Summary
Mammalian development is highly sensitive to Tbx1 gene dosage reduction. Gene function insights can also be learned from increased or ectopic expression. The authors generated a novel mouse transgenic line, named COET, which expresses Tbx1 upon Cremediated recombination. The authors crossed this transgenic line with Tbx1Cre animals to activate expression in the Tbx1-expression domain. Compound mutant COET;Tbx1Cre/+ animals died after birth and showed heart enlargement. At E18.5, compound mutants showed ventricular septal defects and thymic abnormalities. The authors crossed compound mutants into a Tbx1 null background to understand whether this phenotype is caused by gene overdosage. Results showed that gene dosage reduction at the endogenous locus could not rescue heart and thymic defects, although the transgene rescued the loss of function phenotype. Thus, the transgenic phenotype appears to be due to gain of function. Resultant data demonstrate that Tbx1 expression must be tightly regulated to be compatible with normal embryonic development.
Keywords: Tbx1, Fgf8, cardiovascular defects, conditional expression, overexpression, pharyngeal apparatus
INTRODUCTION
Tbx1 encodes a T-box transcription factor of critical importance for the development of the pharyngeal apparatus in vertebrates. Loss of function of this gene has been studied in several species but mostly in the mouse, where gene targeting strategies have been used to generate several alleles (Baldini, 2006; Lindsay, 2001). In humans, TBX1 has been definitively linked to the DiGeorge syndrome phenotype, which includes a broad array of clinical findings related to developmental abnormalities of derivatives of the embryonic pharyngeal apparatus (Yagi et al., 2003; reviewed in Baldini, 2005).
Studies in the mouse have shown that progressive dosage reduction of Tbx1 mRNA is associated with the non-linear increase of phenotypic severity (Zhang and Baldini, 2008). However, little is known regarding the phenotypic consequences of increased or ectopic expression, which can also uncover critical insight into the developmental role of a gene. Reports of Tbx1 locus duplications and BAC transgenics using human TBX1 have shown partially conflicting results (Liao et al., 2004; Lindsay et al., 1999; Merscher et al., 2001). This may be due to the fact that transgenes and genomic rearrangements were not limited to Tbx1 alone, but also included neighboring genes.
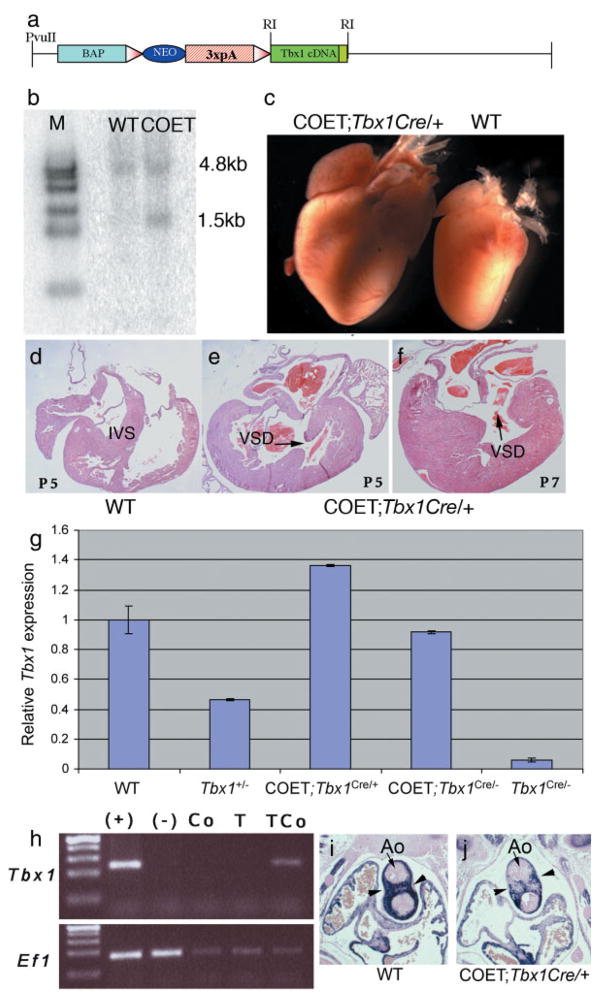
In order to develop a tool useful for controlled spatiotemporal expression of Tbx1, we generated a construct in which a chicken β-actin promoter drives the expression of a loxP-flanked neomycin resistance cassette and triple polyadenylation signal, followed by a mouse Tbx1 cDNA. In this construct, removal of the neo-3xpA cassette by Cre recombinase activates transcription of the Tbx1 cDNA (Fig. 1a). We generated mouse embryonic stem (ES) cells carrying a copy of this construct (example in Fig. 1b). A clone with single copy insertion and in which the transgene had integrated in its entirety (clone FFCOET3) was selected for injection into C57Bl6 blastocysts. Chimeric males were crossed with C57Bl6 wild type females until the transgene was transmitted through the germ line, as determined by the presence of a 270bp transgene-specific amplicon in the progeny. The new transgenic line was named COET (for Conditional Over-Expresson of Tbx1). COET-positive mice were viable and fertile and did not show any gross abnormality (not shown). We crossed COET animals with Tbx1Cre/+ animals expressing Cre in the Tbx1 domain (Huynh et al., 2007). These animals are Tbx1 heterozygous mutants. Most COET;Tbx1Cre/+ animals failed to thrive after birth and died a few days later. Only three COET;Tbx1Cre/+ mice survived weaning from three litters of mice generated from this cross, and of these, only 1 survived to 33 days. Necropsy revealed a severely hypoplastic thymus and enlarged heart (Fig. 1c). Dissection of the heart showed a ventricular septal defect (VSD) (Fig. 1d–f) that might have caused the ventricular enlargement and eventual heart failure. We subsequently examined the phenotype of term embryos at E18.5. Results (summarized in Table 1) indicated that COET;Tbx1Cre/+ embryos exhibited thymus hypoplasia in 79% of the embryos (Fig. 2a–d), while 6/11 embroys sectioned showed VSD (Fig. 2i–j). Of note, none of 3 control COET;Tbx1+/+ or Tbx1Cre/+ embryos examined had VSD, indicating that over-expression or ectopic expression of Tbx1 causes VSD in these mutants. COET;Tbx1Cre/+ mice also showed additional cardiovascular abnormalities such as enlarged pulmonary trunk, enlarged right ventricle (RV) and great artery patterning defects (Fig. 2e–l). However, none of the embryos presented with the substantial heart enlargement observed few days after birth, suggesting that this may be a postnatal consequence of structural heart defects.
FIG. 1.
Generation of the COET transgene and COET transgenic mice. (a) Schematic of the COET construct; BAP: β-actin promoter, 3xpA: three repetitions of a polyadenylation signal; triangles, loxP; RI: EcoRI. (b) Southern blot autoradiography confirms integration of the entire transgene into the ES cell clone used for the generation of the transgenic line. A Tbx1 exon 1 specific probe reveals a 4.8kb fragment from the endogenous allele and a 1.5kb transgenic fragment. Genomic DNA was cut with EcoRI. (c) Hearts dissected from 7-day-old littermates. The hearts of COET;Tbx1Cre/+ are greatly enlarged. (d–f) Transverse sections of P5 and P7 animals. Note the thickened myocardium and VSD in the COET;Tbx1Cre/+ hearts. IVS, interventricular septum. (g) Quantification of Tbx1 expression. Quantitative real-time PCR analysis of the cranial region of 3 mutants per genotype shows that Tbx1 dosage from the COET transgene corresponds to about 1.8x Tbx1. At E10.5 one activated transgene expresses approximately much Tbx1 as 2 wild-type alleles. (h) Reverse transcription PCR of RNA samples from a whole E10.5 wild type embryo (+), Tbx1−/− embryo (−), dissected heart from a E14.5 COET embryo (Co), a Tbx1Cre/+ embryo (T), and a COET;Tbx1Cre/+ embryo (TCo). Note that Tbx1 expression is only detectable in the heart of the latter embryo. Ef1 is a positive control carried out in the same samples. (i,j) Coronal sections from E12.5 embryos (at the level of the cardiac outflow tract) immunostained with an anti α smooth muscle actin antibody. Note the much lighter staining of the outflow tract (arrowheads) in the conditional expression mutant (j) as compared with the wild type control (i). Ao: aortic valve.
Table 1.
Cardiovascular and Thymic Phenotype at E18.5
| CVD |
|||||||
|---|---|---|---|---|---|---|---|
| Normal | AoA only | Enlarged V, PT | Abn OFT | VSD | Thymus: hypo/aplasia | Total | |
| COET; Tbx1Cre/+ | 1 | 4 | 11 | 4 | 6/11 | 15 (79%)* | 19 (95%)** |
| Tbx1Cre/+ | 8 | 3 | - | - | - | 1 (9%) | 11 (27%) |
| COET;Tbx1+/+ | 18 | - | - | - | - | - | 18 |
AoA, aortic arch defects including abnormal origin of right subclavian, interrupted aortic arch; V, PT, ventricle, pulmonary trunk; OFT, outflow tract (includes TGA, DORV and TAC).
Student’s t-test:
P = 0.0002;
P =0.0001.
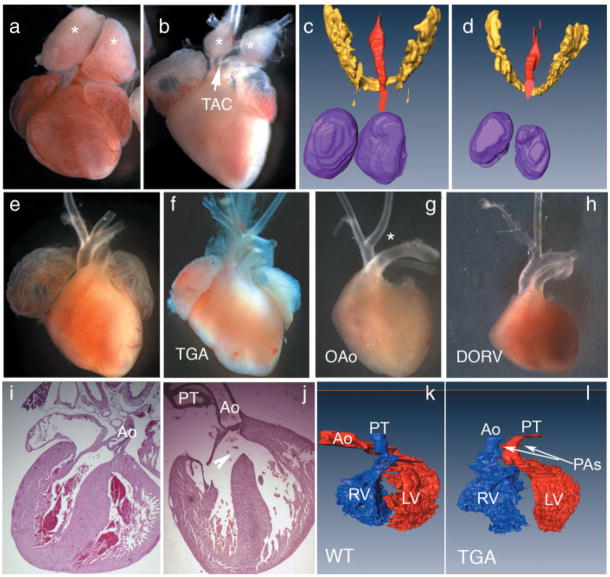
FIG. 2.
COET;Tbx1Cre/+ exhibit cardiovascular and pharyngeal defects. (a–d) Thymus hypolplasia is evident from dissections at E18.5 (a, b) and in 3D reconstructions of sections at E15.5 (c, d, thymus, purple; thyroid, yellow). The ventral view of the isolated hearts show WT (a) vs. COET;Tbx1Cre/+ (b) thymus overlying the great arteries (asterisks). The reconstructed dorsal views compare thymus size in near-term Tbx1Cre/+ (c) and COET;Tbx1Cre/+ (d) embryos. The red color indicates the lumen of the trachea. (e–h) Cardiomegaly is not observed in term embryos, but mutants show a range of OFTseptation defects, including truncus (TAC in b), transposition of the great arteries (TGA in f), overriding of the aorta (OA in G, this mutant also shows IAA-B, asterisk, which is a Tbx1-haploinsufficiency phenotype) and double-outlet right ventricle (DORV in H). (i–j) Histological sections at the same stage show a large VSD (arrowhead) that occurs in ~55% of COET;Tbx1Cre/+ yet not in Tbx1Cre/+ or COET-only mice. (k,l) Ventral view of 3D reconstruction showing transposition of the great arteries from the mutant shown in panel F, vs. a WT littermate. Blue corresponds to the right ventricular cavity, red to the left ventricular cavity. Ao, aorta; PT, pulmonary trunk; PA, pulmonary artery.
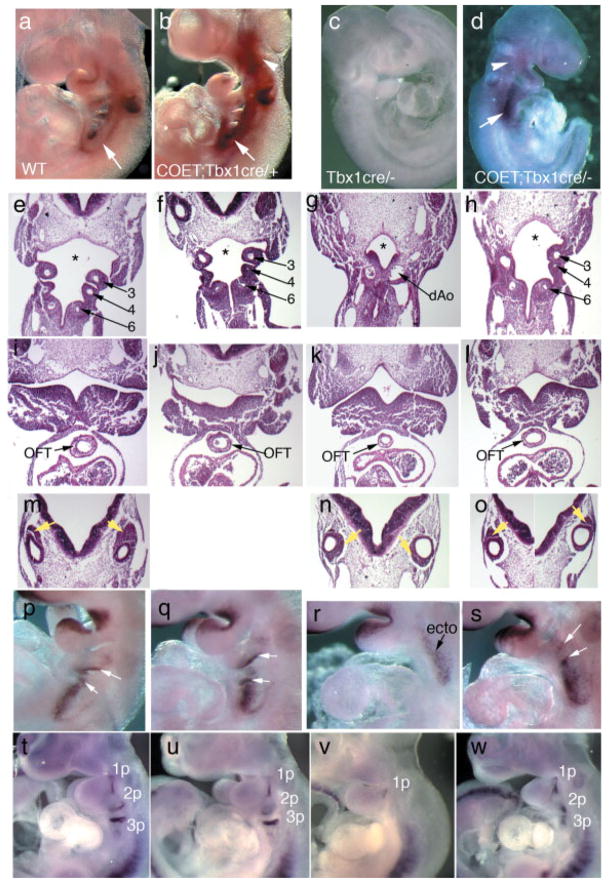
Since Tbx1Cre is a null allele, Tbx1Cre/+ embryos exhibit a haploinsufficiency phenotype that is most commonly represented by great arteries patterning defects at E18.5 (in approx. 30% of the embryos) (Vitelli et al., 2002a). As stated above, COET;Tbx1Cre/+ embryos at E18.5 also show similar vascular abnormalities (Table 1), suggesting that either COET is unable to rescue the haploinsufficiency phenotype or that the abnormalities found in COET;Tbx1Cre/+ embryos are pathogenetically distinct from those found in Tbx1Cre/+ (or Tbx1−/+) embryos. To address this point, we analyzed the 4th pharyngeal arch arteries (PAAs) in COET;Tbx1Cre/+ and Tbx1Cre/+ embryos at E10.5 using intracardiac ink injection. Indeed, defects of the 4th PAAs (which contribute to segments of the mature aortic arch and great arteries) are highly penetrant in heterozygous mutants. We found that only 4/12 (33%) of the COET;Tbx1Cre/+ embryos had defects of the 4th PAAs, while, in the same crosses, most Tbx1Cre/+ embryos examined (4/5, 80%) showed these defects, indicating that COET partially rescued this particular phenotype. These results suggest that the aortic arch and great vessel abnormalities observed at E18.5 in COET;Tbx1Cre/+ are likely of different origin than those observed in heterozygous Tbx1 mutants. Next, we wanted to establish whether the VSD observed in the COET;Tbx1Cre/+ embryos is due to overexpression or to misregulated expression of Tbx1. To this end, we eliminated the expression of the endogenous gene by breeding the COET line into a null Tbx1 background and obtained COET;Tbx1cre/− embryos. Quantitative real time PCR (qRT-PCR) of RNA from embryos at E10.5 showed a dosage of Tbx1 transcript comparable to that of WT embryos at the same stage (Fig. 1g). This mRNA is entirely of COET origin, as these mutants are homozygous null at the endogenous locus. To confirm expression of the transgene, we carried out whole mount in situ hybridization of embryos at E9.5 and found a pattern of Tbx1 expression similar to normal expression, albeit with some differences (Fig. 3a–d). Specifically, in COET;Tbx1cre/− embryos, we observed an extension of the Tbx1-positive domain in the circumpharyngeal region as well as a cranial extension of expression in the head mesenchyme, as compared to WT embryos (Fig. 3a,b). A similar expression pattern was observed in the rescued COET;Tbx1cre/− embryos (Fig. 3d). This pattern is similar to the distribution of Tbx1-fated cells Tbx1Cre/+;R26R embryos at the same stage (Huynh et al., 2007). These similarities are expected because, as is the case for the lacZ reporter in Cre-loxP-based lineage tracing experiments, the COET transgene is irreversibly activated after Cre-induced recombination. Indeed, reverse transcription PCR of dissected hearts from E14.5 embryos revealed Tbx1 expression in COET;Tbx1Cre/+ embryos (Fig. 1h, lane Tco) but not in control embryos (Fig. 1h, lanes Co and T), indicating ectopic expression of the gene in heart tissues of COET;Tbx1Cre/+ mutant embryos. Liao and co-workers (Liao et al., 2008) recently showed that loss of function of Tbx1 is associated with ectopic expression of muscle differentiation markers in the cardiac outflow tract. Consistent with that report, we found reduced expression of α smooth muscle actin in the outflow tract of COET;Tbx1Cre/+ embryos (Fig. 1i–j). However, we found no alteration of the SHF markers Isl1 by immunohistochemistry (data not shown) or Fgf8 by in situ hybridization (Fig. 3p–s) in COET;Tbx1Cre/+ vs. controls.
FIG. 3.
The COET transgene drives Tbx1 expression and partially restores morphological and molecular patterning of the pharyngeal apparatus. (a–d) RNA in situ hybridization of Tbx1 on compound COET;Tbx1Cre/+ mutants shows an extension of Tbx1 staining in the head mesenchyme (arrowhead) and circumpharyngeal area (arrow) compared to control embryos. While Tbx1cre/− embryos lack Tbx1-expression as expected, the introduction of the COET allele on the Tbx1-null background restores the overexpression pattern, corresponding to Tbx1-fated cells. (e–o) Genotypes are as follows: WT: e, i, m; Tbx1Cre/+: f, j; Tbx1cre/−: g, k, n; COET;Tbx1Cre/+: h, l, o. Reinstating Tbx1 expression in COET;Tbx1cre/− (h, l, o) mice reveals a restoration of many defects previously described in Tbx1-null mutants (g, k, n) including pharyngeal apparatus patterning and morphology (e–h), rescue of OFT hypoplasia (i–l), and correct positioning of the cochleo-vestibular ganglion (yellow arrows; compare m–o). Coronal sections at E10.5. 3–6, 3rd, 4th, and 6th pharyngeal arch arteries; dAo, dorsal aorta. (p–s) RNA in situ hybridization at E9.5 of Fgf8, a known Tbx1-target reveals that reinstating Tbx1 expression in COET;Tbx1cre/− embryos restores endodermal Fgf8 expression (white arrows, compare r, s). Ectodermal expression (ecto) is not lost in Tbx1-null mutants (R). (t–w) Pax1 expression at E9.5 shows that the molecular patterning of the pharyngeal endoderm is partially restored in COET;Tbx1cre/− (W) vs. Tbx1cre/− (v) embryos. While the shape of the 1 pharyngeal pouch (1p) seems essentially identical to controls (t, u), pharyngeal pouches 2 and 3 (2p, 3p) do not show normal morphology, suggesting that although the endoderm is specified in the COET;Tbx1cre/− mutants, the pouches do not mature properly.
Restoration of Tbx1 expression in COET;Tbx1cre/− mutants (hereafter referred to as rescued mutants) correlated well with improvement in patterning of the pharyngeal apparatus at mid-gestation, particularly of the caudal pharyngeal arches (Fig. 3e–h), compared to Tbx1 null embryos. Interestingly, the 4th arch and the associated artery remained hypoplastic in rescued embryos (Fig. 3h), despite a full Tbx1 dosage restoration in these mutants. Expression of Tbx1 from the COET allele also improved cardiac outflow tract (OFT) hypoplasia (compare Fig. 3i–l) and abnormal positioning of the cochleovestibular ganglion seen in Tbx1 null mutants (compare Fig. 3m–o). Morphological analyses of rescued embryos at mid-gestation, correlated with molecular findings. In particular, the expression of the Tbx1 target gene Fgf8 (Aggarwal et al., 2006; Brown et al., 2004; Hu et al., 2004; Park et al., 2006; Vitelli et al., 2002b) in the endoderm of the 2nd to 4th pharyngeal pouches was restored in rescued mutants (Fig. 3p–s). At the same developmental stage, the expression of Pax1, a marker of pharyngeal pouches, showed complete restoration of the 1st pharyngeal pouch signal morphology (Fig. 3t,w). However, the 2nd and 3rd pouches appeared shortened. Thus, the COET transgene rescued the specification and formation of the pharyngeal pouches but was not sufficient to support their maturation.
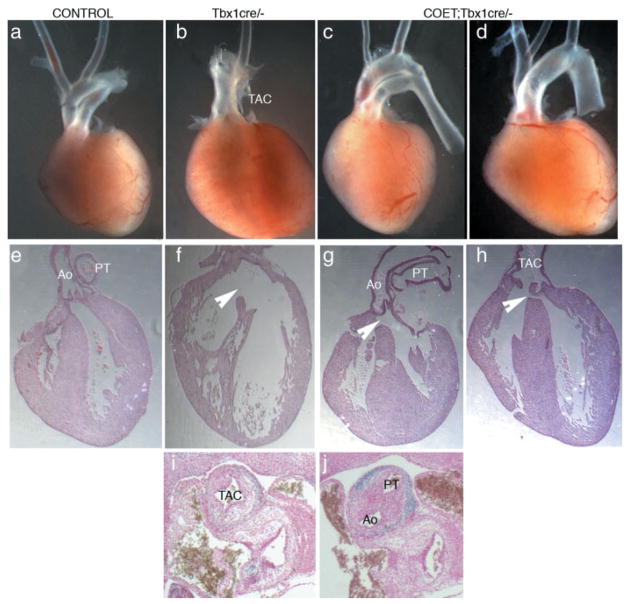
Phenotypic analysis of rescued embryos at a later stage (E18.5) confirmed that many of the abnormalities found in homozygous mutants (Jerome and Papaioannou, 2001; Vitelli et al., 2002a) were ameliorated or rescued by COET, for example the external ear phenotype, cleft palate and OFT septation (Table 2). In particular, we observed different degrees of OFT rescue in these mutants. For example, Figure 4c shows complete septation and proper OFT alignment, also visible at E12.5 (Fig. 4j). At the opposite end of the spectrum, we observed complete distal septation of the great arteries, but these merged proximally into a single OFT trunk and valve (Fig. 4d). While the COET transgene was capable of rescuing most of the phenotypic abnormalities caused by loss of function of Tbx1, it could not rescue some of the defects present in COET;Tbx1Cre/+ embryos. These included VSD (Fig. 4g,h), enlarged pulmonary trunk and thymic aplasia or hypoplasia (Table 2). These results suggest that these defects are not due to increased dosage of Tbx1 mRNA but to inappropriate regulation of transgene expression. Most likely, this is a consequence of the irreversible activation of the transgene by Cre recombinase. Therefore, in contrast to the endogenous gene, COET cannot be dowregulated or turned off, thus potentially causing developmental defects because of persistent expression in tissues where the endogenous gene is normally shut down (e.g., heart tissue, Fig. 1h).
Table 2.
Phenotype of Near-Term COET;Tbx1cre/− Animals
| CVD |
Other |
|||||||
|---|---|---|---|---|---|---|---|---|
| TAC | Restore septation | Restore alignment | VSD | Rescue Palate | Rescue Ear | Normal Thymus | Total | |
| COET;Tbx1cre/− | 1 | 5 | 3 | 6 | 6 | 6 | 6 | 6 |
| Tbx1cre/− | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 4 |
TAC, truncus exiting right ventricle. Septated outflow tract in rescued mutants can occur with or without a corresponding improvement in OFT alignment or aortic arch patterning defects.
FIG. 4.
Variability of cardiovascular rescue in COET Tbx1cre/− mutants. (a–d) Isolated hearts with atria removed at E18.5 show a range of restored OFT septation from complete (c) to distal only (d) as compared to Tbx1-null mutants (b). (e–h) Histological sections show that VSD is found in all COET;Tbx1cre/− term pups examined (arrowheads). Rescue of septation and alignment of the OFT are variable and independent, suggesting that appropriate regulation of Tbx1 expression is required. (i, j) Coronal sections at E12.5 showing the rescue of OFT septation in a COET;Tbx1cre/− embryo (j) compared to Tbx1cre/− (i). TAC, truncus; PT, pulmonary trunk; Ao, aorta.
In summary, the COET transgene expresses Tbx1 mRNA in response to Cre recombination. Quantitative real time PCR indicates that the level of expression of the transgene is comparable to the level of expression of two endogenous alleles. Our genetic crosses demonstrated that the transgene is functional as it can rescue most of the Tbx1 loss of function phenotype and, consistently, can regulate the expression of an endogenous target of Tbx1, Fgf8. Hu and collaborators (Hu et al., 2004) used a β-myosin heavy chain promoter (non-inducible) to express a Tbx1 cDNA in cardiomyocytes. They observed an elongated outflow tract and upregulated and ectopic Fgf10 and Fgf8 expression, but embryos died early, therefore a full analysis could not be carried out.
Our data also uncovered a gain of function phenotype due to COET activation in the Tbx1 expression domain. This phenotype includes structural heart and thymic defects. Thus, these organs are sensitive not only to reduced Tbx1 dosage but also to disregulation of the expression of this gene. We think that COET will be an invaluable tool for further analysis of Tbx1 function in the developing mouse.
METHODS
Mouse Mutants and Breeding
All the experiments involving mice were approved by the IACUC of IBT and in compliance with the US Public Health Service Policy on Humane Care of Laboratory Animals.
The following mutants used in this study have been previously reported Tbx1Cre (Huynh et al., 2007), R26R (Soriano, 1999), and Tbx1− (Lindsay et al., 2001). The COET transgenic mouse line was generated by injecting into blastocyst transgenic mouse embryonic stem cells as detailed below. Founders were backcrossed into the C57/Bl6 strain and maintained in a mixed genetic background C57/Bl6, 129SvEv. COET transgenic mice were crossed with Tbx1+/− mice to generate COET;Tbx1+/− mice for use in timed matings. COET;Tbx1+/− mice were crossed with Tbx1Cre/+ to generate embryos of the appropriate stage and genotype for use in further analyses. Embryos were genotyped with primers spanning the junction of the 3′ end of neopolyA cassette and the 5′ end of the Tbx1 cDNA: (1) 5′-TATCATGTCTGGATCCACTAG-3′ and (2) 5′-ACTGCAGCGCACGGATCGTA-3′.
The COET construct was generated as follows: a full length Tbx1 cDNA (GeneBank accession number AF326960) was cloned 3′ to a loxP-flanked neomycin resistance cassette and 3 polyadenylation-sites included into the pBALNLXGFP backbone vector (a kind gift of Drs. A Simeone and F. Tuorto, Institute of Genetics and Biophysics, Naples). The construct was linearized and electroporated into feeder-free E14Tg2A.4 embryonic stem cells (strain 129/Ola, BayGenomics). Cells were selected with G418, and 40 resistant clones were screened by southern blot. We used two probes: (i) a Tbx1 exon 1-specific probe (350bp) used on an EcoRI restriction digest (to establish that the transgene had integrated in its entirety) and (ii) a probe specific for the neomycin-resistance gene to identify clones with single integration events of the transgene. Of the 2 positive clones, one, FFCOET3, was selected for injection into mouse blastocysts. We obtained nine chimeric mice and of these six gave germline transmission of the COET transgene.
Morphological Analyses
Hearts and aortic arch arteries were dissected and isolated from near-term pups (E17.5, E18.5). Isolated tissues were then embedded in paraffin, sectioned and stained with Hematoxylin-Eosin according to standard protocols. Mid-gestation embryos were collected fixed and embedded in paraffin for histological analysis. Intracardiac ink injections were performed to visualize the developing pharyngeal arch arteries at E10.5. Embryos were fixed O/N at room temperature in 95:1:1 solution of ethanol:acetic acid:choloroform and subsequently cleared in a 1:1 solution of benzyl benzoate: methyl salicylate. When required, these embryos were embedded in paraffin and sectioned for further examination of vessel morphology.
Three-dimensional reconstructions were done with the AMIRA 4.1.2 software (Mercury Computer Systems) using digital images of serial histological 10 μm-thick sections.
Whole mount RNA in situ hybridization with digoxigenin- labeled probes following standard protocols.
Quantitative Real-Time PCR Analysis
E10.5 embryos were used for RNA extraction after removal of the caudal half of the embryo (posterior to the forelimb bud), or from dissected hearts of E14.5 embryos. RNA was prepared using TRIzol (Invitrogen) and processed for cDNA synhesis using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). RNA was quantized by QeneQuant Pro and approximately 60 ng was used for quantitative real time PCR, using the Mx300p apparatus (Strategene). Results are shown as relative quantitation of Tbx1 compared to an internal control (Beta Actin from Applied Biosystems).
Acknowledgments
We wish to thank the transgenic core at the Institute of Biosciences and Technology, and Hedda Leeming, Guilian Li, and Wei Yu for technical support. We thank Drs. Antonio Simeone and Francesca Tuorto for providing the plasmid backbone for the COET construct. A.B. is partially supported by the Italian Telethon Foundation.
Contract grant sponsor: NIH-NHLBI; Contract grant number: HL064832; Contract grant sponsor: EU grant; Contract grant number: AnEUploidy; Contract grant sponsor: American Heart Association; Contract grant number: 0465133Y
LITERATURE CITED
- Aggarwal VS, Liao J, Bondarev A, Schimmang T, Lewandoski M, Locker J, Shanske A, Campione M, Morrow BE. Dissection of Tbx1 and Fgf interactions in mouse models of 22q11DS suggests functional redundancy. Hum Mol Genet. 2006;15:3219–3228. doi: 10.1093/hmg/ddl399. [DOI] [PubMed] [Google Scholar]
- Baldini A. Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev. 2005;15:279–284. doi: 10.1016/j.gde.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Baldini A. The 22q11.2 deletion syndrome: A gene dosage perspective. Sci World J. 2006;6:1881–1887. doi: 10.1100/tsw.2006.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CB, Wenning JM, Lu MM, Epstein DJ, Meyers EN, Epstein JA. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev Biol. 2004;267:190–202. doi: 10.1016/j.ydbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004;131:5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- Huynh T, Chen L, Terrell P, Baldini A. A fate map of Tbx1 expressing cells reveals heterogeneity in the second cardiac field. Genesis. 2007;45:470–475. doi: 10.1002/dvg.20317. [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, Epstein JA, Brown MC, Adams J, Morrow BE. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13:1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- Lindsay EA. Chromosomal microdeletions: Dissecting del22q11 syndrome. Nature Rev Genet. 2001;2:858–868. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Botta A, Jurecic V, Carattini-Rivera S, Cheah Y-C, Rosenblatt HM, Bradley A, Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379– 383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Min Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, St Jore B, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler PJ, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio- facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain [letter] Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Morishima M, Taddei I, Lindsay EA, Baldini A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum Mol Genet. 2002a;11:915–922. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Taddei I, Morishima M, Meyers EN, Lindsay EA, Baldini A. A genetic link between Tbx1 and Fibroblast Growth Factor signaling. Development. 2002b;129:4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S-i, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet. 2008;17:150–157. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]