Abstract
Qualitative and quantitative analysis of phosphorylation continues to be both an important and challenging experimental paradigm in proteomics-based research. Unfortunately researchers face difficulties inherent to the optimization of complex, multivariable methods and their application to the analysis of rare and often experimentally intractable phosphorylated peptides. Here we describe a platform based on manipulation of magnetic beads in a 96-well format that facilitates rapid evaluation of experimental parameters required for enrichment of phosphopeptides. Optimized methods provided for automated enrichment and subsequent LC-MS/MS detection of over 1000 unique phosphopeptides (~1% FDR) from 50 μg of cell lysates. In addition we demonstrate use of this platform for identification of phosphopeptides derived from proteins separated by SDS-PAGE and visualized near the detection limit of silver staining.
Keywords: Quantitative proteomics, mass spectrometry, LC-MS, MS/MS phosphorylation, phosphoproteomics
INTRODUCTION
Techniques that target phosphorylated peptides and proteins continue to proliferate in the proteomics field.1, 2, 3, 4–8 In addition there are many reports that detail various refinements to existing phosphopeptide enrichment protocols; often these describe incremental improvements via modification of resin type, buffer composition, pH, ionic strength, and binding capacity.9–12 The careful control required for success with these multi-variable methods likely reflects a high degree of overlap in the physio-chemical properties of phosphopeptides and their unmodified counterparts. Analysis of phosphopeptides is further exacerbated by well-recognized hurdles associated with the low stoichiometry of phosphorylation in biological systems; in fact these experiments are often performed in low-throughput mode in order to minimize peptide losses related to excessive sample manipulation and non-specific binding to active surfaces of tubes, pipettes, and other apparatus. Moreover, recent literature contains conflicting details regarding specific parameters for optimal performance of phosphopeptide enrichment protocols. For example, several studies4, 11, 13 have reported that conversion of peptide carboxyl groups to their corresponding methyl esters significantly improves selectively for enrichment of phosphopeptides via Fe-IDA IMAC resins; however, an equal body of work, for example Nuhse et al.14 indicated satisfactory results, seemingly under similar experimental conditions, in the absence of methylation.
These observations present a challenge for researchers who wish to compare methods or optimize a published protocol for their own specific application. Thorough evaluation is labor and time intensive, and will likely require relatively large quantities of biological material in order to sample a representative cross-section of phosphorylated peptides. Clearly, analytic platforms that facilitate rapid and reproducible evaluation of experimental variables would be a welcome addition to the phosphoproteomics toolbox. Toward this end, we sought to develop a simple and robust platform for high-throughput exploration of experimental parameters for phosphopeptide enrichment. Our approach is based on automated manipulation of magnetic beads; we found that they facilitated rapid comparison of metal ion, chelating moiety, buffer composition, and sample clean-up conditions. Our results demonstrate that this robotic platform provides for high selectivity, automated enrichment of phosphopeptides from a range of sample sources, including silver stained gel bands and cell lysates.
EXPERIMENTAL METHODS
Materials
Magnetic Ni-NTA-agarose conjugates were obtained from Qiagen (Valencia, CA). Magnetic Ni-IDA-agarose was purchased from Novagen (Gibbstown, NJ). Acetonitrile, EDTA, FeCl3, GaCl3, CuCl2, ZnCl2, AlCl3, ZrOCl, angiotensin I, glu fibrinopeptide, and alpha casein were obtained from Sigma-Aldrich (St. Louis, MO). Trifluoroacetic acid and guanidinium hydrochloride (8M solution) were obtained from Pierce (Rockford, IL).
Cell Culture and preparation of digested lysate
K562 cells were cultured in RPMI 1640 media supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C in 5% CO2. Aliquots of ~5e7 cells were harvested by centrifugation during log phase. After washing twice with 20 mL phosphate buffered saline, the pellet was lysed with 3 mL of 8M urea, 100 mM ammonium bicarbonate, and 30 μL each of Sigma-Aldrich phosphatase inhibitor cocktails I and II. Protein concentration was determined using the Bradford Assay (Bio-rad laboratories, Hercules, CA). Proteins were reduced by adding dithiothreitol (DTT) to a final concentration of 10 mM and incubating for 30 minutes at 60 °C, and alkylated with iodoacetamide (final concentration 20 mM) for 30 minutes in the dark at room temperature. Excess iodoacetamide was quenched by the addition of DTT to a final concentration of 20 mM. This solution was diluted to a final volume of 12 mL in 0.1M ammonium bicarbonate. Trypsin (150 μg, 1:50 enzyme:substrate) was added and digestion was performed at 37 °C overnight. The resulting peptide solution was acidified with 10% TFA, and desalted on a C18 solid phase extraction cartridge. Unless noted otherwise, 25% acetonitrile with 0.1% TFA was used for peptide elution from C18. Eluted peptides (400 μg) were lyophilized by vacuum centrifugation and stored at −80 °C.
Solution Digestion of alpha casein
Alpha casein (400 μg), dissolved in 50 mM ammonium bicarbonate, was added to 20 μg of trypsin (Promega, Madison, WI), and incubated overnight at 37 °C.
In Gel Digestion of Model Proteins
Alpha and beta caseins were dissolved in 100 mM triethylammonium bicarbonate (pH ~8) and stock solutions were made at varying concentrations with LDS loading buffer containing beta mercaptoethanol, and denatured for 10 minutes at 70C. Amounts ranging from 1 μg to 5 ng were loaded onto a NuPAGE 4–12% Bis-Tris 1.5 mm gel. Electrophoresis was performed at 200V (constant voltage) for approximately 50 minutes. Silver staining and in-gel digestion was performed according to established procedures,15 except that after digestion, 2 pmol of enolase digest (Waters, Milford, MA) was added as a carrier, and gel pieces were extracted twice with 50 μL of 80% MeCN/0.1% TFA. Pooled extracts were dried to ~5 μL by vacuum centrifugation and were immediately processed (see below).
Bead Preparation
Ni-NTA and Ni-IDA agarose were supplied as 5% and 50% bead suspensions, respectively. Large aliquots of beads (1 mL Ni-NTA and 100 μL Ni-IDA) were washed 3x with 800 μL water, and treated with 800 μL of 100 mM EDTA, pH 8.0 for 30 minutes with end-over-end rotation. EDTA solution was removed, and beads were then washed 3x with 800 μL water, and treated with 800 μL of 10 mM aqueous metal ion solutions for 30 minutes with end-over-end rotation. After removing excess metal ions, beads were washed 3x with 800 μL water, and resuspended in 1:1:1 acetonitrile:methanol:0.01% acetic acid for aliquotting into 96-well plates for automated phosphopeptide enrichment.
Automated Phosphopeptide Enrichment from Cell Lysates
To enrich phosphopeptides from whole cell extracts, a series of 8 96-well plates (KingFisher shallow 96-well plates, ThermoFisher Scientific, San Jose, CA) were prepared: (1) beads plate; (2) beads wash plate; (3) sample plate; (4) wash plate 1; (5) wash plate 2; (6) wash plate 3; (7) elution plate; (8) tip plate. The beads plate contained metal ion activated NTA (50 μL of the 5% suspension) or IDA beads (5 μL of the 50% suspension) in 200 μL of 1:1:1 acetonitrile:methanol:0.01% acetic acid. The beads wash plate and wash plates 1–3 contained 200 μL of 80% acetonitrile with 0.1% TFA, formic acid, or acetic acid, depending on the experiment. The sample plate contained peptides (100 μg) derived from K562 cells, resuspended in 200 μL of the same buffer used to wash the beads. The elution plate contained 50 μL of 1:1 acetonitrile/1:20 ammonia:water. Elution plates were pre-washed with 200 μL of acetonitrile for 20 minutes before adding the elution buffer. The KingFisher magnetic bead processor (ThermoFisher Scientific, San Jose, CA) was programmed to perform a bead pick up (bind time 1 min, speed medium, 5 collections), and a bead wash (wash time = 1 min, speed medium, 5 collections) before capture of phosphopeptides (release time = 1 min, speed = slow; wash time = 30 min, speed = slow; collections = 5). Beads were then sequentially washed in wash plates 1–3 (release time = 1 min, speed = slow; wash time = 1 min, speed = slow; collections = 5) before phosphopeptide elution (wash time = 1 min, speed = bottom very slow, 5 collections). For most experiments, eluates were transferred to an autosampler plate (96 well PCR plate, Sarstedt, Newton, NC), dried to 5–10 μL by vacuum centrifugation, acidified with 10% TFA, reconstituted to 30 μL with 0.1% TFA, and transferred to a 96-well plate (Sarstedt, Newton, NC). Half (15 μL, corresponding to 50 μg or 5E5 c.eq.) of each sample was analyzed by automated LC/MS as described below. In the experiments intended to test the reproducibility of replicate injections from the same or independent KingFisher enrichments, or the effects of freeze/thaw cycles, eluates were transferred to an autosampler plate, dried to 5–10 μL by vacuum centrifugation, acidified with 10% TFA, reconstituted to 25 μL with 0.1% TFA, and transferred to a 96-well autosampler plate (Sarstedt). Ten μL, corresponding to 40 μg or 4E5 c.eq. of each sample was analyzed by automated LC/MS.
Automated Phosphopeptide Enrichment from Gel Bands
Enrichment of phosphopeptides from in-gel digests was performed as described above, except that the beads plate contained 10 μL of Fe3+-NTA beads in 200 μL 1:1:1 acetonitrile:methanol:0.01% acetic acid, the sample plate contained extracted peptides in 50 μL 80% MeCN/0.1% TFA, and the elution plate contained 50 μL of elution buffer with carriers (1:1 acetonitrile/1:20 ammonia:water with 1.5 mM EDTA, 25 mM guanidinium HCl, 50 fmol/μL [glu-1] fibrinopeptide B, 50 fmol/μL angiotensin I) that was prewashed for 20 minutes with 100 μL of the same buffer. Elutions were transferred to autosampler plates, dried by vacuum centrifugation to 5 μL, acidified with 10% TFA, and reconstituted to a volume of 17.5 μL. The entire sample was analyzed by automated LC/MS as described below.
Automated Phosphopeptide Enrichment of Alpha Casein
Enrichment of phosphopeptides from solution digested alpha casein was performed as described above, with the exception that the beads plate contained 20 μL of Fe3+-NTA beads in 200 μL 1:1:1 acetonitrile:methanol:0.01% acetic acid, the sample plate contained 10 pmol alpha casein digest in 50 μL 80% MeCN/0.1% TFA, and the elution plate contained 50 μL of elution buffer without carriers (to facilitate direct spotting for MALDI analysis). Elutions were transferred to an autosampler plate, dried by vacuum centrifugation to 5 μL, and reconstituted to 20 μL with 0.1% TFA. An amount corresponding to 500 fmol (2 μL) was spotted on an Opti-TOF 384 well plate, and 1 μL of matrix was added (5 mg/mL HCCA in 70% acetonitrile, 0.1% TFA with 120 μg/mL diammonium citrate). Additionally, aliquots corresponding to 125 fmol (0.5 μL) of rows C and H were spotted, and 0.5 μL of matrix was added. After drying, the samples were analyzed by MALDI (see below).
LC/MS Analysis of Phosphorylated Peptides
Samples were loaded onto a precolumn (100 μm I.D.; packed with 4 cm POROS 10R2, Applied Biosystems, Framingham, MA) at a flow rate of 4 μL/min for 15 minutes using a NanoAcquity Sample Manager (20 μL sample loop) and UPLC pump (Waters, Milford, MA). After loading, the peptides were gradient eluted (1–40% B in 60 minutes for lysate analysis; 1–40% B in 40 minutes for gel band analysis; A=0.1% aqueous formic acid, B=0.1% formic acid in acetonitrile) at a flow rate of ~100 nL/min to an analytical column (50 μm I.D. packed with 12 cm Monitor 5 μm C18 from Column Engineering, Ontario, CA), and introduced into an LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific, San Jose, CA) by electrospray ionization (spray voltage = 2200V). The mass spectrometer was programmed to operate in data dependent mode, such that the top 10 (for experiments analyzing cell lysate) or top 8 (for experiments analyzing gel bands) most abundant precursors in each MS scan (detected in the Orbitrap mass analyzer, resolution = 60,000) were subjected to MS/MS (CAD, electron multiplier detection, collision energy = 35%, isolation width = 3.0 Da, threshold = 10,000). Dynamic exclusion was enabled with a repeat count of 1 and a repeat duration of 30 seconds.
MALDI-MS and MS/MS analysis of phosphorylated peptides
Samples were analyzed using a 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA) in reflectron mode averaging 1500 laser shots in a random, uniform pattern (30 sub-spectra, pass or fail, 50 shots/sub-spectrum) with a laser intensity of ~3700. MS/MS experiments were performed in reflectron mode averaging 5000 laser shots in a random uniform pattern (100 sub-spectra, pass or fail, 50 shots/sub-spectrum) with CID gas on and the precursor mass window set to relative with a value of 200 (FWHM).
Database Searching
Orbitrap data files were directly accessed and converted to .mgf using in-house software.16 Files were searched using Mascot version 2.2.1 against a human protein subset of the NCBI nr database (for experiments that utilized K562 extract) or a database of 12 protein standards that included alpha and beta casein (for experiments enriching phosphopeptides from these model proteins). Search parameters specified a precursor ion mass tolerance of 25 ppm, a product ion mass tolerance of 0.8 Da, fixed carbamidomethylation (C, +57 Da), and variable deamidation (NQ, −1 Da), oxidation (M, +16 Da), and phosphorylation (STY, +80 Da). False discovery rates (FDR) were evaluated by performing a search with a reverse database and were calculated using the formula: estimated FDR = reverse database identifications/forward database identifications.
Safety Considerations
Gallium chloride reacts violently with water. Preparation of aqueous solutions of Ga3+ therefore requires the careful, slow addition of the chloride to water in a fume hood with proper safety equipment. Trifluoroacetic acid is corrosive, and should be handled in a fume hood with appropriate protective equipment as described in the manufacturer’s material safety data sheet.
RESULTS AND DISCUSSION
Instrument platform
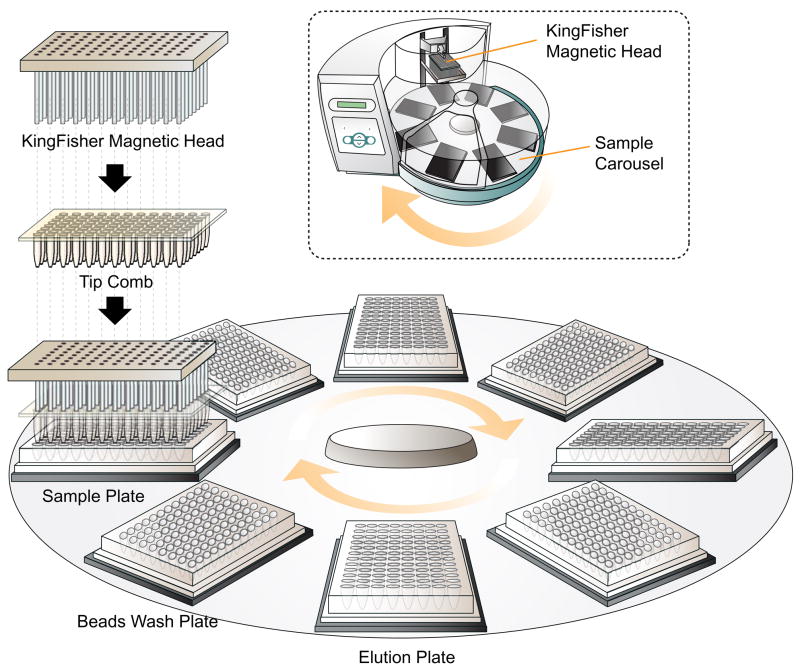
Our high-throughput platform is based on a KingFisher (ThermoFisher Scientific) magnetic bead processor that provides for automated manipulation of magnetic beads. Figure 1 shows a schematic view of the instrument and general principle of operation. Briefly, a rotary deck allows an array of 96 magnetic pins to access and transfer paramagnetic beads between wells of up to 8 plates without manual intervention. User-defined parameters allow construction of flexible methods to accommodate a range of sample preparation tasks. In our experience to date we have found that well-to-well transfer of beads is very efficient, even for working volumes of 50 μL per well. The KingFisher was originally designed for processing of DNA and RNA,17 and has recently been employed for protein and peptide profiling in serum18 and CSF19, 20, 21. Given the experimental challenges inherent to phosphoproteomics, we speculated that the KingFisher may facilitate evaluation and development of protocols for phosphopeptide enrichment.
Figure 1.
(A) Schematic illustration of the KingFisher magnetic bead processor. (B) An array of 96 magnetic pins is used on conjunction with disposable tips to transfer beads between 96-well plates on a rotary platform. The tip comb serves to mix samples when the pins are disengaged.
Evaluation of metal and chelator on peptide yield and specificity
We began with an evaluation of phosphopeptide enrichment as a function of metal ion and chelator moiety. Here again there is some apparent conflict in the literature. For example, Nuhse et al.14 reported that the performance of IDA-Fe was superior to that of NTA-Fe, while recent work from Tsai et al.12 demonstrated that in fact the latter combination provided for very high enrichment specificity. We screened a combination of 6 different metal ions (Fe3+, Ga3+, Al3+, Zn2+, Cu2+, ZrO2+) and two chelators (IDA and NTA), in each case keeping the reconstitution and wash buffers constant. Table 1 shows that IDA provided relatively poor selectivity for each metal/chelator pair analyzed, while the combination of NTA with either iron or gallium yielded a higher number of identified phosphopeptides, and improved enrichment specificity. Interestingly, a significant fraction of peptides identified after enrichment with NTA-Ga3+ were multiply-phosphorylated, suggesting that a combination of NTA-Ga3+/Fe3+ may provide for identification of a more diverse set of phosphopeptides than either used alone. Our results provide compelling evidence that the use of NTA with either iron or gallium provides for greater phosphopeptide selectivity and yield as compared to IDA charged with any of the other metals tested. Importantly, these enrichments were performed in parallel on a 96-well plate and completed in ~45 minutes.
Table 1.
Peptides identified (MASCOT score > 30) and phosphopeptide specificity as a function of metal ion and chelator moiety.
| Chelator | Metal | No. Peptides ID’d | No. Phospho- peptides ID’d | Selectivity | No. Multiply Phosphorylated Peptides ID’d | % Multiply Phosphorylated Peptides ID’d |
|---|---|---|---|---|---|---|
| NTA | Fe | 633 | 589 | 93% | 27 | 4.6% |
| NTA | Ga | 282 | 255 | 90% | 65 | 26% |
| NTA | ZrO | 869 | 62 | 7.1% | 2 | 3.2% |
| NTA | Zn | 12 | 1 | 8.3% | 0 | 0% |
| NTA | Cu | 18 | 8 | 44% | 5 | 63% |
| NTA | Al | 84 | 29 | 35% | 1 | 3.5% |
| IDA | Fe | 935 | 66 | 7.1% | 7 | 11% |
| IDA | Ga | 183 | 78 | 43% | 8 | 10% |
| IDA | Al | 914 | 52 | 5.7% | 0 | 0% |
| IDA | Cu | 317 | 82 | 26% | 5 | 6.1% |
| IDA | ZrO | 509 | 189 | 37% | 24 | 13% |
| IDA | Zn | 281 | 88 | 31% | 9 | 10% |
Peptide yield and specificity as a function of pH
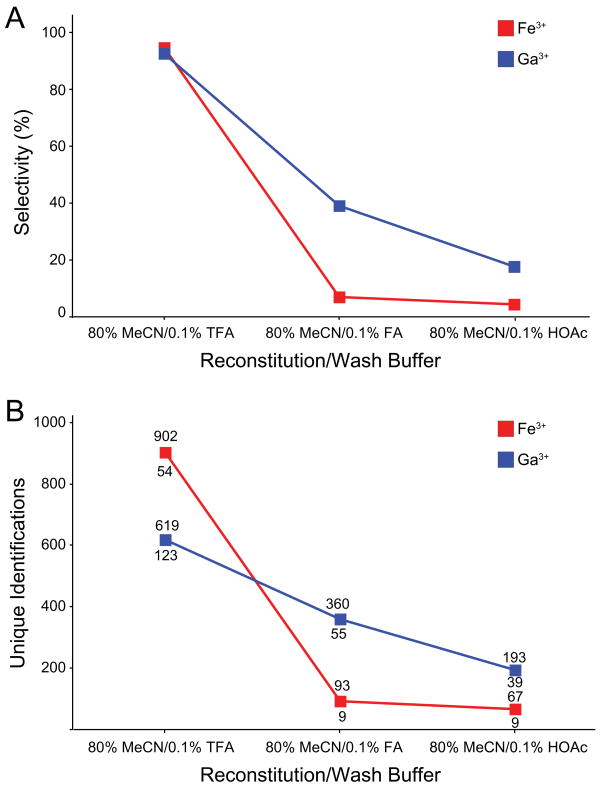
We11 and others12 have shown that overall performance of phosphopeptide enrichment is dependent upon the composition of sample loading, wash, and elution buffers. In particular, careful adjustment of solution pH is crucial for maintenance of protonated carboxyl groups, relative to ionized phosphate groups, and hence minimizes binding by non-phosphorylated peptides. Unfortunately estimation of optimum pH is complicated by the heterogeneous nature of tryptic peptides derived from biological sources, and thus the variation of pI for each acidic (and basic) side chain in a given peptide. To demonstrate the utility of the magnetic bead processor we used NTA-Ga3+/Fe3+ for automated enrichment of phosphopeptides derived from human myeloid K562 cells, under three different combinations of reconstitution and wash buffers. Figure 2a shows that enrichment specificity increases dramatically as the pH is reduced from ~3.5 (0.1% acetic acid) to ~1.5 (0.1% trifluoroacetic acid), for both Ga3+- and Fe3+-charged NTA resin, in agreement with a recent report in which phosphopeptides were enriched by use of NTA-Fe3+ immobilized on silica.12 Similarly (Figure 2b), the total number of unique phosphopeptide sequences varied inversely with solution pH. Based on these data and those in Table 1, we speculate that NTA-Ga3+ exhibits improved selectivity (as compared to NTA-Fe3+) at elevated pH due to its propensity to bind multiply-phosphorylated peptides, which may in-turn correlate with weaker binding of carboxylate groups.
Figure 2.
(A) Phosphopeptide selectivity and (B) identification (MASCOT score >30) as a function of sample reconstitution and wash buffers for Fe3+ and Ga3+-NTA based enrichment. Numbers above and below data points (B) indicate totals for all phosphopeptides (top) and multiply phosphorylated peptides (bottom) identified, respectively.
Phosphopeptide yield and specificity as a function of reversed phase fractionation
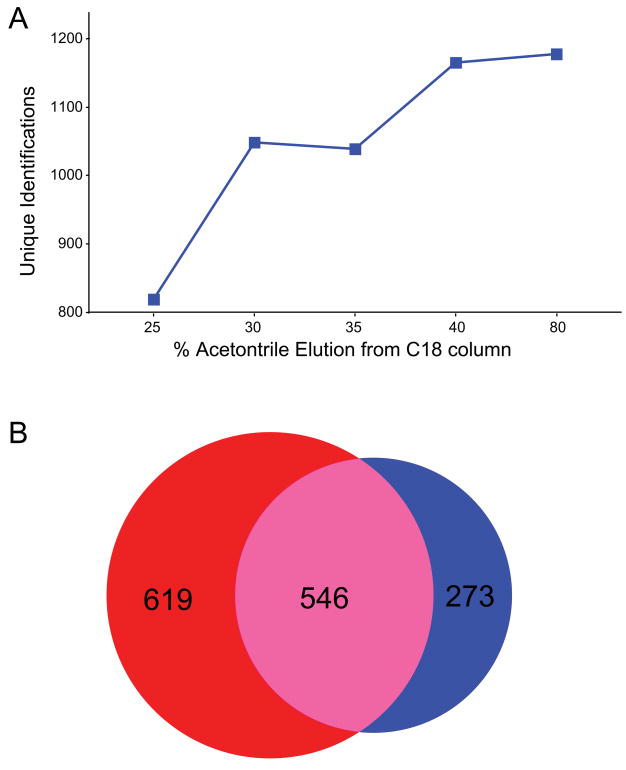
Reversed phase is commonly employed as a concentration step prior to phosphopeptide enrichment. In addition to removing salts that may interfere with phosphopeptide binding, this clean-up step may be used to fractionate complex mixtures, or reduce the instantaneous peptide load presented to the enrichment resin. However, the percentage of acetonitrile used to elute peptides from the reversed phase media can have a strong influence on subsequent steps; low concentrations may not elute all phosphopeptides, while higher concentrations may elute larger and more hydrophobic peptides that could interfere with phosphopeptide binding and selectivity. To study this parameter, we performed reversed phase clean-up of tryptic peptides derived from K562 cells in a 96-well SPE plate. Peptides from equal aliquots of cell lysate were eluted with varying concentrations of acetonitrile, and then subjected to automated NTA-Fe3+ phosphopeptide enrichment on the KingFisher platform. Figure 3A shows that the higher-organic fractions (corresponding to 40% and 80% acetonitrile elutions) contain nearly 45% more unique phosphopeptide sequences as compared to the 25% acetonitrile elution. Moreover, the improved yield was accompanied by only a modest decrease in specificity (94% to 89%). A Venn diagram (Figure 3B) representing the overlap of unique phosphopeptide sequences identified in the 25% and 40% fractions demonstrates that a large majority of the phosphopeptides detected in the 25% fraction are also detected in the 40% fraction. These results suggest higher organic elutions can be utilized for increased phosphoproteome coverage without sacrificing enrichment specificity.
Figure 3.
(A) Number of unique phosphopeptides identified (MASCOT score >30; ~1% FDR) as a function of acetonitrile concentration utilized for elution during reversed phase sample clean-up. (B) Venn diagram illustrates overlap in phosphopeptide identifications for the 25% and 40% organic elutions.
Platform Reproducibility
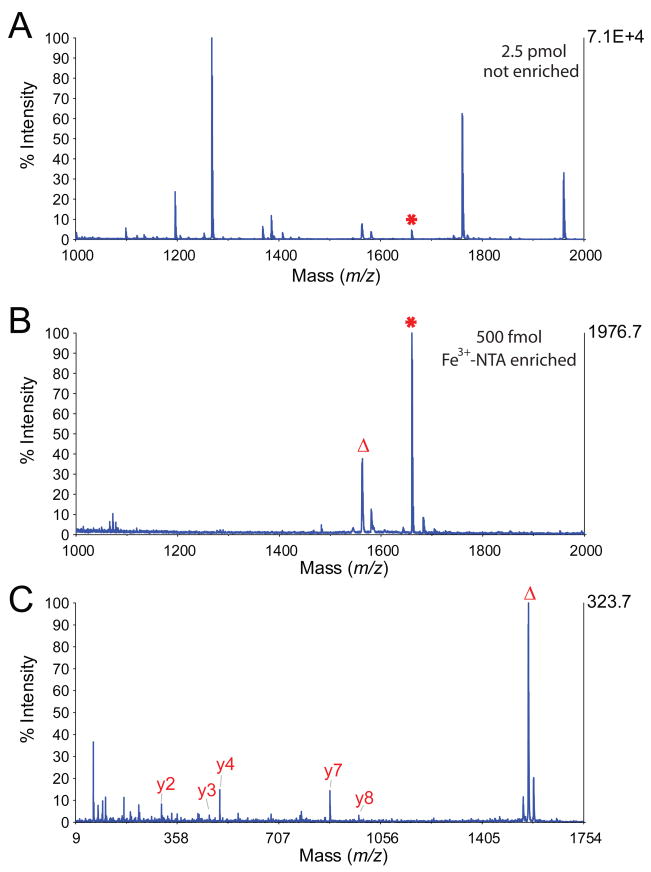
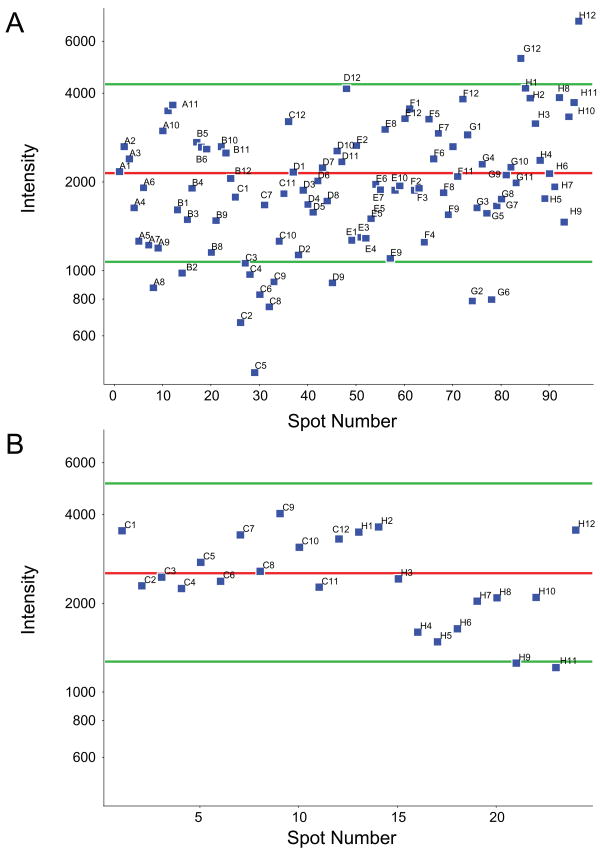
Parallel enrichment of phosphopeptides in a 96-well format provides the potential for high throughput processing, for example to support identification of phosphorylated peptides as biomarkers of disease onset, clinical prognosis, or drug efficacy. To establish basic analytical figures of merit for reproducibility, we first explored parallel enrichment across a 96-well plate. A tryptic digest of alpha casein was distributed in aliquots of 10 pmol per well. Enrichment of phosphopeptides was performed on the KingFisher with eluates corresponding to 500 fmol from each well analyzed by MALDI. Figure 4 shows representative spectra for input (A), post-enrichment (B) and MS/MS of the phosphopeptide VPQLEIVPNpSAEER (C). Figure 5A shows precursor signal intensity for VPQLEIVPNpSAEER in each of the 96 spots. We observed poor crystallization in several spots along Row C. Otherwise, we observed that only 7 spots exhibited precursor signal intensity that varied more than 2-fold relative to the mean value of ~2100 counts. To verify that the deviation in signal intensity observed in Row C was primarily related to MALDI, we re-crystallized the spots in Rows C and H (as a control) and then re-analyzed them by MALDI. In this experiment (Figure 5B), phosphopeptide signal intensity from only 2 of 24 wells deviated by more than 2-fold from the mean intensity value. These results indicated that the KingFisher platform provides for reproducible and parallel enrichment across a 96-well plate.
Figure 4.
MALDI mass spectra of tryptic α-casein peptides before (A) and after (B) enrichment by Fe3+-NTA using the KingFisher platform. (C) MS/MS spectrum of the phosphopeptide VPQLEIVPNpSAEER observed at m/z 1660.8. *, peak corresponding to VPQLEIVPNpSAEER. Δ, peak corresponding to phosphate loss.
Figure 5.
(A) MALDI-MS signal intensities for VPQLEIVPNpSAEER observed in 96 separate Fe3+-NTA enrichments of alpha casein digest. (B) MALDI-MS signal intensities (average of 5 experiments) for VPQLEIVPNpSAEER observed after recrystallization of target plate rows C and H. Mean intensities are depicted as a red line, while green lines denote a two-fold deviation from the mean.
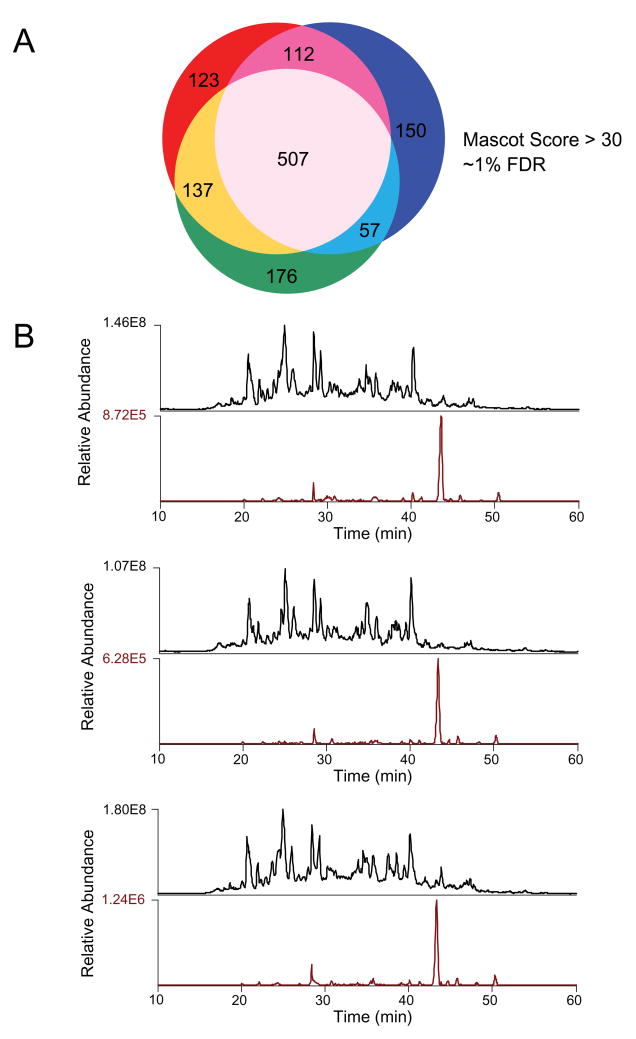
Next we examined reproducibility in the context of large-scale phosphopeptide analyses. Towards this end, we enriched phosphopeptides from 100 μg aliquots of K562 cell lysates and analyzed 50 μg of each by LC-MS/MS; we included blank gradients between each run to minimize bias associated with peptide carry-over. Retention times for commonly identified phosphopeptides were reproducible, varying by an average of only 10.7 seconds across all three analyses. In addition, the Venn diagram in Figure 6a illustrates that a high degree of reproducibility in phosphopeptide identification was observed across the replicate analyses. Out of 1262 unique phosphopeptide sequences (MASCOT score >30; FDR ~1%), 507 (40%) were observed in all three analyses, while 813 (64%) were detected in at least two analyses. For commonly detected phosphopeptides, we observed a median coefficient of variation in detected peptide (RIC) peak area of 34%. Figure 6b shows total ion chromatograms (TIC) for each LC-MS/MS run and reconstructed ion chromatograms (RIC) for the peptide ATNEpSEDEIPQLVPIGK, which exhibited a CV for peak area (33.5%) near the median value. To evaluate the relative contribution of enrichment variability versus that due to the stochastic nature of data-dependent MS/MS, we performed replicate LC-MS/MS analyses of K562 phosphopeptides derived from a single Kingfisher enrichment (i.e. from the same well), and compared these data to those derived from an independent enrichment (i.e. from a separate well). We observed (Figure 7A) a similar degree of overlap (53%) even for analysis of peptides from the same well, suggesting that unique identifications are most likely due to MS/MS undersampling rather than variability in enrichment between wells.
Figure 6.
(A) Venn diagram shows overlap in peptide identifications for triplicate analyses of phosphopeptides derived from 50 μg of K562 cell lysate. (B) Total ion chromatograms (TIC) and extracted ion chromatograms (XIC) for the phosphopeptide ATNEpSEDEIPQLVPIGK detected in 3 independent enrichments performed on the KingFisher.
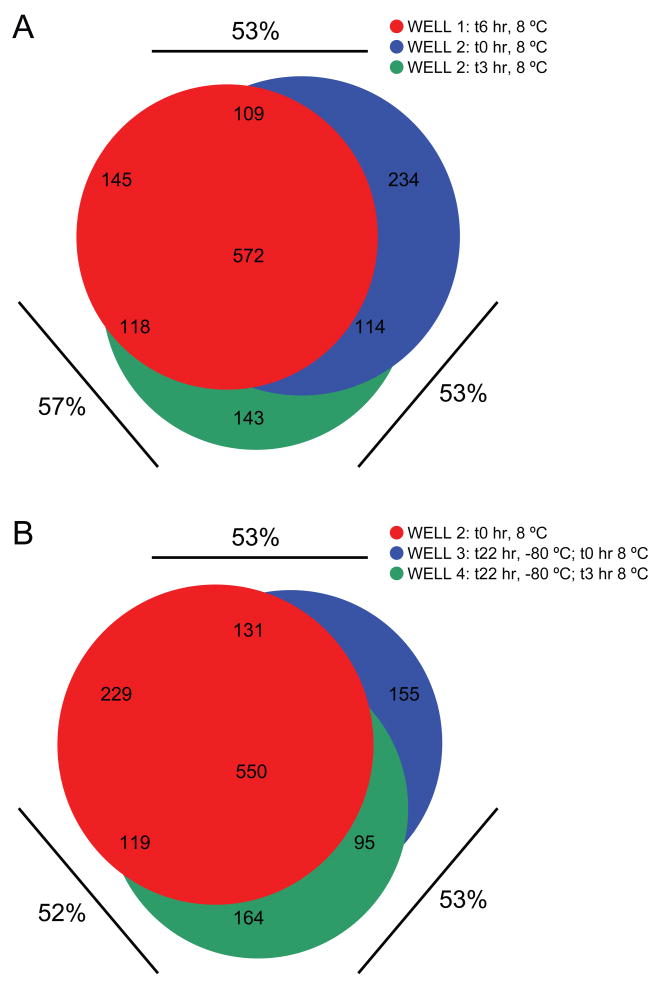
Figure 7.
Venn diagrams illustrating overlap in peptide identifications between (A) replicate analyses of the same enrichment and an independent enrichment analyzed the same day, (B) replicate analyses of enriched phosphopeptide samples that were frozen and one that was not. Percentages refer to overlap between indicated analyses.
Although the KingFisher can perform phosphopeptide enrichment from up to 96 samples in parallel, the analysis time of typical “discovery” mode LC-MS/MS analyses dictates that phosphopeptides remain in solution for extended periods of time while awaiting injection and analysis. It is easy to envision that such a scenario may lead to excessive sample losses to the plastic surfaces of 96-well plates. One potential solution would be to freeze batches of enriched samples, thawing them in appropriate numbers based on available instrument time and injection-to-injection duty cycle. As a brief test of this experimental paradigm, we performed equivalent enrichments of phosphopeptides from K562 lysate in four separate wells. Peptides from two enrichments were analyzed without further manipulation (Figure 7, WELL 1 and WELL 2), while the remaining eluates were frozen at −80 °C overnight, and analyzed the following day (Figure 7, WELL 3 and WELL 4). We observed that overall reproducibility was nearly identical for replicate analyses from the same well, or adjacent wells analyzed in series (Figure 7A), as well as between replicate analyses of wells frozen for 24 hrs., or between wells analyzed before and after one freeze-thaw cycle (Figure 7B). Collectively, our reproducibility studies demonstrate that the KingFisher coupled with nanoflow LC-MS/MS provides a reproducible platform for phosphopeptide enrichment and identification from complex biological samples. Moreover, storage of enriched samples at −80C may provide a convenient means to impedence match total throughput with available LC-MS/MS instrument capacity.
Analysis of phosphopeptides from silver stained gel bands
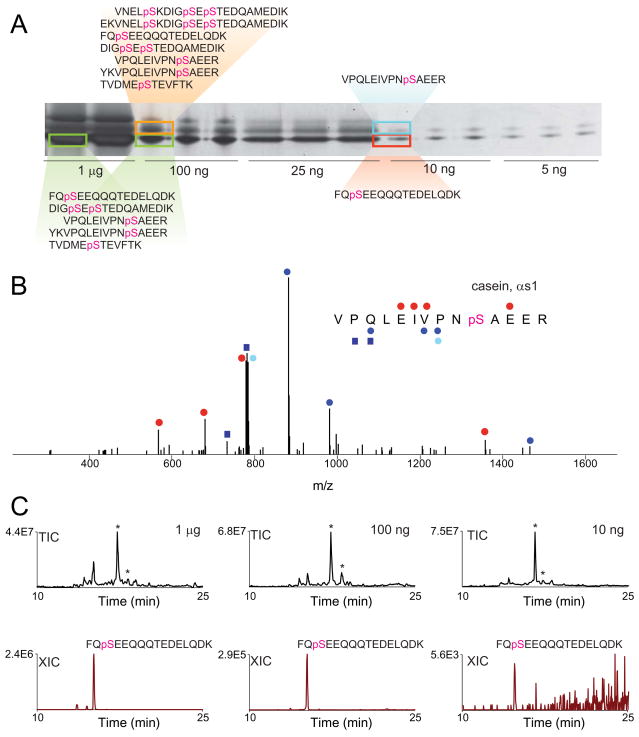
To evaluate the ability of the platform to effectively enrich phosphopeptides from quantities of protein amenable to proteomics studies (i.e. low nanogram amounts), we employed the model proteins alpha and beta casein. Protein amounts ranging from 1 μg to 5 ng were subjected to SDS-PAGE separation, with bands visualized by silver stain (Figure 8A). Several bands were excised, digested in gel with trypsin, with phosphopeptides enriched with Fe-NTA on the KingFisher. Eluted peptides were then placed in a 96 well autosampler plate and analyzed by nanoscale-LC/MS. Major phosphopeptides from both standards (FQpSEEQQQTEDELQDK from beta casein and VPQLEIVPNpSAEER from alpha casein) were detected from bands that contained as little as 10 ng of protein (Figure 5A and 5B). For example, Figure 5B shows an MS/MS spectrum for the peptide VPQLEIVPNpSAEER (MASCOT score 35), derived from analysis of 10 ng of alpha casein. The phosphorylation site was unambiguously identified. Analysis of increasing amounts of protein led to the identification of additional phosphorylation sites for alpha casein (Figure 5A), and we generally observed correspondingly higher signals for individual phosphopeptides (Figure 5C). Notably, in all experiments, only a single non-phosphorylated casein-derived peptide, FQSEEQQQTEDELQDK, was detected, indicating the selective nature of the enrichment. Taken together, these results suggest that this methodology is compatible with high-throughput determination of phosphorylation sites on proteins separated by gel electrophoresis.
Figure 8.
(A) Varying quantities of alpha and beta casein separated by SDS-PAGE and visualized by silver stain. Bands corresponding to data displayed in (B) and (C) are indicated with boxes, along with detected phosphopeptides. (B) MS/MS spectrum of VPQLEIVPNpSAEER derived from in-gel digestion and automated phosphopeptide enrichment of the 10 ng band of alpha casein shown in (A). Blue and red circles denote singly-charged y- and b-type ions, respectively, with doubly-charged fragments indicated with squares. Neutral loss of phosphate from y7 with a lighter shade. (C) Total ion chromatograms (TIC) and extracted ion chromatograms (XIC) for the peptide FQpSEEQQQTEDELQDK detected in analyses of 1 μg, 100 ng, and 10 ng of beta casein. *, identifies elution times of carrier peptides [glu-1] fibrinopeptide B and angiotensin I.
CONCLUDING REMARKS
The use of proteomics technologies for systematic production of data in support of large-scale studies requires the continued development and refinement of standardized protocols for sample preparation, data acquisition, post-processing, and data analysis. In fact a recent program sponsored by the National Cancer Institute established technology assessment centers whose mission is in-part to implement standard operating procedures and explore performance boundaries for various proteomics workflows22. Development of universally applicable methods for phosphoproteomics represents a particularly challenging case given the generally low stoichiometry of phosphorylation in biological systems and the complex, multivariable protocols typically employed for phosphopeptide enrichment. The work described herein demonstrates that use of magnetic bead-based methods in conjunction with the KingFisher platform facilitates the evaluation of various parameters for phosphopeptide enrichment in a high throughput manner. We rapidly optimized variables for metal/chelator combinations, buffer composition, and pre-enrichment peptide clean-up. In addition our data demonstrate that the KingFisher magnetic bead processor provides an economical and powerful tool for phosphopeptide identification from a wide range of samples, including cell lysates and low nanogram quantities of proteins separated by SDS-PAGE.
Acknowledgments
Generous support for this work was provided by the Dana-Farber Cancer Institute. The authors acknowledge financial support from the W.M. Keck Foundation and the National Human Genome Research Institute (P50HG004233).
References
- 1.Posewitz MC, Tempst P. Anal Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 2.Kinoshita-Kikuta E, Kinoshita E, Yamada A, Endo M, Koike T. Proteomics. 2006;6:5088–5095. doi: 10.1002/pmic.200600252. [DOI] [PubMed] [Google Scholar]
- 3.Ficarro SB, Parikh JR, Blank NC, Marto JA. Analytical Chemistry. 2008;80:4606–4613. doi: 10.1021/ac800564h. [DOI] [PubMed] [Google Scholar]
- 4.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Nat Biotech. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 5.Kweon HK, Hakansson K. Anal Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 6.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Watts JD, Aebersold R. Nature Biotechnology. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 9.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Molecular & Cellular Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Molecular & Cellular Proteomics. 2007;6:1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Ndassa YM, Orsi C, Marto JA, Chen S, Ross MM. Journal of Proteome Research. 2006;5:2789–2799. doi: 10.1021/pr0602803. [DOI] [PubMed] [Google Scholar]
- 12.Tsai CF, Wang YT, Chen YR, Lai CY, Lin PY, Pan KT, Chen JY, Khoo KH, Chen YJ. Journal of Proteome Research. 2008;7:4058–4069. doi: 10.1021/pr800364d. [DOI] [PubMed] [Google Scholar]
- 13.Kim JE, Tannenbaum SR, White FM. Journal of Proteome Research. 2005;4:1339–1346. doi: 10.1021/pr050048h. [DOI] [PubMed] [Google Scholar]
- 14.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Molecular & Cellular Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Shevchenko A, Wilm M, Vorm O, Mann M. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 16.Askenazi M, Parikh JR, Marto JA. Nat Methods. 2009;6:240–241. doi: 10.1038/nmeth0409-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makinen J, Marttila H, Viljanen MK. Elsevier Science; Bv: 2001. pp. 134–137. [Google Scholar]
- 18.Jimenez Connie R, ZEFJCKKHFAEKGGABSKWL PROTEOMICS - Clinical Applications. 2007;1:598–604. doi: 10.1002/prca.200600483. [DOI] [PubMed] [Google Scholar]
- 19.Jimenez CR, Koel-Simmelink M, Pham TV, van der Voort L, Teunissen CE. Proteomics Clinical Applications. 2007;1:1385–1392. doi: 10.1002/prca.200700330. [DOI] [PubMed] [Google Scholar]
- 20.Portelius E, Hansson SF, Tran AJ, Zetterberg H, Grognet P, Vanmechelen E, Höglund K, Brinkmalm G, Westman-Brinkmalm A, Nordhoff E, Blennow K, Gobom J. Journal of Proteome Research. 2008;7:2114–2120. doi: 10.1021/pr7008669. [DOI] [PubMed] [Google Scholar]
- 21.Portelius E, Tran AJ, Andreasson U, Persson R, Brinkmalm G, Zetterberg H, Blennow K, Westman-Brinkmalm A. Journal of Proteome Research. 2007;6:4433–4439. doi: 10.1021/pr0703627. [DOI] [PubMed] [Google Scholar]
- 22.Tao F. Expert Review of Proteomics. 2008;5:17–20. doi: 10.1586/14789450.5.1.17. [DOI] [PubMed] [Google Scholar]