Abstract
The conversion of adenosine-to-inosine within RNA transcripts is regulated by the ADAR family of enzymes. Little is known regarding the developmental expression of ADAR family members or the mechanisms responsible for the specific patterns of editing observed for ADAR substrates. We have examined the spatiotemporal expression patterns for ADAR1 and ADAR2 in mouse forebrain. ADAR1 and ADAR2 are broadly distributed in most regions of the mouse forebrain by P0, including the cerebral cortex, hippocampus, and diencephalon. High expression levels were maintained into adulthood. Co-localization studies demonstrated ADAR1 and ADAR2 expression in neurons but not astrocytes. Editing for specific ADAR mRNA targets precedes high expression of ADAR proteins, suggesting that region-specific differences in editing patterns may not be mediated solely by ADAR expression levels.
Keywords: prenatal, neuron, serotonin, inosine, adenosine, mouse, ADAR
INTRODUCTION
RNA editing is a prevalent post-transcriptional processing event that can involve the substitution or deletion of RNA sequences or the modification of specific nucleoside bases. These sequence alterations serve to generate genomic diversity in the transcriptome by producing multiple protein isoforms from a single genomic locus [1]. In this process, adenosine residues undergo a site-selective hydrolytic deamination to inosine (A-to-I editing) in pre- and mature mRNA transcripts that is catalyzed by a family of double-stranded RNA-specific adenosine deaminases referred to as ADARs [1, 2].
The mammalian ADAR family has two catalytically active family members, ADAR1 and ADAR2 [3]. Each of these enzymes contains multiple copies of a double-stranded RNA-binding motif that are required for the binding of ADARs to their target transcript(s) and an adenosine deaminase domain near the carboxyl terminus that is required for catalytic activity [4–9]. ADAR1 and ADAR2 have been shown to edit all of the identified mammalian adenosine to inosine editing sites, including transcripts encoding multiple subunits of the ionotropic glutamate receptors, i.e GluR-2, Q/R and R/G sites; GluR-3, R/G site; GluR-4, R/G site; GluR-5, Q/R site; GluR-6, Q/R, I/V and Y/C sites. The 2C-subtype of serotonin receptor (5HT2C, sites A–E), the α3 subunit of the GABAA receptor (Gabra3, I/M site) and a voltage-gated potassium channel (Kv1.1, I/M site) are also ADAR substrates [10–13].
RNA editing can alter the amino acid coding potential of the mRNA and thereby alter the function of the encoded protein product. Such changes include modulation of Ca2+ permeability (GluR-2, GluR-5, GluR-6) [14–17], alteration of recovery from receptor desensitization (GluR-2, GluR-3, GluR-4) [18], channel inactivation (Kv1.1) [12], and modulation of constitutive activity and G-protein coupling efficacy (5HT2C receptor) [19–21]. In addition to alterations in coding potential, RNA editing can also affect the structure, stability, translation efficiency and splicing patterns of edited transcripts [1, 3]. Moreover, environmental factors, such as early life stress and antidepressant drug administration, have been shown to alter the editing patterns of ADAR substrates [22, 23]. Alterations in RNA editing patterns for the 5HT2C receptor have been observed in numerous human neuropsychiatric diseases, including schizophrenia, depression and suicide [24–26], whereas a decrease in editing at the Q/R site of GluR-2 mRNAs has been associated with the selective death of spinal motor neurons in sporadic amyotrophic lateral sclerosis [27, 28].
Both ADAR1 and ADAR2 are known to be highly expressed in the brain, yet limited studies have been performed to characterize region-specific patterns of expression during development. One such study used in situ hybridization analyses to determine the developmental pattern of ADAR2 expression in the rat [29]. This study reported the earliest expression of ADAR2 in the ventral thalamus during late embryonic development, while at birth expression was still restricted to thalamic nuclei. ADAR2 expression was detected more broadly and near peak levels by postnatal day (P)21 and leveled off into adulthood with the greatest hybridization signal detected in hippocampus, olfactory bulb and thalamus. This restricted pattern of ADAR2 expression during development was quite surprising given the broad expression pattern for GluR-2 transcripts in the central nervous system and previous observations regarding the high efficiency (>99%) by which the Q/R site is edited within whole brain-derived GluR-2 mRNAs during development [14]. Recent findings by the GENSAT Consortium (http://www.gensat.org/index.html) and the Allen Brain Atlas (http://www.brainatlas.org/aba/) have suggested a more widespread distribution of the ADAR enzymes.
Developmental and regional mapping of ADAR1 in the brain have not been reported, and homozygous ADAR1-null animals die at E11.5–12.5 [30,31]; the phenotype is characterized by slowed development, an altered fetal liver structure and impaired hematopoiesis, as well as widespread apoptosis in heart, liver and vertebra. Examination of the neuroepithelium did not display any abnormalities [30] raising questions regarding what role(s) ADAR1 may play, if any, during neurological development.
There is limited data describing not only developmental regulation of ADAR enzymes, but also of editing patterns for ADAR substrates, particularly the 5HT2C receptor. Characterization of 5HT2C receptor expression in developing mice is limited to a single survey demonstrating receptor immunoreactivity as early as E10–10.5 in the mesenchyme underlying the mesencephalic flexure of the brain [32]. Immunohistochemical as well as Northern and western blotting analyses performed during rat brain development have been more extensive, indicating that 5HT2C receptor mRNA levels increase 5-fold between E17 and P27 and reach peak levels by P13 [33]; however, 5HT2C receptor protein levels demonstrate a more attenuated developmental profile, increasing only 2-fold during the same time period [33]. Thus, our current study examined the temporal and spatial regulation of ADAR1 and ADAR2 in mouse forebrain, as well as developmental regulation of RNA editing for multiple ADAR substrates. Our data suggest that ADAR1 and ADAR2 are expressed broadly across nearly all brain regions by P0. Furthermore, these enzymes appear to be expressed in neurons, but not in glial cells, supporting a potential role for RNA editing in neuronal development across a wide variety of forebrain regions.
EXPERIMENTAL PROCEDURES
Mice
129S6 male mice (Taconic) from developmental stages E15, E19, P0, P21, P100 (plug day=E0, n=6), were used for this study. To determine the gender of embryonic and early postnatal animals, a PCR-based strategy was developed to detect the genes encoding Smcy and its X-chromosome homolog Smcx. Both Y and X chromosome homologs are widely expressed in all male tissues and SMCX escapes X-inactivation in females [34]. PCR amplification using primers (sense: 5′-CCGCTGCCAAATTCTTTGG-3′, antisense: 5′-TGAAGCTTTTGGCTTTGAG-3′) designed to amplify both X- and Y-copies generated amplicons of 290 and 330 bp, respectively [35].
Immunohistochemistry
At E15, dams were deeply anesthetized with sodium pentobarbital and embryos were obtained by Caesarean section. Whole embryonic heads were immersion-fixed in 4% paraformaldehyde. For postnatal ages, mice were deeply anesthetized with sodium pentobarbital and transcardially perfused with 4% paraformaldehyde. Brains were removed and postfixed overnight at 4°C. All brains were immersed in a series of sucrose (10%, 20%, 30%) solutions prior to sectioning for immunohistochemistry. Coronal sections were cut on a freezing microtome at 40 μm, or on a cryostat (20 μm, E15 only), and stained using modifications of previously published protocols [36, 37]. Briefly, sections were treated with 0.1 M glycine (pH 7.4) to reduce nonspecific labeling, blocked in 4% nonfat dried milk, 0.2% Triton-X 100 in PBS (pH 7.4) and incubated at 4°C for 3 days with antibodies directed against ADAR1 [guinea pig polyclonal; 1:1000, [38]] and ADAR2 [sheep polyclonal; 1:500, [11, 39]], respectively. Following five washes in blocking solution at room temperature, sections were incubated with biotinylated anti-mouse, anti-rabbit, anti-sheep or anti-guinea pig IgG (Jackson, 1:1000) for 60 min. Avidin-biotin amplification (Vectastain ABC Standard) and 3-3′-diaminobenzidine reactions were used to visualize labeled proteins. Sections from different ages were processed in parallel to minimize variability across developmental time points. Adjacent sections were stained with 0.5% cresyl violet to aid in anatomical identification of regions of interest. Negative controls, in which primary antibodies were omitted, revealed no specific labeling. Background labeling of blood vessels was observed in the E15-derived samples.
Immunofluorescent techniques were used for colocalization analyses where mouse monoclonal antibodies directed against either NeuN (Chemicon MAB377; 1:100), glial fibrillary acidic protein (Chemicon MAB360; 1:250), or S100β (DakoCytomation Z0311, 1:4000) were used in combination with guinea pig anti-ADAR1 and sheep anti-ADAR2 antisera; appropriate Cy2- and Cy3-labeled secondary antibodies were utilized (1:1000, Jackson). Sections were mounted onto gelatin/poly-L-lysine-coated slides, dehydrated, coverslipped with DPX (EMS, Hatfield PA) and imaged using an Axioplan II microscope and Axiocam HR (Carl Zeiss, Thornwood, NY). Images were digitally captured using Axiovision software (version 4.1) and exported as TIFF files. Images were merged in Adobe Photoshop where brightness was adjusted, as necessary. Fields within the cerebral cortex and hippocampus were collected for cell counting in a subset of the adult animals (n=3–4 animals per immunohistochemistry condition).
RNA characterization
At each developmental age, mice (n ≥ 6) were rapidly sacrificed, and whole brain or brain regions (adult only: choroid plexus, cortex, hippocampus, hypothalamus, olfactory bulb, striatum) were immediately frozen in liquid nitrogen. RNA was isolated and purified using the PerfectPure RNA tissue kit (5 PRIME, Gaithersburg, MD) according to the manufacturer’s instructions.
For isolation of specific brain regions, dissections were performed as described in Glowinski and Iversen [40]. Briefly, a coronal cut was made on the ventral surface of the brain at the level of the optic chiasm (Bregma ~−0.3 mm). The region generated from this first cut was used to harvest cortex and striatum; olfactory bulb was removed, and striatum was separated from cortex with corpus callosum and lateral ventricles defining the boundaries. The residual white matter was removed, and remaining tissue was defined as cortex. Choroid plexus was removed from both lateral ventricles and the third ventricle. In the remaining tissue sample, the hippocampus was removed, and then a second coronal cut was made in front of the cerebellum (Bregma ~3.2 mm). Hypothalamus was separated from thalamus using the anterior commissure and mammillary bodies as reference boundaries.
To determine 5HT2C receptor editing patterns during development, a Pyrosequencing™ (Biotage AB) strategy was utilized with individual cDNA clones at each age, as described [41]. To determine the extent of site-selective editing for other ADAR substrates, first-strand cDNA was synthesized and amplified using gene-specific primers and assessed using a modified primer-extension analysis [42]. The extension products were resolved by denaturing polyacrylamide gel electrophoresis and quantified using phosphorimager analysis (GE Healthcare).
To quantify levels of ADAR1 and ADAR2 mRNA expression, first-strand cDNA was synthesized as described above and subjected to real-time PCR analysis using Taqman-based probes (Applied Biosystems); all primers andprobes were products of Assay-On-Demand from Applied Biosystems (ADAR1, assay ID Mm00508001_m1; ADAR2, assay ID Mm00504621_m1). Eukaryotic 18S rRNA (product no. 4319413E; Applied Biosystems) was included in each multiplex PCR as aninternal control. Real-time PCR and subsequent analysis were performed with an ABI Prism 7900HT sequence detection system (SDS v2.3; Applied Biosystems). Quantification of target gene expression in all samples was normalized to 18S rRNA expression by the equation CT(target) − CT(18S) =ΔCT, where CT is the threshold cycle number. Differences between each developmental time point, including individual variation, were calculated by the equation ΔCT(individual age) − ΔCT(mean adult) = ΔΔCT. Changes in target gene expression (n-fold) in each sample were calculated by 2−(ΔΔCT) from which the means and standard errors of themean (SEM) were derived.
Statistical analyses
All statistical analyses were performed using GraphPadPRISM (GraphPad Software, Inc.). Values are reported as mean ± SEM, and post-hoc P values of <0.05 were considered significant.
RESULTS
ADAR1 and ADAR2 Transcripts
ADAR1 and ADAR2 mRNA levels were determined over a developmental time course in whole mouse brain using quantitative real-time RT-PCR (qPCR). Overall, the steady-state levels of ADAR1 and ADAR2 transcripts increased gradually during development. At E15, expression of each of these mRNAs was relatively low, and expression continued to increase for each ADAR-encoding mRNA species through adulthood (P100; fig. 1).
Fig. 1. Temporal regulation of ADAR1 and ADAR2 expression.
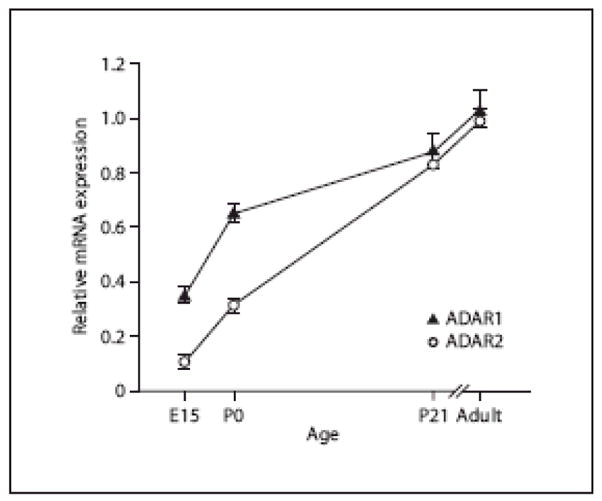
ADAR1 and ADAR2 mRNA expression in whole mouse brain over a developmental time course was determined (mean ± SEM) by qPCR. Expression levels of each ADAR were normalized to the adult mRNA level and varied significantly in developing mouse brain by one-way ANOVA (p<0.001). Dunnett’s post-hoc tests revealed that each age differed significantly from adult ADAR levels (p<0.01).
To further assess the expression of ADAR1 and ADAR2 transcripts in dissected brain regions from adult mice, we again used qPCR to quantify steady-state mRNA levels in frontal cortex, hippocampus, hypothalamus, olfactory bulb, striatum, thalamus and choroid plexus; values from dissected brain regions were normalized to whole brain expression levels (fig. 2). ADAR1 displayed relatively low levels of expression in the choroid plexus, frontal cortex and hippocampus (p<0.001 for each region as compared to whole brain; fig. 2A). In contrast, ADAR1 appeared to be relatively enriched in the hypothalamus and thalamus (fig. 2A), while ADAR1 levels in the olfactory bulb and striatum were comparable to those from whole brain.
Fig. 2. Region-specific expression of ADARs in the adult mouse.
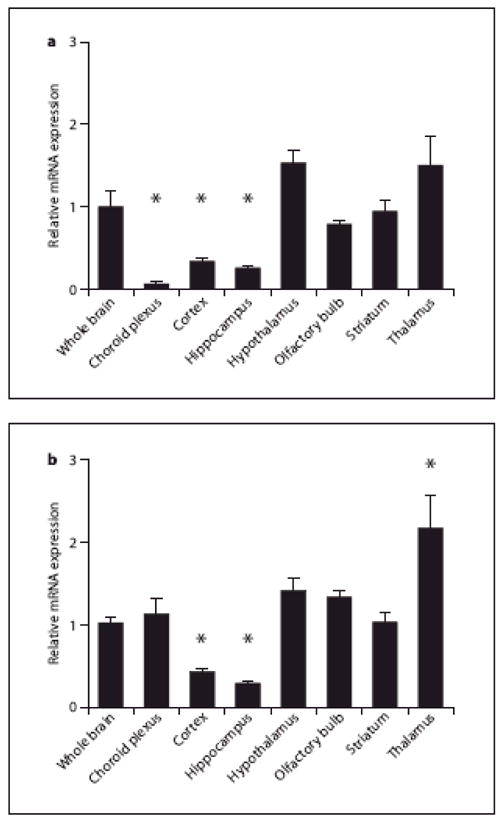
The relative expression of ADAR1 (A) and ADAR2 (B) mRNA was quantified by qPCR in mouse whole brain, and in dissected brain regions. For each region, values are expressed as mean ± SEM and normalized to the expression value observed in whole brain samples. A) Choroid plexus, cortex and hippocampus ADAR1 mRNA levels are significantly different that whole brain mRNA (individual t-test, Bonferroni post-hoc analyses). B) Cortical, hippocampal and thalamic ADAR2 transcripts are significantly different than whole brain transcripts (individual t-test, Bonferroni post-hoc analyses), *: p<0.001.
The general expression pattern for ADAR2 in most brain regions was similar to that for ADAR1, with the lowest relative expression levels observed in frontal cortex and hippocampus (Fig. 2B). ADAR2 mRNA expression was the highest in the thalamus, with a two-fold increase relative to whole brain (fig. 2B). ADAR2 expression in the choroid plexus, hypothalamus, olfactory bulb and striatum was comparable to that of whole brain.
ADAR1 and ADAR2 Protein
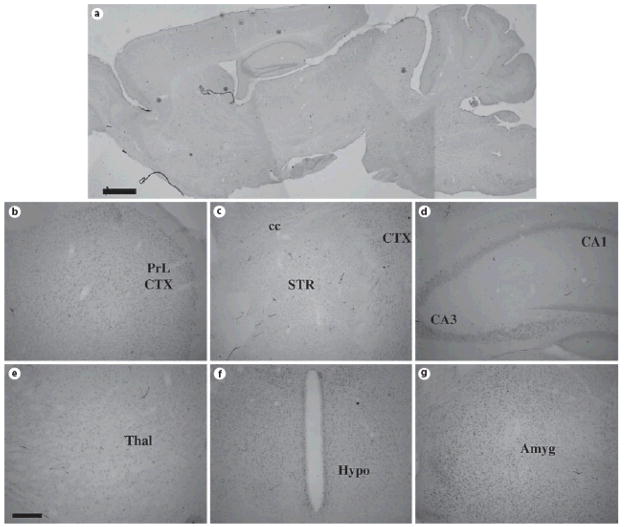
To examine the spatial pattern of ADAR1 and ADAR2 protein expression, we used an immunohistochemical strategy with previously characterized ADAR-specific antisera [11, 38, 39]. In adult mice, expression of both ADAR1 and ADAR2 protein was detected throughout the forebrain (fig. 3, 4). ADAR1 labeling was prominent in the nuclei of cells contained within nearly all grey matter brain regions, including the cerebral cortex (fig. 3B and 3C), striatum (fig. 3C), hippocampus (fig. 3D), thalamus (fig. 3E), hypothalamus (fig. 3F) and amygdala (fig. 3G).
Fig. 3. ADAR1 protein expression in the adult mouse forebrain.
Sagittal (A) and coronal (B–G) sections of adult (P100) mouse brain were stained with specific anti-ADAR1 antisera; note the widespread expression in cell nuclei in most brain regions. Scale bars = 500 μm (A) and 200 μm (B–G). Abbreviations: PrL CTX = prelimbic cortex; cc = corpus callosum; STR = striatum; CA1 = CA1 subregion of hippocampus; CA3 = CA3 subregion of hippocampus; HYPO = hypothalamus. The sagittal section was reconstructed as overlays from images captured at higher power in Adobe Photoshop, thus resulting in different apparent levels of background on different areas of the section.
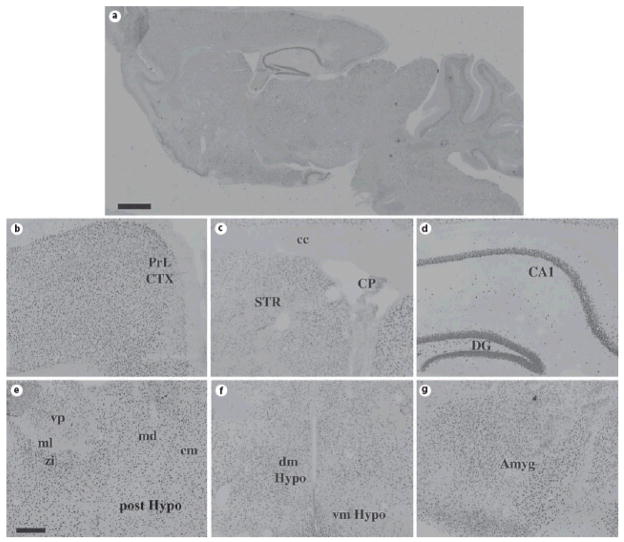
Fig. 4. ADAR2 protein expression in the adult mouse forebrain.
Sagittal (A) and coronal (B–G) sections of adult (P100) mouse brain were stained with specific anti-ADAR2 antisera; note the widespread expression in cell nuclei in most brain regions. Scale bars = 500 μm (A) and 200 μm (B–G). Abbreviations: PrL CTX = prelimbic cortex; cc = corpus callosum; STR = striatum; CP = choroids plexus; CA1 = CA1 subregion of hippocampus; DG = dentate gyrus of hippocampus; zi = zona incerta; ml = medial lemniscus; md = mediodorsal thalamic nucleus; vp = ventroposterior thalamic nucleus; dm Hypo = dorsomedial hypothalamus; vm Hypo = ventromedial hypothalamus; post Hypo = posterior hypothalamic nucleus; Amyg = amygdala. The sagittal section was reconstructed as overlays from images captured at higher power in Adobe Photoshop, thus resulting in different apparent levels of background on different areas of the section.
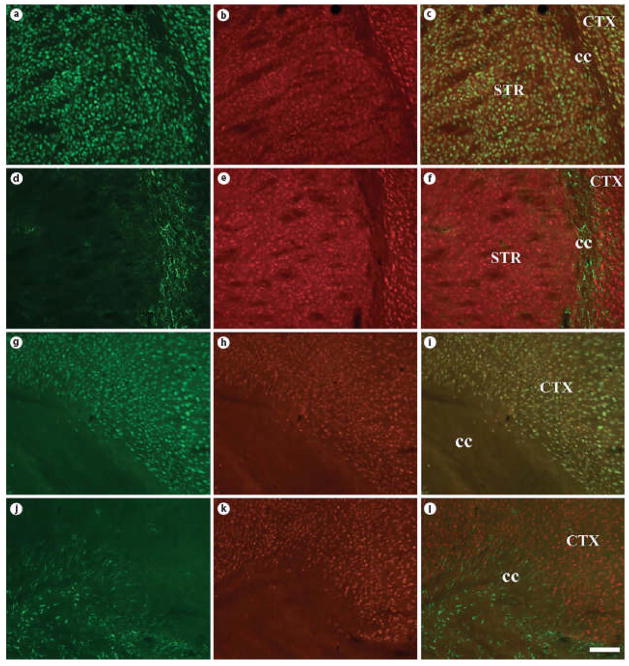
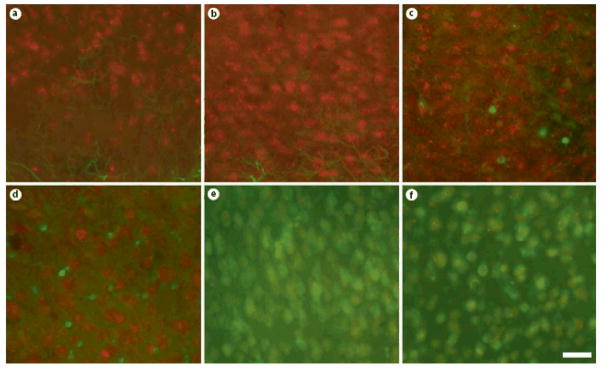
Even more intense labeling was observed using antisera derived against ADAR2 (fig. 4) in nearly all forebrain regions examined. Qualitatively, both ADAR1- and ADAR2-immunoreactivity appeared to be restricted to neurons, but expressed ubiquitously across neuronal subtypes. Colocalization studies using specific markers of neurons (NeuN) and astrocytes (GFAP, S100β) confirmed these findings [fig. 5 (low power) & 6 (high power)]. We also quantified the extent of colocalization in two brain regions, the cerebral cortex and hippocampus. The vast majority of ADAR1- and ADAR2-immunoreactive cells were positive for NeuN (97.6% ± 1.58% and 99.0% ± 0.333%, respectively). In contrast, very few ADAR1- and ADAR2-immunoreactive cells were positive for GFAP (0.513% ± 0.230% and 0.550% ± 0.268%, respectively) or S100β (1.32% ± 0.178% and 3.02% ± 1.41%, respectively). Similar results were obtained in the hippocampus, with NeuN detected in the majority of the ADAR1- and ADAR2-immunoreactive cells (96.9% ±0.515% and 92.6% ± 2.03%, respectively), but very little co-expression of ADAR1 or ADAR2 with GFAP (0% ± 0% and 1.67% ± 1.67%, respectively) or S100β (3.67% ± 1.41% and 6.00% ± 1.79%, respectively).
Fig. 5. ADAR1 and ADAR2 protein colocalizes with NeuN, but not GFAP in the adult mouse forebrain.
Coronal sections of adult (P100) mouse brain were stained with specific antibodies against NeuN (A, G), GFAP (D, J), ADAR1 (B, E), and ADAR2 (H, K). Overlayed images (C, F, I, L) demonstrate colocalization of ADAR1 and ADAR2 with NeuN, but not GFAP. Scale bar = 200 μm.
We then assessed ADAR1 and ADAR2 protein expression developmentally, examining labeling patterns at E15, P0 and P21. At E15, the earliest stage examined, no specific immunoreactivity for ADAR2 was detected within the forebrain (fig. 7b, d, f). ADAR1-immuno-reactive cells were seen only occasionally, with no clear delineation of cell type (fig. 7a, c, e). By P0, robust expression of both ADAR1 and ADAR2 protein was detected (fig. 8, 9). For example, ADAR1 was observed within superficial and deep layers of the still developing cerebral cortex (fig. 8A), hippocampus (fig. 8B), thalamus (fig. 8D) and hypothalamus (fig. 8D). Labeling in the striatum was limited, however (fig. 8B). Nearly identical patterns of ADAR2 protein expression were observed at P0 (fig. 9). Co-localization studies again revealed expression of each editing enzyme with NeuN, but not GFAP (online suppl. fig. 1, www.karger.com/doi/10.1159/000210185). Expression patterns nearly indistinguishable from the adult were observed at P21 (fig. 10, 11), consistent with qPCR analyses of ADAR1 and ADAR2 transcripts (fig. 1).
Fig. 7. Paucity of ADAR1 or ADAR2 protein expression in E15 mouse forebrain.
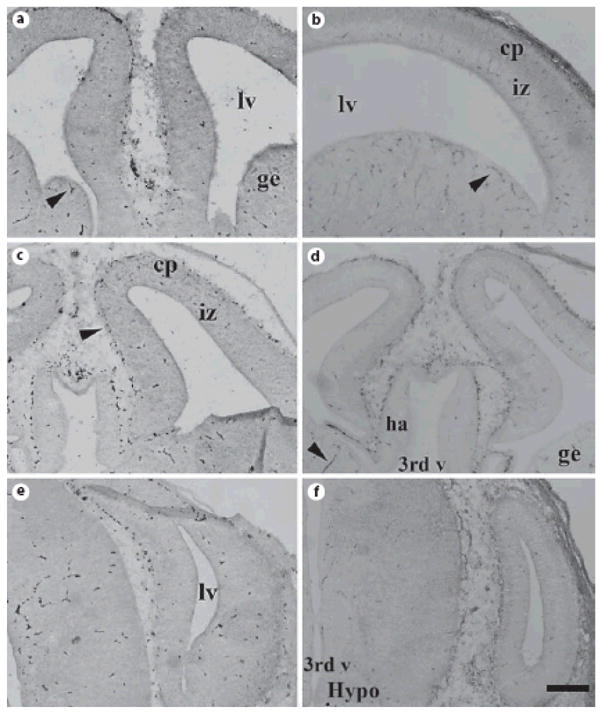
Coronal sections of E15 mouse brain were stained with specific anti-ADAR1 (A, C, E) and anti-ADAR2 (B, D, F) antisera, but no specific cellular labeling was observed. Scale bar = 200 μm.
Fig. 8. ADAR1 protein expression in mouse brain at P0.
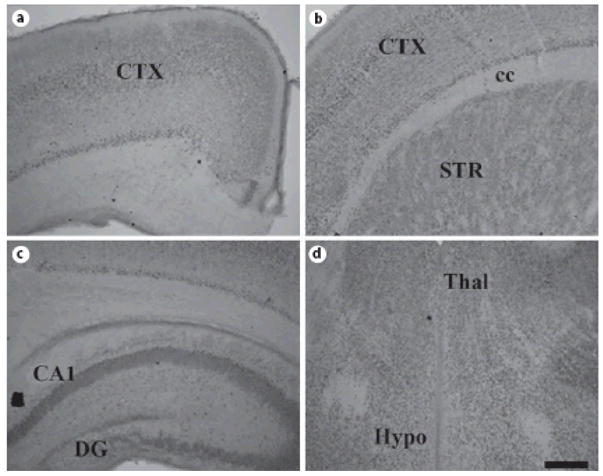
Coronal sections of P0 mouse brain were stained for ADAR1; note the widespread expression in deep and superficial layers of the still developing cerebral cortex (A, B), hippocampus (C), thalamus (D), and hypothalamus (D). Labeling in the striatum is fairly low (B). Scale bar = 200 μm.
Fig. 9. ADAR2 protein expression in mouse brain at P0.
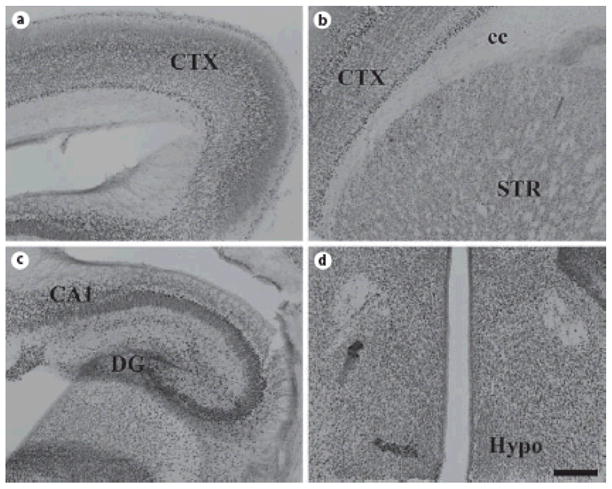
Coronal sections of P0 mouse brain were stained for ADAR2. Similar to ADAR1, widespread expression in deep and superficial layers of the still developing cerebral cortex (A, B), hippocampus (C), thalamus (D), and hypothalamus (D) is readily observed. Labeling in the striatum is fairly low (B). Scale bar = 200 μm.
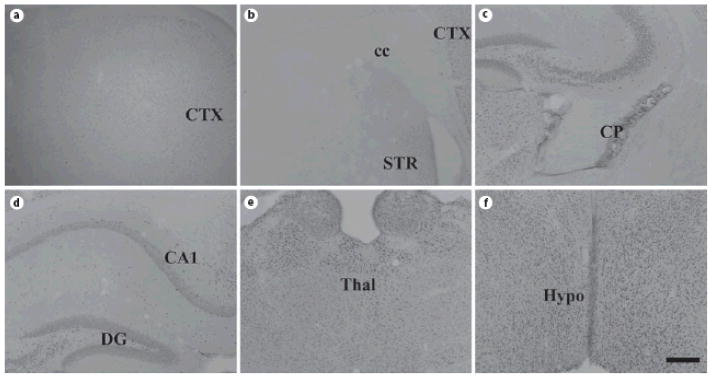
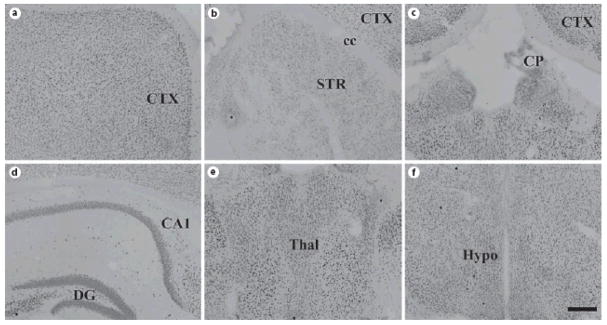
Fig. 10. ADAR1 protein expression in mouse brain at P21.
Coronal sections of P21 mouse brain were stained for ADAR1, revealing widespread expression similar to that observed in the adult. Abbreviations: CTX = cortex; cc = corpus callosum; STR = striatum; CP = choroid plexus; CA1 = CA1 subregion of hippocampus; DG = dentate gyrus of hippocampus; thal = thalamus; hypo = hypothalamus; Scale bar = 200 μm.
Fig. 11. ADAR2 protein expression in mouse brain at P21.
Coronal sections of P21 mouse brain were stained for ADAR2, revealing widespread expression similar to that observed in the adult. Abbreviations: CTX = cortex; cc = corpus callosum; STR = striatum; CP = choroid plexus; CA1 = CA1 subregion of hippocampus; DG = dentate gyrus of hippocampus; thal = thalamus; hypo = hypothalamus; Scale bar = 200 μm.
ADAR1 and ADAR2 Substrate RNAs
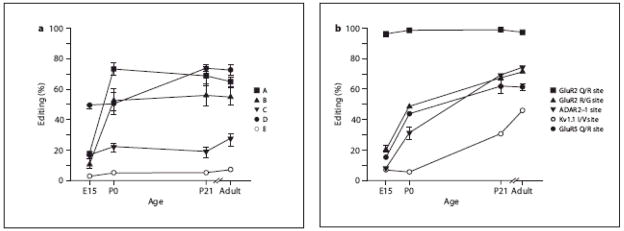
We next determined the editing patterns for several known ADAR1 and ADAR2 substrates in RNA isolated from whole brain at specific developmental time points (fig. 12). The 5HT2C receptor contains five editing sites (termed sites A–E) that can alter amino acids 157, 159 and 161, within the putative second intracellular loop of the receptor [20, 21]. These edited isoforms have distinct functional properties affecting constitutive activity and the efficacy and specificity of receptor coupling to G proteins [19, 21, 43]. When we examined the extent of RNA editing at each of the five individual sites within mouse 5HT2C receptor transcripts, most appeared to follow similar developmental patterns of editing. With the exception of the D site, the other edited sites display low levels (~10–15%) of editing at E15 and reach adult levels of editing by P0 (fig. 12a). The D site, however, was already edited in 50% of the transcripts at E15 and increased at P21, when it reached adult levels (~75%). Editing at the D site changes a genomically-encoded isoleucine codon (AUU) to a valine codon (IUU), yet this amino acid alteration has little effect on the signaling properties of the 5HT2C receptor [19].
Fig. 12. Developmental alterations in editing efficiency for multiple ADAR substrates.
(A) The extent of RNA editing for the five 5HT2C receptor editing sites (A–E) was determined by Pyrosequencing analysis from mouse whole brain transcripts isolated over a developmental time course. With exception of the D site, each site displayed low levels of editing at E15 that gradually and significantly increased to maximal levels by P0 (p<0.01), with very little increase observed between P0 and adulthood. (B) The extent of RNA editing for several other ADAR substrates was determined by primer-extension analysis in mouse whole brain transcripts isolated over a developmental time course. With exception of the GluR-2 Q/R site, which was maximally edited at the earliest age examined, each substrate displayed low levels of editing at E15 that gradually and significantly increased to maximal levels in adult animals (p<0.01).
The Q/R site within GluR-2 transcripts is edited to near completion by E15 and maintains this level of editing throughout adulthood (fig. 12b), as previously described [44]. Other substrates, including the GluR-2 R/G site, the GluR-5 Q/R site and the ADAR2 auto-editing site (−1 site) demonstrated modest editing at E15, with increases at P0 and P21, at which point the adult pattern of editing was achieved (fig. 12b). Interestingly, significant editing of the Kv1.1 I/V site appears to occur sometime after P0, substantially later than the other transcripts studied (fig. 12b).
DISCUSSION
Using multiple experimental strategies, we have examined the spatiotemporal expression patterns for ADAR1 and ADAR2 in the mouse forebrain. Quantitative RT-PCR analyses of ADAR1 and ADAR2 mRNA detected very low expression at E15, but robust and widespread expression by P0. ADAR1 and ADAR2 mRNA expression each appeared to increase steadily throughout development, with peak expression observed in adulthood. In adult animals, we examined ADAR1 and ADAR2 levels in whole brain, as well as specific brain regions, to determine whether these editing enzymes demonstrated a preferential pattern of expression. ADAR1 and ADAR2 mRNA appear to be expressed at somewhat lower levels in the cortex and hippocampus, with message levels ~50% lower than that observed in whole brain. ADAR1 transcripts are most highly expressed in the hypothalamus and thalamus. ADAR2 also has higher expression in the thalamus than in other brain regions. High levels of RNA editing for several ADAR2-specific substrates have been observed in the thalamus including the Q/R site of the GluR-5 subunit in rat (90%) [45], the D site of the 5HT2C receptor in human (85%) [46] and the I/M site of the α3 subunit of the GABAA receptor (90%) [13], suggesting that increased expression of ADAR2 in the thalamus can lead to maximal levels of RNA editing.
A previous study of ADAR2 expression using in situ hybridization with oligonucleotide probes [29] concluded that the ADAR2 transcript is not present at birth and appears gradually over the first week of life in a region-dependent manner. In contrast, our present study using quantitative RT-PCR revealed that ADAR2 is widely distributed in mouse brain at birth and is expressed in all brain regions examined where editing substrates are expressed. Our data agree with the previous study [29] in that expression was highest in the thalamus, suggesting that differences in assay sensitivity may have contributed to the observed differences between these studies. Like ADAR2, ADAR1 also shows widespread expression in the mouse brain and also suggests an extensive overlap between expression of editing substrates and the ADAR proteins. The Allen Brain Atlas and GENSAT databases present images of ADAR expression that are largely in agreement with our study, demonstrating that ADARs are widely distributed throughout the mouse forebrain by birth, with the possible exception of the striatum. The functional relevance of abundant expression of both enzymes during development is unknown, but may relate to intrinsic differences in their substrate specificity.
Further, we used double-label immunohistochemistry to determine the patterns of expression for ADAR1 and ADAR2 in neuronal and glial cells. Co-labeling studies performed with GFAP or NeuN indicated coexpression of ADAR1 and ADAR2 with NeuN. Only very limited ADAR1 or ADAR2 labeling was observed in GFAP- or S100β-positive cells, indicating that ADARs are not expressed in most glial cells in the mouse forebrain. Although we did not use a direct marker of oligodendrocytes, we did not observe ADAR1 or ADAR2 labeling in any white matter tracts (fig. 3, 4, 8, 9, 10, 11), suggesting that these ADAR proteins may be largely restricted to neurons in the mouse forebrain. In contrast, low levels of ADAR transcripts and edited glutamate receptor subunits have been previously reported in white matter from humans [46]; thus, there may be pertinent species differences.
Similar to previous reports [44, 48], we observed extensive editing of glutamate receptor transcripts prenatally. Data generated from knockout animals, single cell RT-PCR studies and in vitro editing activity assays have shown that the Q/R site is preferentially edited by ADAR2 [49,50]. Our study reveals Q/R site editing to be greater than 99% by E15 when ADAR2 mRNA is quite low and ADAR2 protein is barely detectable. Additionally, we find that the D-site of the 5HT2C receptor, a site that is preferentially edited by ADAR2 [20, 21], is edited in 50% of the transcripts by E15, highlighting another discordant relationship between ADAR2 expression and RNA editing levels. It is possible that the Q/R site of GluR-2 and D-site of the 5HT2C receptor are kinetically favored and do not require abundant ADAR2 protein for efficient deamination, yet previous in vitro competition analyses revealed that the 5HT2C receptor duplex is particularly weak in its ability to compete for ADAR2-mediated editing [51]. Alternatively, ADAR2 expression could be selectively increased in neurons expressing GluR-2 and 5HT2C receptor mRNAs; yet given the broad expression of AMPA receptors across neuronal subpopulations in the central nervous system [52], it is unclear whether spatial patterns of A-to-I conversion in the brain reflect region-specific differences in ADAR expression. By contrast, the major 5HT2C receptor mRNA isoforms in the choroid plexus are either non-edited or edited solely at the D-site to encode isoleucine, asparagine and isoleucine (INI) or isoleucine, asparagine and valine (INV) in the second intracellular loop of the receptor, respectively, a pattern consistent with low levels of ADAR1 and increased levels of ADAR2 in the adult rat and mouse brains (21; Jacobs and Emeson, unpubl obs). While no catalytic activity for ADAR3 has been observed with known ADAR substrates or synthetic dsRNAs [53, 54], it is possible that this protein may also be involved in the region-specific regulation of editing patterns given its robust and restricted expression in the brain [53]. Previous studies have suggested that ADAR3 may interfere with the activities of ADAR1 and ADAR2 by acting as a competitive inhibitor, thus decreasing the editing of specific substrates in those regions where it is preferentially expressed (e.g. thalamus) [54].
We have demonstrated that ADARs are expressed at low levels in fetal brain, and increase gradually over time. ADAR1 mRNA has its lowest expression in choroid plexus, cortex and hippocampus, while ADAR2 mRNA is least expressed in cortex and hippocampus, with increased expression in the thalamus. We have also reported complete ADAR colocalization with NeuN, indicating that ADAR expression is restricted to neuronal populations. ADAR1 and ADAR2 expression do not necessarily correlate with the extent of editing activity for identified substrates, raising questions regarding the regulatory mechanisms that may be necessary to maintain normal spatiotemporal patterns of editing in the mammalian brain. While in vitro studies of A-to-I editing have not demonstrated a requirement for any proteins other than ADARs for the site-selective editing of specific RNA substrates, the discordance between ADAR expression levels and editing patterns in mouse brain regions suggests that additional regulatory factors may be involved in the formation of protein-RNA complexes to modulate site-selective A-to-I conversion.
Supplementary Material
Fig. 6. ADAR1 and ADAR2 protein is expressed in neurons but not glial cells.
Coronal sections of adult (P100) mouse brain were double labeled with specific antibodies against GFAP (green - A, B), S100β (green - C, D) or NeuN (green - E, F), as well as ADAR1 (red - A, C, E) or ADAR2 (red - B, D, E). High power photomicrographs demonstrate prominent colocalization of ADAR1 and ADAR2 with NeuN, but not GFAP or S100β. Scale bar = 25 μm.
Acknowledgments
Support was provided by U.S Public Health Service Grants NS33323 and MH078028 (RBE), NICHD core grant P30HD15052, and the Vanderbilt Kennedy Center. We thank Dr. Aurea Pimenta for assistance with thalamic dissections and Dr. Pat Levitt for critical reading of the manuscript.
Abbreviations
- 5HT
serotonin, 5-hydroxytryptamine
- 5HT2C
serotonin 2C receptor, HTR2C
- ADAR
adenosine deaminase that acts on RNA
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- CNS
central nervous system
- E
embryonic day
- EDTA
ethylenedinitrilotetraacetic acid
- G
guanosine
- GFAP
glial fibrillary acidic protein
- GluR-2
glutamate receptor, ionotropic, AMPA 2, GRIA2
- GluR-3
glutamate receptor, ionotropic, AMPA 3, GRIA3
- GluR-4
glutamate receptor, ionotropic, AMPA 4, GRIA4
- GluR-5
glutamate receptor, ionotropic, kainate 1, GRIK1
- GluR-6
glutamate receptor, ionotropic, kainate 2, GRIK2
- Kv1.1
potassium voltage-gated channel, Shaker-related subfamily, member 1, KCNA1
- NeuN
neuronal nuclei
- P
postnatal day
- PBS
phosphate-buffered saline
- qPCR
quantitative real-time RT-PCR
References
- 1.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 2.Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O’Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 3.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hough RF, Bass BL. Purification of the Xenopus laevis double-stranded RNA adenosine deaminase. J Biol Chem. 1994;269:9933–9939. [PubMed] [Google Scholar]
- 5.Kim KS, Teixeira SM, Kirchhoff LV, Donelson JE. Transcription and editing of cytochrome oxidase II RNAs in Trypanosoma cruzi. J Biol Chem. 1994;269:1206–1211. [PubMed] [Google Scholar]
- 6.Kim U, Garner TL, Sanford T, Speicher D, Murray JM, Nishikura K. Purification and characterization of double-stranded RNA adenosine deaminase from bovine nuclear extracts. J Biol Chem. 1994;269:13480–13489. [PubMed] [Google Scholar]
- 7.Maas S, Melcher T, Herb A, Seeburg PH, Keller W, Krause S, Higuchi M, O’Connell MA. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 8.O’Connell MA, Keller W. Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc Natl Acad Sci U S A. 1994;91:10596–10600. doi: 10.1073/pnas.91.22.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rueter S, Emeson R. Adenosine-to-inosine conversion in mRNA. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 343–361. [Google Scholar]
- 11.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 12.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nature structural & molecular biology. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 13.Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 15.Kohler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: Diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 16.Sailer A, Swanson GT, Perez-Otano I, O’Leary L, Malkmus SA, Dyck RH, Dickinson-Anson H, Schiffer HH, Maron C, Yaksh TL, Gage FH, O’Gorman S, Heinemann SF. Generation and analysis of GluR5(Q636R) kainate receptor mutant mice. J Neurosci. 1999;19:8757–8764. doi: 10.1523/JNEUROSCI.19-20-08757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egebjerg J, Heinemann SF. Ca2+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc Natl Acad Sci U S A. 1993;90:755–759. doi: 10.1073/pnas.90.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 19.Niswender C, Copeland S, Herrick-Davis K, Emeson R, Sanders-Bush E. RNA editing silences serotonin 2c receptor constitutive activity. J Biol Chem. 1999;274:9742–9751. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 20.Niswender CM, Sanders-Bush E, Emeson RB. Identification and characterization of RNA editing events within the 5-HT2C receptor. Ann N Y Acad Sci. 1998;861:38–48. doi: 10.1111/j.1749-6632.1998.tb10171.x. [DOI] [PubMed] [Google Scholar]
- 21.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2c receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 22.Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C. Modulation of serotonin 2c receptor editing by sustained changes in serotonergic neurotransmission. J Neurosci. 2002;22:10529–10532. doi: 10.1523/JNEUROSCI.22-24-10529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhansali P, Dunning J, Singer SE, David L, Schmauss C. Early life stress alters adult serotonin 2c receptor pre-mRNA editing and expression of the alpha subunit of the heterotrimeric G-protein Gq. J Neurosci. 2007;27:1467–1473. doi: 10.1523/JNEUROSCI.4632-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-HT2C receptor. Alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 25.Sodhi MS, Burnet PW, Makoff AJ, Kerwin RW, Harrison PJ. RNA editing of the 5-HT(2c) receptor is reduced in schizophrenia. Mol Psychiatry. 2001;6:373–379. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- 26.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2c receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 27.Kawahara Y, Sun H, Ito K, Hideyama T, Aoki M, Sobue G, Tsuji S, Kwak S. Underediting of GluR2 mRNA, a neuronal death inducing molecular change in sporadic ALS, does not occur in motor neurons in ALS1 or SBMA. Neurosci Res. 2006;54:11–14. doi: 10.1016/j.neures.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 29.Paupard MC, O’Connell MA, Gerber AP, Zukin RS. Patterns of developmental expression of the RNA editing enzyme rADAR2. Neuroscience. 2000;95:869–879. doi: 10.1016/s0306-4522(99)00431-5. [DOI] [PubMed] [Google Scholar]
- 30.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 32.Lauder JM, Wilkie MB, Wu C, Singh S. Expression of 5-HT(2A), 5-HT(2B), and 5-HT(2C) receptors in the mouse embryo. Int J Dev Neurosci. 2000;18:653–662. doi: 10.1016/s0736-5748(00)00032-0. [DOI] [PubMed] [Google Scholar]
- 33.Roth BL, Hamblin MW, Ciaranello RD. Developmental regulation of 5-HT2 and 5-HT1c mRNA and receptor levels. Brain Res Dev Brain Res. 1991;58:51–58. doi: 10.1016/0165-3806(91)90236-c. [DOI] [PubMed] [Google Scholar]
- 34.Agulnik AI, Mitchell MJ, Mattei MG, Borsani G, Avner PA, Lerner JL, Bishop CE. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum Mol Genet. 1994;3:879–884. doi: 10.1093/hmg/3.6.879. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez A, Fernandez R, Madrid-Bury N, Moreira PN, Borque C, Pintado B, Gutierrez-Adan A. Experimental demonstration that pre- and post-conceptional mechanisms influence sex ratio in mouse embryos. Mol Reprod Dev. 2003;66:162–165. doi: 10.1002/mrd.10345. [DOI] [PubMed] [Google Scholar]
- 36.Stanwood GD, Parlaman JP, Levitt P. Anatomical abnormalities in dopaminoceptive regions of the cerebral cortex of dopamine D1 receptor mutant mice. J Comp Neurol. 2005;487:270–282. doi: 10.1002/cne.20548. [DOI] [PubMed] [Google Scholar]
- 37.Stanwood GD, Washington RA, Levitt P. Identification of a sensitive period of prenatal cocaine exposure that alters the development of the anterior cingulate cortex. Cereb Cortex. 2001;11:430–440. doi: 10.1093/cercor/11.5.430. [DOI] [PubMed] [Google Scholar]
- 38.George CX, Wagner MV, Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem. 2005;280:15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- 39.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci U S A. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glowinski J, Iversen LL. Regional studies of catecholamines in the rat brain. I. The disposition of [3H]norepinephrine, [3H]Dopamine and [3H]Dopa in various regions of the brain. J Neurochem. 1966;13:655–669. doi: 10.1111/j.1471-4159.1966.tb09873.x. [DOI] [PubMed] [Google Scholar]
- 41.Sodhi MS, Airey DC, Lambert W, Burnet PW, Harrison PJ, Sanders-Bush E. A rapid new assay to detect RNA editing reveals antipsychotic-induced changes in serotonin-2c transcripts. Mol Pharmacol. 2005;68:711–719. doi: 10.1124/mol.105.014134. [DOI] [PubMed] [Google Scholar]
- 42.Feng Y, Sansam CL, Singh M, Emeson RB. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol Cell Biol. 2006;26:480–488. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitzgerald LW, Iyer G, Conklin DS, Krause CM, Marshall A, Patterson JP, Tran DP, Jonak GJ, Hartig PR. Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology. 1999;21:82S–90S. doi: 10.1016/S0893-133X(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 44.Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- 45.Paschen W, Dux E, Djuricic B. Developmental changes in the extent of RNA editing of glutamate receptor subunit GluR5 in rat brain. Neurosci Lett. 1994;174:109–112. doi: 10.1016/0304-3940(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, O’Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2c receptors. J Neurochem. 2000;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- 47.Kawahara Y, Ito K, Sun H, Kanazawa I, Kwak S. Low editing efficiency of GluR2 mRNA is associated with a low relative abundance of ADAR2 mRNA in white matter of normal human brain. The European journal of neuroscience. 2003;18:23–33. doi: 10.1046/j.1460-9568.2003.02718.x. [DOI] [PubMed] [Google Scholar]
- 48.Paschen W, Schmitt J, Gissel C, Dux E. Developmental changes of RNA editing of glutamate receptor subunits GluR5 and GluR6: In vivo versus in vitro. Brain Res Dev Brain Res. 1997;98:271–280. doi: 10.1016/s0165-3806(96)00193-9. [DOI] [PubMed] [Google Scholar]
- 49.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 50.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 51.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J Biol Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 52.Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 53.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. Red2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 54.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.