564 | 183±25* | n.d. | 2.93±0.31 | 13.4±1.21 | |
| Heading | C6001 | 288±21 | 0.97±0.08 | 4.54±1.24 | 26.8±1.05 |
| H4564 | 562±41** | 0.54±0.04** | 7.55±2.08* | 35.5±2.10** | |
| Milky | C6001 | 523±41 | 0.63±0.05 | 10.20±1.68 | 90.4±5.07 |
| H4564 | 933±81** | 0.37±0.03** | 14.49±2.30* | 131.3±8.94** |
n.d. means not detected. Data represent means ±standard deviation (n=15 for stem strength and lodging index, n=4 for lignin content and COMT activity). Significant differences at P0.05 and P0.01 levels between the two cultivars in the same stage are marked by * and **, respectively.
Lignin accumulations in relation to wheat stem development
To determine whether lignin synthesis was associated with stem strength and lodging resistance, lignin content was analysed in the basal second internodes at different developmental stages in the two wheat cultivars (Table 1). From elongation to the milky stage, the content of Klason lignin increased accordingly, reflecting active lignin synthesis during wheat stem maturity. The Klason lignin content of H4564 was significantly higher than that of C6001 at the heading and milky stages, more than 66.3% and 42.1%, respectively. By contrast, there was no remarkable difference for lignin content between the two cultivars at the elongation stage.
Expression of the TaCM gene at different developmental stages in wheat stem
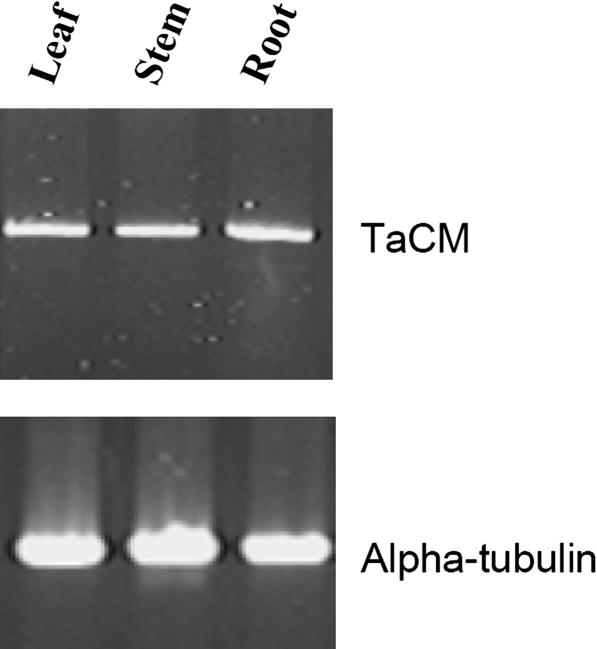
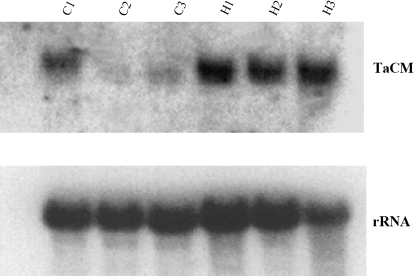
RT-PCR analysis showed that the TaCM gene was constitutively expression in stem, leaf, and root tissues (Fig. 1), indicating that TaCM is associated with the constitutive lignification process in various vegetative growing tissues. TaCM gene expression was analysed in the basal second internodes at different stem developmental stages by Northern blot hybridization (Fig. 2). At the elongation stage, the TaCM mRNA levels in C6001 and H4564 were similar high. At the heading and milky stages, the TaCM mRNA levels in H4564 stem still remained very high, while these mRNA levels declined very markedly in C6001. Quantification of the TaCM1 mRNA levels with a Phosphor Image with 18S rRNA normalization confirmed these results. By contrast to the stem tissues, the TaCM mRNA levels in leaf tissues of the two wheat cultivars did not show much difference at different developmental stages (data not shown).
Fig. 1.
RT-PCR analyses of TaCM gene expression in various wheat tissues. RT-PCR was performed using the gene-specific primers. Alpha-tubulin RT-PCR was included as a control.
Fig. 2.
RNA gel blot analysis of TaCM gene expression at different developmental stages of wheat basal second internodes. Ten μg total RNA were loaded in each lane. C1, C2, and C3: C6001 elongation, heading, and milky stages; H1, H2, and H3: H4564 elongation, heading, and milky stages. Hybridization with an 18S rDNA probe has been included to confirm that the RNA preparations are undegraded and to serve as an internal control of variations in gel loading and blotting.
COMT enzyme activity at different developmental stages in wheat stems
COMT enzyme activity was analysed in the basal second internodes at different developmental stages of the wheat stem (Table 1). The results showed that the COMT activity increased continuously from the elongation to the milky stage, consistent with active lignin synthesis during stem development in wheat. COMT activity showed little difference between C6001 and H4564 at the elongation stage. However, COMT activity in H4564 was much higher than that in C6001 at the heading and milky stages, more than 32.5% and 45.2%, respectively.
TaCM enzyme protein at different developmental stages in wheat stems
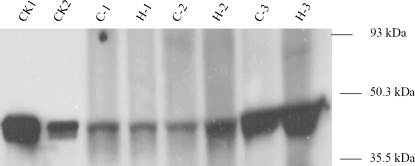
TaCM protein levels in the basal second internodes of wheat stems were determined by protein gel blot analysis by using alfalfa monospecific polyclonal antisera against COMT (Fig. 3). A single band was detected from wheat stem samples; indicating that only one COMT protein isozyme was present in the stem tissue of wheat. This is in accordance with the genetic mapping results (Fig. 4), suggesting that TaCM exists as a single-copy gene in the wheat genome. The immunological reactivity of COMT protein from wheat stem extracts was similar to that of recombinant TaCM protein expressed in E. coli. This further supports a single COMT protein relating to the TaCM gene is present in stem tissues of wheat.
Fig. 3.
Protein gel blot analysis of TaCM protein in wheat basal second internodes. Protein extracted from wheat stem tissues was separated by SDS-PAGE, blotted to nitrocellulose membranes and probed with monospecific polyclonal antisera raised against alfalfa COMT protein. Molecular marker is indicated on right of figure. CK1 and CK2, 6 ng and 0.6 ng of recombinant TaCM protein expressed in E. coli. C1, C2, and C3: C6001 elongation, heading, and milky stages; H1, H2, and H3: H4564 elongation, heading, and milky stages. 1.5 μg protein was loaded on each of these lanes.
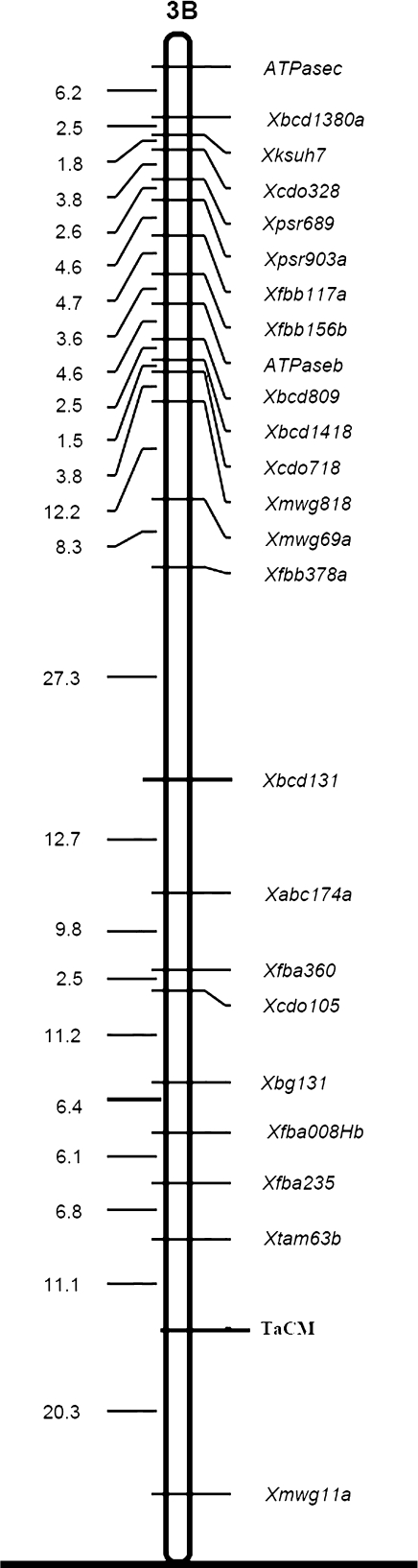
Fig. 4.
Linkage maps of TaCM in the chromosome of wheat. CentiMorgan (cM) distances are indicated at the left of the chromosome and marker loci to the right. The map positions of TaCM are indicated in bold face.
The TaCM protein levels increased with the maturity of the wheat stem tissues, from the elongation to the milky stages. By comparison between H4564 and C6001, the TaCM protein levels were similar at the elongation stage. However, these levels were higher in H4564 than in C6001 at the heading and milky stages (Fig. 3). This is in good agreement with the results of COMT activity.
Chromosome location of TaCM
Chromosome location of the TaCM gene was investigated by genetic mapping. Southern analysis with TaCM cDNA inserts produced two bands with EcoRI digestion. Nulli-tetrasomic analyses localized TaCM to group-3 chromosmes. Linkage mapping using ITMI mapping population indicated that the TaCM locus was mapped to the proximal region of the distal region of chromosome 3B at a position between markers Xtam63b and Xmwg11a (Fig. 4). These results suggest that TaCM is present in a single locus in the wheat genome B, consistent with the data from RT-PCR, Northern hybridization, and protein gel blot analyses.
Discussion
Lodging resistance in crops represents a quantitative trait that is suggested to be involved in the actions of many genes and is difficult to assess on a phenotypic basis (Keller et al., 1999; Berry et al., 2004). Therefore, to link the actions of a single gene directly to the lodging resistant phenotype will not only be very useful in elucidating this complex agronomic trait but also helpful in developing lodging-resistant cultivars in crop-breeding programmes.
In theory, lignin determines the physical strength of plants, as a lower lignin content causes stems to be soft and brittle. Some mutants called bm were obtained which were shown to be associated with low lignin quantity and low stem strength. The first bm mutant was described in maize as early as 1924. Based on morphological and genetic analyses, four mutants named bm1, bm2, bm3, and bm4 were described in maize (Cherney et al., 1991; Barrière et al., 2004). These mutations were found to be located on chromosomes 1, 4, 5, and 9, respectively (Kuc et al., 1968). However, until recently, the bm3 mutation was demonstrated to occur in the gene encoding COMT (Vignols et al., 1995). Since the COMT protein is encoded by a single gene in maize, the results indicated that the bm3 mutation had arisen from a retrotransposon event or a deletion of part of the COMT gene that had been cloned previously (Collazo et al., 1992). As the maize COMT gene is known to be strongly expressed in roots and to a lesser extent in the stems and leaves, the reason why this mutation leads to the visible phenotype in stem and leaf tissues is still questionable. Another mutant, bm1, was demonstrated to be closely associated with, if not identical to, the gene encoding CAD (Halpin et al., 1998). CAD activity was reduced in bm1 mutant plants but the strong reduction was observed in root (90–97% reduction) and leaf midrib (70% reduction) tissues, not in stem tissues. Furthermore, two bands were detected in Western and Northern blot hybridization although Southern hybridization suggested that there was only a single CAD gene in the maize genome. Clearly, the issue of lignin synthesis with plant tissue strength and lodging resistance, particularly in stems, still needs intensive investigation.
The natural mutation for lignin synthesis in wheat is still lacking. For this reason, it was hoped to use another approach to explore the relationship between lignin synthesis and the wheat stem lodging-resistant phenotype. A COMT cDNA was cloned from wheat, namely TaCM. Its biochemical identification as an authentic COMT enzyme involved in lignin biosynthesis was confirmed by an analysis of enzyme activity and a transgenic approach (Ma and Xu, 2008). Genetic mapping indicated that the TaCM gene was present as a single locus in the wheat genome (Fig. 4). In contrast to maize, the wheat TaCM gene was actively expressed in stem tissues, suggesting that it played a role in lignin biosynthesis and stem development (Fig. 1). The TaCM gene expression was kept fairly high from the elongation to the milky stages in the basal second internodes of the lodging-resistant cultivar (H4564), but it declined markedly at the heading and milky stages in the lodging-sensitive cultivar (C6001) (Fig. 2). In agreement with these results, TaCM protein levels (Fig. 3) and COMT activity (Table 1) exhibited the significant differences between H4564 and C6001 at the heading and milky stages. The TaCM gene exists as a single copy in the wheat genome. Furthermore, only a single band was detected in wheat stem tissues using an anti-alfalfa COMT antibody, and this band exhibited a similar immunological reaction with the recombinant TaCM protein that was expressed in E. coli. These results suggest that the COMT protein and activity detected in wheat basal second internodes is likely to corresponding to the TaCM gene. Therefore, high TaCM gene expression will lead the more TaCM protein and enzyme activity, which results in more lignin accumulation in the basal second internodes of the H4564 cultivar and, subsequently, makes these internodes have strong strength and exhibit high lodging resistance.
The comparison of the different parameters between C6001 to H4564 cultivars revealed that stem strength and lodging index showed good consistency. The difference in the lignin content between two cultivars was in proportion to that of stem strength and lodging index, and this difference was remarkable at the heading stage rather than at the milky stage. The total increase in lignin content from C6001 to H4564 was a little lower than that of the stem strength and lodging index, which may reflect that a small increase in lignin content will result in a high increase in stem strength and lodging resistance. TaCM enzyme activity, however, exhibited a slightly different trend with the other three parameters. In one aspect, TaCM activity showed more increase at the milky stage than at the heading stage. In another aspect, the total increase of TaCM activity from C6001 to H4564 was less than these other parameters, for example, a 32.5% increase in TaCM activity versus 95.65%, 78.6%, and 66.3% in stem strength, lodging index, and lignin content, respectively, at the heading stage. This reflects that other genes may also be involved in lignin biosynthesis and contribute to the stem strength and lodging resistance. In fact, of two CCR genes that have been analysed in wheat, CCR1 has also been shown to contribute to lignin biosynthesis and stem strength (Ma and Tian, 2005; Ma, 2007). What and how CCR1 and TaCM genes influence this process is an interesting issue and will warrant further investigation. It suggests that different genes will together regulate the whole lignin biosynthesis pathway, and then lead to lignin accumulation in certain tissues at the proper developmental time, subsequently making the stem have high strength and a lodging-resistant phenotype. TaCM will be a regulating point or marker for what takes place in the whole lignin biosynthesis pathway.
In general, lignin or cellulose in the cell wall determines the physical strength of plant tissues, particular for lignin. Cellulose has good tensile strength, but it is soft and flexible in nature, while lignin is much like a polyether network, with a large molecular weight, random structure, and an enormous capacity to cross-link with cellulose together with the process of thickening plant secondary walls, which makes the plant tissue's mechanical intensity and supportable ability. In practice, however, the roles of lignin and cellulose in crop lodging resistance are not consistent. In maize, evidence from bm mutants have shown that lignin is involved in lodging resistance (Vignols et al., 1995; Halpin et al., 1998). In rice, the brittle culm1 (bc1) mutant, which altered the expression of a COBRA-like protein, causes a reduction in cell wall thickness and mechanical strength. The bc1 mutant, however, exhibits a reduction in cellulose content but an increase in lignin level, suggesting that cellulose instead of lignin plays an important role in the cell wall mechanical strength (Li et al., 2003). By contrast, the maize brittle stalk2 (bk2) mutant, which is involved in a COBRA-like protein similar to rice bc1, exhibits a similar cellulose content as the wild type in developing stems and a loss of staining lignin in the vascular bundles and sclereid layers (Sindhu et al., 2007). Furthermore, two loci have been identified that are responsible for the higher starch accumulation and, consequently, for the higher lodging resistance during typhoons in the lower and upper rice stems, respectively, which show that a high accumulation of starch in the stem instead of structural carbohydrates such as lignin and cellulose, plays the vital role for lodging resistance in rice (Kashiwagi and Ishimaru, 2004; Kashiwagi et al., 2006; Ishimaru et al., 2008). The different roles of lignin in lodging resistance between rice versus maize and wheat are unclear at present. It is possible that the aquatic versus the terrestrial environments that are used to cultivate these crops may affect the contribution of lignin and cellulose to mechanical strength in crop stems.
Analysis of quantitative trait loci (QTLs) can reveal the genetic basis of relationships among traits, this is particularly important for comprehensive understanding of complex traits such as lodging (Cuthbert et al., 2008). Comparisons among QTLs for lodging traits have been made in several crop plants. In barley, one QTL has been associated with grain yield, plant height, and lodging severity (Spaner et al., 1999). Two loci on chromosome 5, namely prl5 and lrt5, have been identified for the improved lodging resistance of the lower and upper stems, respectively, in rice (Kashiwagi and Ishimaru, 2004; Ishimaru et al., 2008). In addition, multiple QTLs for stem diameter are required for the best pushing resistance but only a single QTL (namely sdm8) has been identified to contribute to thicker stems towards lodging resistance in rice (Kashiwagi et al., 2008). Nine lodging resistant QTLs have been identified, but five of them have been shown to be coincident with QTLs for plant height in a population between wheat and spelt (Keller et al., 1999). In another report using the doubled haploid population derived from two Canadian wheat cultivars, three QTLs have been identified for lodging resistance in chromosome 4D, 5A, and 6D in wheat. One of three QTLs for lodging coincides with a plant height QTL which is located on chromosome 4DS. Only QTL QLdg.crc-5A detected in this report is in a similar position to a QTL for lodging on chromosome 5AL identified by Keller et al. (1999). This QTL is also associated with a yield QTL (Huang et al., 2006). Using a population of 132 F12 RILs derived by single-seed descent from a cross between the Chinese facultative wheat Ning7840 and the US soft red winter wheat Clark, three QTLs for lodging score were identified in linkage groups 1B, 4AL, and 5A (Marza et al., 2006). The QTL in 5A is in the similar location as QTL of lodging resistance reported by Keller et al. (1999) and Huang et al. (2006). Using a population of 96 doubled haploid lines from the cross ‘Milan’בCatbird’ wheat, QTL for lodging and associated traits were found on chromosomes IB, ID, 2B, 2D, 4B, 4D, 6D, and 7D (Verma et al., 2005). A doubled-haploid population was generated from the cross RL4452בAC Domain’ spring wheat, and three QTLs were identified to control plant lodging in chromosomes 4B, 4D, and 3D, respectively (McCartney et al., 2005). A doubled-haploid population was derived from anther culture of the wheat cross CA9613 and H1488 and stem strength and related basal internode traits were measured at the milk stage in this population. A total of six QTLs for stem strength, culm wall thickness, pith diameter, and stem diameter were identified on chromosomes 1A, 2D, 3A, and 3B, respectively (Hai et al., 2005). These data indicate that multiple genes are involved in lodging resistance, consistent with physiological analysis. As there is low resolution power of QTL experiments, it is difficult from the above data to detect the real number of genes that regulate the lodging resistant trait. The TaCM gene identified is the first genetic locus with a clear biochemical function that links with lodging phenotype in wheat. It is interesting that the TaCM locus is located on chromosome 3B, which is not overlapped by any other lodging QTLs that have been reported previously. Chromosome 3B is also mentioned as one of the locations of a stem solidness gene (Lanning et al., 2006) and two QTLs for stem strength and diameter (Hai et al., 2005), although they do not overlap with the TaCM locus. Therefore, TaCM plus stem strength, diameter, and solidness loci in chromosome 3B will be useful for the rapid development of lodging-resistant genotypes using a marker-assisted selection approach. It would be an interesting further experiment to test the relationship among these loci and whether they co-segregates with a stem lodging phenotype.
In conclusion, a COMT gene, namely TaCM, was characterized from wheat. Its mRNA levels were examined in lodging-resistant (H4564) and lodging-sensitive (C6001) cultivars in the basal second internodes at different stem developmental stages. TaCM mRNA levels in H4564 remained constantly high from the elongation to the milky stages, while they declined markedly in C6001 at the heading and milky stages. TaCM protein levels, determined by protein gel blot, were higher in H4564 than that in C6001 at the heading and milky stages. COMT activity was also 33–45% higher in H4564 than that in C6001 at the heading and milky stages. These corresponded to the higher Klason lignin content (about 42–66%), higher stem strength (about 78–95%), and lower lodging index (about 70–80%) in H4564 than that in C6001 during these developmental stages. This suggests that the regulation on TaCM gene expression is involved in lignin synthesis and subsequently contributes to stem strength and the lodging-resistant trait in wheat C6001 and H4564 cultivars. The TaCM locus was genetically mapped to chromosome 3B which would be useful for the rapid development of lodging-resistant genotypes using a marker-assisted selection approach.
Acknowledgments
This work was supported by grants from the National High Technology and Research Development Program of PR China (‘863’ programme, No. 2007AA10Z101), the Chinese National Special Foundation for Transgenic Plant Research and Commercialization (No. 2008ZX08002-003), the National Natural Science Foundation of China (No. 30671134, No. 30671043, No. 30570133), the Natural Science Foundation of Beijing (No. 6082018), and the Innovation Project of the Chinese Academy of Sciences. The author thanks Dr Richard Dixon of the Noble Foundation (USA) for providing the anti-alfalfa COMT serum.
References
- Barrière Y, Ralph J, Méchina V, Guillaumie S, Grabber JH, Argillier O, Chabbert B, Lapierre C. Genetic and molecular basis of grass cell-wall degradability. II. Lessons from brown-midrib mutants. Comptes Rendus Biologies. 2004;327:847–860. doi: 10.1016/j.crvi.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Berry PM, Sterling M, Spink JH, Baker CJ, Sylvester-Bradley R, Mooney SJ, Tams AR, Ennos AR. Understanding and reducing lodging in cereals. Advances in Agronomy. 2004;84:217–271. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Review of Plant Biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Cherney JH, Cherney DJR, Akin DE, Axtell JD. Potential of brown-midrib, low-lignin mutants for improving forage quality. Advances in Agronomy. 1991;46:157–198. [Google Scholar]
- Collazo P, Montoliu L, Puigdomenech P, Rigau J. Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Molecular Biology. 1992;20:857–867. doi: 10.1007/BF00027157. [DOI] [PubMed] [Google Scholar]
- Cuthbert JL, Somers DJ, Brule-Babel AL, Brown PD, Crow GH. Molecular mapping of quantitative trait loci for yield and yield components in spring wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2008;117:595–608. doi: 10.1007/s00122-008-0804-5. [DOI] [PubMed] [Google Scholar]
- Dong Q, Wang AP, Liang SM. Study on the architectural characteristics of wheat stalks. Journal of Shanxi Agricultural University. 2003;2003:188–191. [Google Scholar]
- Faris JD, Haen KM, Gill BS. Saturation mapping of a gene rich recombinant hot spot region in wheat. Genetics. 2000;154:823–835. doi: 10.1093/genetics/154.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham JE, Borner A, Worland AJ, Gale MD. Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. Journal of Agricultural Science, Cambridge. 1997;128:11–25. [Google Scholar]
- Geng S, Hills FG. Biometrics in agriculture science. Dubuque, Iowa: Kendell Hunt Publisher; 1989. [Google Scholar]
- Hai L, Guo HJ, Xiao SH, Jiang GL, Zhang XY, Yan CS, Xin ZY, Jia JZ. Quantitative trait loci (QTL) of stem strength and related traits in a doubled-haploid population of wheat (Triticum aestivum L.) Euphytica. 2005;141:1–9. [Google Scholar]
- Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakats AA, Foxon GA. Brown-midrib maize (bm1): a mutation affecting the cinnamyl alcohol dehydrogenase gene. The Plant Journal. 1998;14:545–553. doi: 10.1046/j.1365-313x.1998.00153.x. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Cloutier S, Lycar L, Radovanovic N, Humphreys DG, Noll JS, Somers DJ, Brown PD. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.) Theoretical and Applied Genetics. 2006;113:753–766. doi: 10.1007/s00122-006-0346-7. [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Togawa E, Ookawa T, Kashiwagi T, Madoka Y, Hirotsu N. New target for rice lodging resistance and its effect in a typhoon. Planta. 2008;227:601–609. doi: 10.1007/s00425-007-0642-8. [DOI] [PubMed] [Google Scholar]
- Kashiwagi T, Ishimaru K. Identification and functional analysis of a locus for improvement of lodging resistance in rice. Plant Physiology. 2004;134:676–683. doi: 10.1104/pp.103.029355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi T, Madoka Y, Hirotsu N, Ishimaru K. Locus prl5 improves lodging resistance of rice by delaying senescence and increasing carbohydrate reaccumulation. Plant Physiology and Biochemistry. 2006;44:152–157. doi: 10.1016/j.plaphy.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Kashiwagi T, Togawa E, Hirotsu N, Ishimaru K. Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.) Theoretical and Applied Genetics. 2008;117:749–757. doi: 10.1007/s00122-008-0816-1. [DOI] [PubMed] [Google Scholar]
- Keller C, Karutz M, Schmid JE, Stamp P, Winzeler M, Keller B, Messmer MM. Quantitative trait loci for lodging resistance in a segregating wheat x spelt population. Theoretical and Applied Genetics. 1999;98:1171–1182. [Google Scholar]
- Kersey R, Inoue K, Schubert KR, Dixon RA. Immunolocalization of two lignin O-methyltransferases in stems of alfalfa (Medicago sativa L.) Protoplasma. 1999;209:46–57. doi: 10.1007/BF01415700. [DOI] [PubMed] [Google Scholar]
- Kinoshita T. Linkage mapping using mutant genes in rice. Rice Genetics Newsletter. 1998;15:13–74. [Google Scholar]
- Kirk TK, Obst JR. Lignin determination. Methods in Enzymology. 1988;161:87–101. [Google Scholar]
- Kokubo A, Sakuri N, Kuraishi S, Takeda K. Culm brittleness of barley (Hordeum vulgare L.) mutants is caused by smaller number of cellulose molecules in cell wall. Plant Physiology. 1991;97:689–701. doi: 10.1104/pp.97.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuc J, Nelson OE, Flanagan P. Degradation of abnormal lignins in the brown-midrib mutants and double mutants of maize. Phytochemistry. 1968;7:1435–1436. [Google Scholar]
- Lanning SP, Fox P, Elser J, Martin JM, Blake NK, Talbert LE. Microsatellite markers associated with a secondary stem solidness locus in wheat. Crop Science. 2006;46:1701–1703. [Google Scholar]
- Li YH, Qian Q, Zhou YH, et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. The Plant Cell. 2003;15:2020–2031. doi: 10.1105/tpc.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QH. Characterization of a cinnamoyl-CoA reductase that is associated with stem development in wheat. Journal of Experimental Botany. 2007;58:2011–2021. doi: 10.1093/jxb/erm064. [DOI] [PubMed] [Google Scholar]
- Ma QH, Tian B. Biochemical characterization of a cinnamoyl-CoA reductase from wheat. Biological Chemistry. 2005;386:553–560. doi: 10.1515/BC.2005.065. [DOI] [PubMed] [Google Scholar]
- Ma QH, Xu Y. Characterization of a caffeic acid 3-O-methyltransferase from wheat and its function in lignin biosynthesis. Biochimie. 2008;90:515–524. doi: 10.1016/j.biochi.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Ma QH, Xu Y, Lin ZB, He P. Cloning of cDNA encoding COMT from wheat which is differentially expressed in lodging-sensitive and -resistant cultivars. Journal of Experimental Botany. 2002;53:2281–2282. doi: 10.1093/jxb/erf102. [DOI] [PubMed] [Google Scholar]
- Marino CL, Nelson JC, Lu YH, Sorrells ME, Leroy P, Tuleen NA, Lopes CR, Hart GE. Molecular genetic maps of the group 6 chromosomes of hexaploid wheat (Triticum aestivum L. em Thell) Genome. 1996;39:359–366. doi: 10.1139/g96-046. [DOI] [PubMed] [Google Scholar]
- Marza F, Bai GH, Carver BF, Zhou WC. Quantitative trait loci for yield and related traits in the wheat population Ning7840×Clark. Theoretical and Applied Genetics. 2006;112:688–698. doi: 10.1007/s00122-005-0172-3. [DOI] [PubMed] [Google Scholar]
- McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N, Noll J, Cloutier S, McCallum BD. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452בAC Domain’. Genome. 2005;48:870–883. doi: 10.1139/g05-055. [DOI] [PubMed] [Google Scholar]
- Ni W, Sewalt VJH, Korth KL, Blount JW, Balance GM, Dixon RA. Stress responses in alfalfa (Medicago sativa L.). XXI. Activation of caffeic acid 3-O-methyltransferase and caffeonyl CoA 3-O-methyltransferase genes does not contribute changes in metabolite accumulation in elicitor-treated cell suspension cultures. Plant Physiology. 1996;112:717–729. doi: 10.1104/pp.112.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi K, Chen F, Guo DJ, Blount JW, Dixon RA. Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. The Plant Journal. 2001;25:193–202. doi: 10.1046/j.1365-313x.2001.00956.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, Carpita NC, Johal G. Maize brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiology. 2007;145:1444–1459. doi: 10.1104/pp.107.102582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaner D, Rossnagel BG, Legge WG, et al. Verification of a quantitative trait locus affecting agronomic traits in two-row barley. Crop Science. 1999;39:248–252. [Google Scholar]
- Sterling M, Baker CJ, Berry PM, Wade A. An experimental investigation of the lodging of wheat. Agricultural and Forest Meteorology. 2003;119:149–165. [Google Scholar]
- Verma V, Worland AJ, Sayers EJ, Fish L, Caligari PDS, Snape JW. Identification and characterization of quantitative trait loci related to lodging resistance and associated traits in bread wheat. Plant Breeding. 2005;124:234–241. [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capeliades M, Puigdomenech P. The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. The Plant Cell. 1995;7:407–416. doi: 10.1105/tpc.7.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li QQ. Evaluation method of stem lodging resistance in wheat. Acta Agriculturae Boreall Sinica. 1995;10:84–88. [Google Scholar]
- Wu TY, Xie LQ, Yang XJ, Zhang CY, Chen RF. Study on the correlations between the components of the plant height and the yield and other traits of wheat. Journal of Agricultural University of Hebei. 2002;25:10–18. [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Research. 1974;14:415–421. [Google Scholar]
- Zuber U, Winzeler H, Messmer MM, Keller B, Schmid JE, Stamp P. Morphological traits associated with lodging resistance of spring wheat (Triticum aestivum L.) Journal of Agronomy and Crop Science. 1999;182:17–24. [Google Scholar]