Abstract
Sonic Hedgehog (SHH) plays an important instructional role in vertebrate development, as exemplified by the numerous developmental disorders that occur when the SHH pathway is disrupted. Mutations in the SHH gene are the most common cause of sporadic and inherited Holoprosencephaly (HPE), a developmental disorder that is characterized by defective prosencephalon development. SHH HPE mutations provide a unique opportunity to better understand SHH biogenesis and signaling, and to decipher its role in the development of HPE. Here, we analyzed a panel of SHH HPE missense mutations that encode changes in the amino-terminal active domain of SHH. Our results show that SHH HPE mutations affect SHH biogenesis and signaling at multiple steps, which broadly results in low levels of protein expression, defective processing of SHH into its active form and protein with reduced activity. Additionally, we found that some inactive SHH proteins were able to modulate the activity of wt SHH in a dominant negative manner, both in vitro and in vivo. These findings show for the first time the susceptibility of SHH driven developmental processes to perturbations by low-activity forms of SHH. In conclusion, we demonstrate that SHH mutations found in HPE patients affect distinct steps of SHH biogenesis to attenuate SHH activity to different levels, and suggest that these variable levels of SHH activity might contribute to some of the phenotypic variation found in HPE patients.
Keywords: Hedgehog, holoprosencephaly, developmental disorder
Introduction
Sonic Hedgehog (SHH) is a morphogen that plays an important patterning role during metazoan development (Ingham and McMahon 2001). SHH is initially produced as a ~45 kDa preprotein (SHH-FL) that is comprised of a signal peptide, an amino-terminal domain (SHHN) and a carboxy-terminal domain (SHHC) (Guerrero and Chiang 2007; Ingham and McMahon 2001; Singh et al. 2006). During maturation of the active ligand the SHHC domain acts as a cholesterol transferase, catalyzing the covalent addition of a cholesterol moiety to the newly generated carboxy-terminus of SHHN (Bumcrot et al. 1995; Lee et al. 1994; Porter et al. 1996a; Porter et al. 1996b). The cholesterol modified SHHN is further palmitoylated (Pepinsky et al. 1998) at its amino-terminus by the product of the SKINNY HEDGEHOG gene (Chamoun et al. 2001; Chen et al. 2004). This dually lipid modified SHHNp is considered the active ligand, whose release from producing cells is actively regulated by the DISPATCHED gene product (Burke et al. 1999; Caspary et al. 2002; Gallet et al. 2003; Guerrero and Chiang 2007; Kawakami et al. 2002; Ma et al. 2002; Tian et al. 2005). The palmitoylated active ligand exists as a part of some extracellular multiprotein complex (Callejo et al. 2006; Gallet et al. 2006; Goetz et al. 2006; Panakova et al. 2005; Zeng et al. 2001), which is implicated in regulating SHH signaling many cell diameters away from its site of synthesis (Chuang and Kornberg 2000; Eaton 2006; Guerrero and Chiang 2007). Upon binding to its receptor PATCHED (PTCH) (Marigo et al. 1996; Stone et al. 1996), SHH relieves the PTCH mediated repression of SMOOTHENED (SMO), activating the SHH pathway through regulation of the GLI family of transcription factors (Robbins and Hebrok 2007; Ruiz i Altaba et al. 2007).
Underscoring the important role of SHH during development, deregulation of the SHH pathway is associated with a variety of developmental patterning defects. One common congenital developmental disorder that results from deregulation of SHH signaling is Holoprosencephaly (HPE), which is characterized by the incomplete separation of left and right cerebral hemispheres and various midline facial defects (Dubourg et al. 2007). HPE is prevalent in early pregnancy affecting approximately 1 in 250 conceptuses (Roessler and Muenke 2003), with a subsequent decrease in prevalence as embryonic development progresses. The expressivity of the HPE associated phenotypes vary, from spontaneous termination of the fetus in the most severe cases, to various cranio-facial abnormalities such as cyclopia and midfacial clefting, or hypotelorism in relatively less severe cases (Dubourg et al. 2007).
Both genetic and environmental factors have been implicated in the development of HPE (Dubourg et al. 2007). Twelve genetic loci (HPE 1-12) have been associated with an inherited predisposition to HPE, with mutations in SHH (HPE3) being the most commonly observed (Dubourg et al. 2007). A number of animal models supporting the important role of the SHH pathway in HPE development have been described (Chiang et al. 1996; Hardcastle et al. 1998; Ma et al. 2002; Milenkovic et al. 1999; Zhang et al. 2001). The targeted disruption of genes encoding components of the SHH pathway in mice, such as SHH (Chiang et al. 1996), DISPATCHED A (DISPA) (Ma et al. 2002), SMO (Zhang et al. 2001) and GLI2 (Hardcastle et al. 1998) results in phenotypes typically found in severe forms of HPE, exhibiting for example cyclopia. However, the phenotype of mice lacking GLI2 is quite mild, showing a single maxillary incisor; a microform of HPE. Similarly, transgenic mice engineered to over express PTCH in the presumptive forebrain, which should attenuate SHH signaling, display a fusion of lateral ventricles characteristic of HPE patients (Milenkovic et al. 1999). While these mouse models have been quite informative, there are some differences between these mouse models and human HPE. For example, humans harboring only a single functional copy of SHH exhibit HPE with varying penetrance (Belloni et al. 1996; Nanni et al. 1999; Roessler et al. 1996; Roessler et al. 1997) whereas mice that are heterozygous for SHH appear quite normal (Chiang et al. 1996).
The wide variability of phenotype associated with different SHH mutations (Dubourg et al. 2007; Hehr et al. 2004; Heussler et al. 2002; Lazaro et al. 2004; Nanni et al. 1999; Odent et al. 1999), and the occurrence of large numbers of such missense SHH mutations in the population, provide a unique opportunity to further dissect SHH biogenesis. We hypothesized that the various SHH HPE point mutations found in HPE patients would result in a disruption of distinct steps in SHH biogenesis, which might in turn contribute to the variability of outcome observed in these HPE patients. Here, we present results showing that SHH HPE mutations can affect SHH biogenesis at various steps, attenuating SHH activity via distinct mechanisms. In brief, these mutations differentially affected SHH stability, SHH processing from its full-length form, secretion into conditioned medium and its inherent activity. Furthermore, some relatively inactive SHH mutants appeared to attenuate wt SHH activity in a dominant negative manner, suggesting a mechanistic insight into the interplay of wt SHH with mutant SHH in heterozygous HPE patients. These findings suggest that mutations of SHH that result in HPE attenuate SHH activity through a variety of distinct mechanisms.
Materials and Methods
Expression and processing of human SHH mutants
All SHH mutant expression constructs were made by site directed mutagenesis of a full-length human SHH cDNA (a gift from Dr. Cliff Tabin) cloned into pcDNA3.1, using the QuickChange II kit (Stratagene Inc.) as per the manufacturer's instructions. The sequence of the mutagenic primers used for the construction of the various SHH mutants will be provided upon request. Bosc cells, a line of human embryonic kidney cells (Pear et al. 1993), were transfected with plasmids expressing mutant or wt SHH using Lipofectamine 2000 (Invitrogen Inc.). The following day, the growth medium was replaced with serum free medium (RPMI-1640). Twenty-four hours later the conditioned serum free medium (CM) was collected and cleared of cell debris by centrifugation at 3000 × g (10 min at 4°C). The expression and processing status of the various SHH mutants was analyzed by immunoblotting the cell lysate and CM with anti-SHH antibodies (H-160, Santa Cruz Biotechnology) essentially as described earlier (Goetz et al. 2006). The immunoblots were also analayzed using ImageJ software (NIH) to quantify the levels of different forms of SHH in both cell lysate and CM.
SHH activity assays
A determination of freely diffusible SHH activity was made by quantitating the ability of CM from SHH mutant expressing Bosc cells to induce alkaline phosphatase production in C3H10T½ cells (Kinto et al. 1997), by estimating alkaline phosphatase activity in the cell lysate (Nakamura et al. 1997; Zeng et al. 2001). All activity assays were done in triplicate and each experiment was repeated at least three times. The data shown is representative and the error bars represent ±S.D. Mean activity was compared between mutants using repeated measures analysis of variance, and student's two tailed t-test was used to compare each mutant to the positive control (WT).
SHH palmitoylation assay
The palmitoylation status of the various SHH mutant proteins was assessed as described earlier (Chen et al. 2004). In brief, HEK 293T cells were transfected with plasmids expressing various SHH mutants or a SHH-C24S mutant that lacked the palmitate acceptor amino acid, which served as a negative control. Twenty four hours later these cells were incubated in 1.5 ml of growth medium supplemented with ~400 μCi/ml [9,10-3H] palmitic acid (31 Ci/mmol; Perkin Elmer) for 4 hrs at 37°C. SHH was then immunoprecipitated from cell lysate using anti-SHH antibodies (H-160, Santa Cruz Biotechnology), resolved by SDS-PAGE, transferred to PVDF membrane and then exposed to film for 7 days. The same immunoprecipitate was also immunoblotted using anti-SHH antibody (H-160, Santa Cruz Biotechnology).
Chick embryo neural tube assay
The ability of SHH mutants to induce the expression of different SHH dependent target genes in the chick neural tube was determined as previously described (Schell-Apacik et al. 2003). In brief, certified pathogen free fertilized eggs from White Leghorn chicken were incubated at 38°C for 36 hours. At Hamburger and Hamilton stage 10-12 (Hamburger 1951), the lumen of the neural tube of chicken embryo was injected with plasmids expressing wt or mutant SHH along with a GFP expression construct (pCAGGS-IRES-GFP) as transfection control. Electroporation of the DNA into one side of the neural tube was carried out by applying current across the neural tube (3 pulses of 25V, 50ms, 1s interval, using a TSS20 Ovodyne electroporator). Electroporated eggs were further incubated for 24 or 48 hours, then harvested, dissected, fixed with 4% paraformaldehyde, cryoprotected in 30% sucrose containing PBS, embedded in OCT medium and cryosectioned (12-16 μm thick). The embryo sections were then probed for the expression of SHH and HNF3β using the anti-SHH mAb 5E1 or anti-HNF3β mAb 4C7 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, Iowa) respectively, or processed for in situ hybridization as previously described (Dahmane et al. 1997), for the expression of Nkx2.2 and Pax6, using previously characterized digoxigenin-labeled antisense RNA probes.
Results
SHH HPE mutations affect its processing and activity
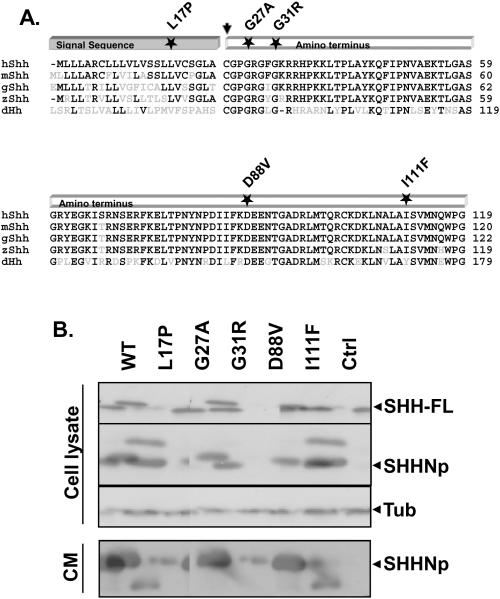
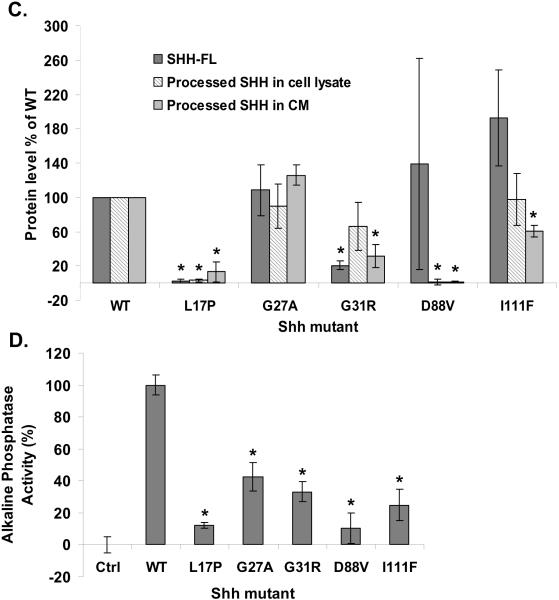
Despite the identification of a large number of SHH mutations in HPE, relatively little work has been done to characterize the effect of these mutations on SHH's biogenesis and activity (Maity et al. 2005; Schell-Apacik et al. 2003; Traiffort et al. 2004). We began our analyses of such mutations by examining the processing and activity of a set of SHH HPE mutant proteins in comparison to wt SHH (Hehr et al. 2004; Kato et al. 2000; Nanni et al. 1999; Nanni et al. 2001; Roessler et al. 1996): SHH-L17P, SHH-G27A, SHH-G31R, SHH-D88V and SHH-I111F. These mutations affect highly conserved amino acids in the amino-terminal domain of SHH, which is thought to be responsible for all SHH signaling activity (Fig. 1A). Plasmids expressing these SHH mutants or wt SHH were transfected into Bosc cells, and the expression and processing of the various forms of the mutant proteins was determined; from their full-length form (Fig. 1B, top panel and 1C), to SHHNp in cell lysate (Fig. 1B, second panel and 1C), and SHHNp in conditioned medium (Fig. 1B, bottom panel and 1C). SHH-L17P encodes a mutation within its signal peptide, which appears to severely affect the expression or intracellular stability of SHH-L17P relative to wt SHH (Fig. 1B and 1C). The processing or stability of SHH-D88V also appeared to be severely impaired, resulting in very little SHHNp in cell lysate, and consequently little SHHNp secreted into the conditioned medium relative to wt SHH. The processing of SHH-I111F was similar to wt SHH, but the secretion of this mutant protein, or its stability in conditioned medium, appeared attenuated relative to wt SHH. The expression and processing of SHH-G27A and SHH-G31R to SHHNp inside the cell was similar to wt SHH (Fig. 1B, second panel and 1C), except for the appearance of an additional higher molecular weight species of SHH-G31R, as has been observed previously (Traiffort et al. 2004). This higher molecular weight SHHG31R species in the cell lysate migrates in a manner similar to SHHN (data not shown), which is not cholesterol modified. We do not know why SHH-G31R would form such a species. The stability of SHH-G31R in conditioned medium was also compromised, resulting in what appeared to be a proteolysed faster migrating protein, as has been reported previously (Maity et al. 2005; Traiffort et al. 2004).
Figure 1. SHHHPE mutations affect its expression, processing and activity.
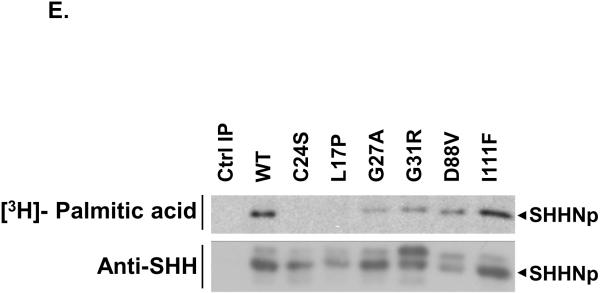
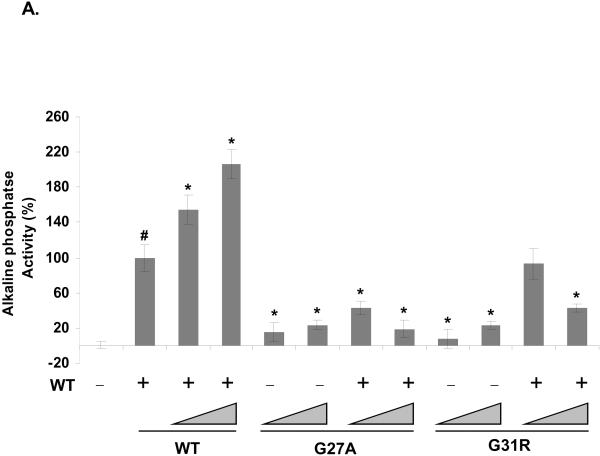
The amino-terminus of SHH is highly conserved across phyla, as shown by the multiple sequence alignment of human SHH (hSHH) with homologs found in Mus musculus (mSHH), Gallus gallus (gSHH), Danio rerio (zSHH) and Drosophila melanogaster (dHh) (A). The residues identical to that of hSHH are in black while others are in grey. For simplicity, only the signal-peptide and amino-terminal signaling domain (SHHN) are shown. The position of amino acid residues altered in SHH as a result of missense mutations in SHH, which were investigated in this paper, are marked by a star. The arrow indicates the palmitoylation site in the signaling domain after removal of the signal peptide. Bosc cells expressing wt SHH (WT), SHH-L17P (L17P), SHH-G27A (G27A), SHH-G31R (G31R), SHH-D88V (D88V), SHH-I111F (I111F) or a control (Ctrl) vector, were analyzed for the expression of SHH by immunoblotting cell lysate (top panel) for full-length SHH (SHH-FL), processed SHH (SHHNp) and tubulin (B) or conditioned medium (CM) (bottom panel) for SHHNp. Note: SHH-G31R is prone to proteolysis in the conditioned medium and the higher molecular weight species of SHH-G31R in the cell lysate migrates in a manner similar to SHHN (data not shown), which is not cholesterol modified. The immunoblots were analayzed using ImageJ software to quantify the level of different forms of SHH for indicated mutants in both cell lysate and CM (C). The protein levels are presented as the mean of three independent experiments and error bars represent ±S.D. The values for the processed form of SHH included both higher molecular weight and lower molecular weight immuno-reactive bands wherever present. The level of the different forms of SHH mutants generally differed from that of wt SHH (P values < 0.05 are indicated by *). Note: The level of SHH-FL form of SHH-D88V mutant was quite variable in different experiments for some unknown reason, as indicated by large S.D. error bar. Conditioned medium from cells expressing the indicated mutant was assayed for its ability to induce alkaline phosphatase activity in C3H10T ½ cells (D). Activity assays were done in triplicate and the error bars represent ±S.D. The activity of all the mutants was significantly different than wt SHH (P values < 0.05 are indicated by *). The palmitate modification of the indicated SHH mutants was estimated by immunoprecipitating SHH from the lysate of HEK 293T cells growing in medium supplemented with [9,10-3H] palmitic acid (E). The mutant SHH-C24S which lacked the palmitate acceptor amino acid acted as a negative control for palmitate labeling and the control immunoprecipitation (Ctrl IP) served as a control for nonspecific immunoprecipitation in this experiment. Note: Mutations near palmitate modification site affected modification more severely.
The activity of the SHH mutants in conditioned medium (Fig. 1D) generally correlated with their protein level in conditioned medium (Fig. 1B, bottom panel and 1C). However, SHH-G27A was found at a similar level as wt SHH, but was significantly less active. Similar results were obtained for SHH mutant proteins in a co-culture assay, in which the SHH mutant transfected Bosc cells were co-cultured with SHH-Light2 cells, a SHH dependent reporter cell line (Taipale et al. 2000) (Fig. S1). The palmitoylation of SHH is known to enhance its activity (Chen et al. 2004; Pepinsky et al. 1998). We investigated these SHH mutants for their differential palmitoylation status, as a possible cause for their altered activity. In general, the palmitate labeling of the mutant proteins was attenuated as compared to wt SHH though at varying degrees (Fig. 1E). The differential palmitoylation may be a contributing factor to the lower activity of some of these SHH mutants (Fig. 1D and S1). In general, the amino-terminal SHH HPE mutants highlight the importance of these residues in the processing and activity of SHH, and illustrate the many ways SHH activity may be attenuated in HPE patients.
After analyzing the above panel of SHH mutations we decided to choose SHH-G27A and SHH-G31R for further studies. The choice of SHH-G27A was based on the observation that though its secretion into conditioned medium and expression level was comparable to wt SHH (compare Fig. 1D with Fig. 1B bottom panel and 1C), its activity seemed to be significantly attenuated. On the other hand, the reduced activity of SHH-G31R in the conditioned medium, along with its apparent altered processing or stability (Fig. 1D and 1B) made it an interesting candidate for further investigation.
SHH mutants SHH-G27A and SHH-G31R have impaired activity in vivo
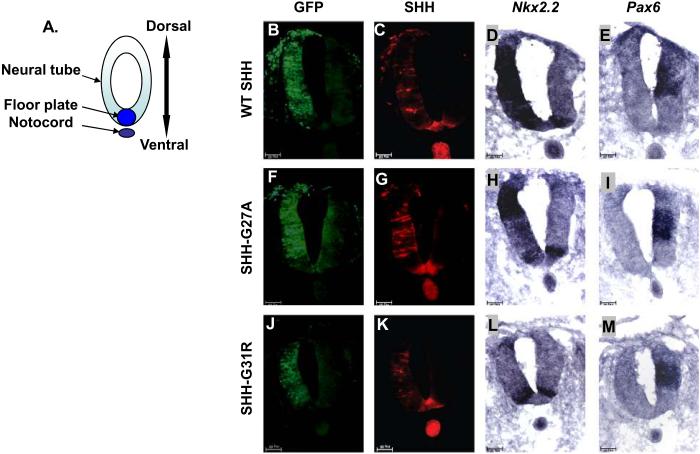
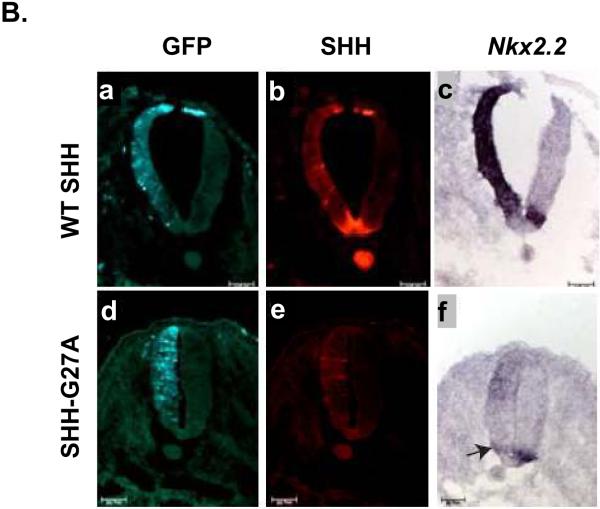
As the role of SHH in the patterning of the forebrain is complex and difficult to manipulate in the early chick embryo, we chose to compare SHH activity in the chick neural tube, where it is well defined, conserved and easier to manipulate (Bourikas et al. 2005; Goodrich et al. 1996; Itasaki et al. 1999; Schell-Apacik et al. 2003). During embryonic development, SHH, which is initially produced by the notochord and later also by the floor plate, is a major determinant of the fate of neural tube cells along the dorsoventral axis. We assessed the neural tube patterning activity of SHH mutants by electroporating SHH mutant expressing plasmids on one side of the neural tube. The untransfected contralateral side served as a negative control for these assays. The cells ectopically expressing SHH were identified by GFP expression (Fig. 2B, 2F and 2J). Ectopic expression of wt SHH in neural tube cells induced the expression of Nkx2.2, a ventral marker (Dessaud et al. 2008) (Fig. 2D), and repressed the expression of the more dorsally expressed transcription factor Pax6 (Fig. 2E). This demonstrates the ability of ectopically expressed human SHH to act functionally in chick neural tube development, as has been shown earlier (Schell-Apacik et al. 2003). We tested the activity of SHH-G27A and SHH-G31R in this neural tube differentiation assay and found that both mutants were able to induce the expression of the SHH target genes Nkx2.2 (compare Fig. 2H and 2L with 2D) and HNF3β (Fig. S2), albeit at levels lower than wt SHH. The ability of SHH-G27A and SHH-G31R to repress Pax6 expression was similar to wt SHH (compare Fig. 2I and 2M with 2E), consistent with repression of Pax6 requiring less SHH activity than activation of Nkx2.2 and HNF3β (Briscoe and Ericson 1999; Dessaud et al. 2008). These observations support our in vitro results, suggesting that SHH-G27A and SHH-G31R have reduced activity compared to wt SHH.
Figure 2. SHH-G27A and SHH-G31R have attenuated activityin vivo.
Panel (A) shows a schematic representation of the neural tube and notochord. (B-M) The activity of wt SHH, SHH-G27A and SHHG31R was determined by electroporating plasmids expressing them, along with a GFP expressing plasmid, on one side of the chick embryonic neural tube. Induction of Nkx2.2 expression and repression of Pax6 expression (dark blue) was then determined by in situ hybridization, while SHH expression was determined using anti-SHH antibodies (red). GFP expression served as an electroporation control in these experiments (light green). Electroporation of SHH-G27A (n=37) and SHH-G31R (n=27) induced Nkx2.2 expression at a lower level than wt SHH (n=52) (compare panel H, L and D to the amount of SHH expressed in panel G, K and C, respectively), but were comparable in their ability to repress Pax6 expression (wt SHH, n=45; SHH-G27A, n=29; SHH-G31R, n=22), whose repression is considered an indicator of low-level SHH activity (panel I, M and E).
SHH HPE mutants attenuate wt SHH activity in a dominant negative manner
Studies using recombinant forms of SHHN have previously suggested that some inactive forms of SHH can act in a dominant negative manner (Williams et al. 1999). We therefore hypothesized that a likely source of phenotypic variability resulting from SHH HPE mutations is that SHH mutants with attenuated activity may affect wt SHH activity in a dominant negative manner. To determine whether SHH-G27A and SHH-G31R might function in a dominant negative manner, we cotransfected different amounts of plasmids expressing SHH-G27A or SHH-G31R, with or without wt SHH (Fig. 3A), into Bosc cells and assayed the SHH activity in conditioned medium. The coexpression of these SHH mutations with wt SHH caused a dose dependent decrease in SHH activity in conditioned medium, with SHH-G27A suppressing wt SHH activity more than SHH-G31R (Fig. 3A). We also noted that expression of SHH-G27A could repress the induction of Nkx2.2 by endogenous SHH in the chick neural tube (arrow in Fig. 3B panel f). This repression of endogenous SHH activity was observed approximately 50% of the time (Table 1). Although we do not know the reason for this variation, we speculate that the relative level of SHH mutant expression, its location of expression within the neural tube, or the presence of other genetic and environmental factors may contribute to this variation (Dubourg et al. 2007; Nanni et al. 1999; Odent et al. 1999; Odent et al. 1998)). Under similar conditions, we were not able to observe the same dominant negative effect for SHH-G31R (Table 1 and data not shown), underscoring the specificity of SHH-G27A inhibitory activity. We speculate that the lack of effect of SHH-G31R on endogenous Shh activity in vivo is due to it having weaker dominant negative activity than SHH-G27A (see Fig. 3A), or that this protein is less stable in vivo than SHH-G27A. Taken together, these results suggest that SHH HPE mutants can attenuate wt SHH in a dominant negative manner.
Figure 3. SHH-G27A dominantly inhibits endogenous SHH activity.
The ability of SHH mutants SHH-G27A and SHH-G31R to affect wt SHH activity was evaluated by testing conditioned medium from Bosc cells expressing wt SHH (WT), SHH-G27A (G27A) and SHH-G31R (G31R) alone or in combination with wt SHH, at different molar ratios (1x:1x and 5x:1x respectively), for its ability to induce alkaline phosphatase activity in C3H10T½ cells (A). In this experiment, 1x corresponded to 80 ng of variant DNA and a total of 500 ng of DNA (equalized using vector DNA) was transfected into Bosc cells plated in 60 mm plates. Activity assays were done in triplicate and the error bars represent ±S.D. Both SHHG27A and SHH-G31R attenuated wt SHH activity (1x wt SHH, indicated by #) in a dose dependent manner. The activity of these mutants alone or when expressed in combination with wt SHH was significantly different from 1x wt SHH as were higher doses of wt SHH (P values < 0.05 are indicated by *). SHH-G27A also attenuated endogenous SHH activity in vivo (B). The ability of SHH-G27A to attenuate endogenous SHH in the chick neural tube was assessed by electroporating a SHH-G27A expressing plasmid (n=16), along with a GFP expressing plasmid, on one side of the chick embryonic neural tube. SHH expression was determined using anti-SHH antibodies (red). Induction of Nkx2.2 expression (dark blue) was determined by in situ hybridization. GFP expression (light-green) served as an electroporation control in these experiments. SHH-G27A attenuated the induction of Nkx2.2 expression by endogenous SHH (arrow in panel f) validating our in vitro observation that some SHH mutants can act in a dominant negative manner. Under these conditions, endogenous SHH activity was attenuated 50% (8 out of 16) of the time embryos were electroporated with SHH-G27A expressing plasmid (see Table 1).
Table 1. SHH-G27A attenuates endogenous SHH activity.
SHH HPE mutants SHH-G27A and SHHG31R were analyzed for their ability to attenuate endogenous SHH activity in the chick neural tube as described in the text. This table summarizes our in vivo results analyzing the activity of the SHH isoforms in each electroporated embryo. SHH-G27A attenuated the expression of endogenous Nkx2.2 in 50% of the electroporated embryos while SHH-G31R did not exhibit such activity under similar conditions, which may be due to its weaker dominant negative activity (see Fig. 3A).
| SHH variant expressed | Attenuation of endogenous Nkx2.2 expression (number of positive embryos) |
|---|---|
| WT SHH | 0% (0/14) |
| SHH-G27A | 50% (8/16) |
| SHH-G31R | 0% (0/7) |
Discussion
Here, we show that SHH mutations found in HPE patients are defective at distinct steps of SHH biogenesis. The SHH mutations included in this study change residues that are highly conserved among different phyla, and encode amino acids throughout SHH's amino-terminal signaling domain. Consistent with the important role of evolutionarily conserved amino acids in protein function, changes in the conserved SHH residues strongly affected its biogenesis and signaling (see Fig. 1). In general, the SHH HPE mutations resulted in low protein expression (SHH-L17P), defective processing to SHHNp (SHHD88V), decreased stability or secretion (SHH-G31R and SHH-I111F) and decreased potency (SHHG27A). Additionally, we provide evidence for the first time that a subset of SHH HPE mutants can act in a dominant negative manner to attenuate the activity of wt SHH. Combined, our results highlight the diverse mechanisms that may underlie the loss of SHH activity in HPE.
SHH is an important regulator of central nervous system (CNS) development. The haploinsufficiency of SHH in human CNS development is associated with HPE development (Belloni et al. 1996; Nanni et al. 1999; Roessler et al. 1996; Roessler et al. 1997). SHH HPE mutations are inherited in an autosomal dominant fashion, and exhibit a range of phenotypes (Nanni et al. 1999; Roessler et al. 1996). The cause of this variable penetrance or expressivity of the HPE phenotype is speculated to be linked to the presence of various modifier genes and environmental factors (Dubourg et al. 2007; Nanni et al. 1999; Odent et al. 1999; Odent et al. 1998). Although there is not a clear link between the processes affected by SHH mutations and the development of HPE, presumably due to masking by these other genetic or environmental factors (Dubourg et al. 2007; Nanni et al. 1999; Odent et al. 1998), we speculate that these various classes of SHH HPE mutations might contribute to the severity of phenotype observed in HPE patients. We suggest that relatively inactive SHH mutants (e.g. SHH-G27A) might attenuate SHH activity below that normally found in a SHH heterozygous state, giving rise to a more severe phenotype than that expected as a result of haploinsufficiency. Conversely, SHH mutants in which the processing of SHH-FL is compromised or overall production of the SHHNp form is attenuated, without affecting its activity per se, might give rise to less severe phenotypes. Based on our results with this subset of SHH HPE mutants, we suggest that in addition to modification by other genetic and environmental factors as proposed previously (see above) the weak correlation between genotype and phenotype of SHH HPE patients may also result from the various mutations linked to HPE affecting SHH biogenesis and activity via distinct mechanisms. Our work also highlights the biological insight to be gained by analyzing these naturally occurring SHH mutants, as they can provide a greater understanding of the SHH pathway itself, as well as provide a mechanistic insight into the variability of SHH associated HPE phenotype.
Supplementary Material
Acknowledgments
We are grateful to the members of the Robbins laboratory for thoughtful discussion during the course of this work. This work was supported by NIH grant GM64011 (DJR), an award from the American Lung Association/LUNGevity Foundation (DJR), a GANN pre-doctoral fellowship in nanomedicine (RT) and grants from the V foundation, The W.W. Smith Charitable Trust and the Albert R.Taxin Brain Tumor Research Center at the Wistar Insitute (ND).
References
- Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet. 1996;14:353–6. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- Bourikas D, Pekarik V, Baeriswyl T, Grunditz A, Sadhu R, Nardo M, Stoeckli ET. Sonic hedgehog guides commissural axons along the longitudinal axis of the spinal cord. Nat Neurosci. 2005;8:297–304. doi: 10.1038/nn1396. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Ericson J. The specification of neuronal identity by graded Sonic Hedgehog signalling. Semin Cell Dev Biol. 1999;10:353–62. doi: 10.1006/scdb.1999.0295. [DOI] [PubMed] [Google Scholar]
- Bumcrot DA, Takada R, McMahon AP. Proteolytic processing yields two secreted forms of sonic hedgehog. Mol Cell Biol. 1995;15:2294–303. doi: 10.1128/mcb.15.4.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–15. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Callejo A, Torroja C, Quijada L, Guerrero I. Hedgehog lipid modifications are required for Hedgehog stabilization in the extracellular matrix. Development. 2006;133:471–83. doi: 10.1242/dev.02217. [DOI] [PubMed] [Google Scholar]
- Caspary T, Garcia-Garcia MJ, Huangfu D, Eggenschwiler JT, Wyler MR, Rakeman AS, Alcorn HL, Anderson KV. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr Biol. 2002;12:1628–32. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, Basler K. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science. 2001;293:2080–4. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18:641–59. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–13. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Chuang PT, Kornberg TB. On the range of hedgehog signaling. Curr Opin Genet Dev. 2000;10:515–22. doi: 10.1016/s0959-437x(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–81. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V. Holoprosencephaly. Orphanet J Rare Dis. 2007;2:8. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S. Release and trafficking of lipid-linked morphogens. Curr Opin Genet Dev. 2006;16:17–22. doi: 10.1016/j.gde.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Gallet A, Rodriguez R, Ruel L, Therond PP. Cholesterol modification of hedgehog is required for trafficking and movement, revealing an asymmetric cellular response to hedgehog. Dev Cell. 2003;4:191–204. doi: 10.1016/s1534-5807(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Gallet A, Ruel L, Staccini-Lavenant L, Therond PP. Cholesterol modification is necessary for controlled planar long-range activity of Hedgehog in Drosophila epithelia. Development. 2006;133:407–18. doi: 10.1242/dev.02212. [DOI] [PubMed] [Google Scholar]
- Goetz JA, Singh S, Suber LM, Kull FJ, Robbins DJ. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J Biol Chem. 2006;281:4087–93. doi: 10.1074/jbc.M511427200. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–12. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- Guerrero I, Chiang C. A conserved mechanism of Hedgehog gradient formation by lipid modifications. Trends Cell Biol. 2007;17:1–5. doi: 10.1016/j.tcb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Hamburger VaH HL. A series of normal stages in the development of the chick embryo. J. Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hardcastle Z, Mo R, Hui CC, Sharpe PT. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–11. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- Hehr U, Gross C, Diebold U, Wahl D, Beudt U, Heidemann P, Hehr A, Mueller D. Wide phenotypic variability in families with holoprosencephaly and a sonic hedgehog mutation. Eur J Pediatr. 2004;163:347–52. doi: 10.1007/s00431-004-1459-0. [DOI] [PubMed] [Google Scholar]
- Heussler HS, Suri M, Young ID, Muenke M. Extreme variability of expression of a Sonic Hedgehog mutation: attention difficulties and holoprosencephaly. Arch Dis Child. 2002;86:293–6. doi: 10.1136/adc.86.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–87. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Bel-Vialar S, Krumlauf R. 'Shocking' developments in chick embryology: electroporation and in ovo gene expression. Nat Cell Biol. 1999;1:E203–7. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- Kato M, Nanba E, Akaboshi S, Shiihara T, Ito A, Honma T, Tsuburaya K, Hayasaka K. Sonic hedgehog signal peptide mutation in a patient with holoprosencephaly. Ann Neurol. 2000;47:514–6. [PubMed] [Google Scholar]
- Kawakami T, Kawcak T, Li YJ, Zhang W, Hu Y, Chuang PT. Mouse dispatched mutants fail to distribute hedgehog proteins and are defective in hedgehog signaling. Development. 2002;129:5753–65. doi: 10.1242/dev.00178. [DOI] [PubMed] [Google Scholar]
- Kinto N, Iwamoto M, Enomoto-Iwamoto M, Noji S, Ohuchi H, Yoshioka H, Kataoka H, Wada Y, Yuhao G, Takahashi HE, Yoshiki S, Yamaguchi A. Fibroblasts expressing Sonic hedgehog induce osteoblast differentiation and ectopic bone formation. FEBS Lett. 1997;404:319–23. doi: 10.1016/s0014-5793(97)00014-8. [DOI] [PubMed] [Google Scholar]
- Lazaro L, Dubourg C, Pasquier L, Le Duff F, Blayau M, Durou MR, de la Pintiere AT, Aguilella C, David V, Odent S. Phenotypic and molecular variability of the holoprosencephalic spectrum. Am J Med Genet A. 2004;129:21–4. doi: 10.1002/ajmg.a.30110. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Autoproteolysis in hedgehog protein biogenesis. Science. 1994;266:1528–37. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- Maity T, Fuse N, Beachy PA. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc Natl Acad Sci U S A. 2005;102:17026–31. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–9. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- Milenkovic L, Goodrich LV, Higgins KM, Scott MP. Mouse patched1 controls body size determination and limb patterning. Development. 1999;126:4431–40. doi: 10.1242/dev.126.20.4431. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Aikawa T, Iwamoto-Enomoto M, Iwamoto M, Higuchi Y, Pacifici M, Kinto N, Yamaguchi A, Noji S, Kurisu K, Matsuya T. Induction of osteogenic differentiation by hedgehog proteins. Biochem Biophys Res Commun. 1997;237:465–9. doi: 10.1006/bbrc.1997.7156. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Bocian M, Steinhaus K, Bianchi DW, Die-Smulders C, Giannotti A, Imaizumi K, Jones KL, Campo MD, Martin RA, Meinecke P, Pierpont ME, Robin NH, Young ID, Roessler E, Muenke M. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet. 1999;8:2479–88. doi: 10.1093/hmg/8.13.2479. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Du Y, Hall RK, Aldred M, Bankier A, Muenke M. SHH mutation is associated with solitary median maxillary central incisor: a study of 13 patients and review of the literature. Am J Med Genet. 2001;102:1–10. doi: 10.1002/1096-8628(20010722)102:1<1::aid-ajmg1336>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Odent S, Atti-Bitach T, Blayau M, Mathieu M, Aug J, Delezo de AL, Gall JY, Le Marec B, Munnich A, David V, Vekemans M. Expression of the Sonic hedgehog (SHH) gene during early human development and phenotypic expression of new mutations causing holoprosencephaly. Hum Mol Genet. 1999;8:1683–9. doi: 10.1093/hmg/8.9.1683. [DOI] [PubMed] [Google Scholar]
- Odent S, Le Marec B, Munnich A, Le Merrer M, Bonaiti-Pellie C. Segregation analysis in nonsyndromic holoprosencephaly. Am J Med Genet. 1998;77:139–43. [PubMed] [Google Scholar]
- Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–6. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepinsky RB, Zeng C, Wen D, Rayhorn P, Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski K, Taylor FR, Wang EA, Galdes A. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273:14037–45. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- Porter JA, Ekker SC, Park WJ, von Kessler DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV, Beachy PA. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996a;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996b;274:255–9. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Hebrok M. Hedgehogs: la dolce vita. Workshop on Hedgehog-Gli Signaling in Cancer and Stem Cells. EMBO Rep. 2007;8:451–5. doi: 10.1038/sj.embor.7400959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–60. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Roessler E, Muenke M. How a Hedgehog might see holoprosencephaly. Hum Mol Genet. 2003;12(Spec No 1):R15–25. doi: 10.1093/hmg/ddg058. [DOI] [PubMed] [Google Scholar]
- Roessler E, Ward DE, Gaudenz K, Belloni E, Scherer SW, Donnai D, Siegel-Bartelt J, Tsui LC, Muenke M. Cytogenetic rearrangements involving the loss of the Sonic Hedgehog gene at 7q36 cause holoprosencephaly. Hum Genet. 1997;100:172–81. doi: 10.1007/s004390050486. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–47. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell-Apacik C, Rivero M, Knepper JL, Roessler E, Muenke M, Ming JE. SONIC HEDGEHOG mutations causing human holoprosencephaly impair neural patterning activity. Hum Genet. 2003;113:170–7. doi: 10.1007/s00439-003-0950-4. [DOI] [PubMed] [Google Scholar]
- Singh S, Goetz JA, Robbins DJ. Sonic hedgehog. 2006 UCSD-Nature Molecule Pages doi:10.1038/mp.a002208.01. [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–34. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–9. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Tian H, Jeong J, Harfe BD, Tabin CJ, McMahon AP. Mouse Disp1 is required in sonic hedgehog-expressing cells for paracrine activity of the cholesterol-modified ligand. Development. 2005;132:133–42. doi: 10.1242/dev.01563. [DOI] [PubMed] [Google Scholar]
- Traiffort E, Dubourg C, Faure H, Rognan D, Odent S, Durou MR, David V, Ruat M. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J Biol Chem. 2004;279:42889–97. doi: 10.1074/jbc.M405161200. [DOI] [PubMed] [Google Scholar]
- Williams KP, Rayhorn P, Chi-Rosso G, Garber EA, Strauch KL, Horan GS, Reilly JO, Baker DP, Taylor FR, Koteliansky V, Pepinsky RB. Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J Cell Sci. 1999;112(Pt 23):4405–14. doi: 10.1242/jcs.112.23.4405. [DOI] [PubMed] [Google Scholar]
- Zeng X, Goetz JA, Suber LM, Scott WJ, Jr., Schreiner CM, Robbins DJ. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 2001;411:716–20. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–92. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.