Abstract
Proteolytic enzymes are necessary for the hardening of dental enamel during development, and mutations in the kallikrein 4 (KLK4) and enamelysin (MMP20) genes cause autosomal recessive amelogenesis imperfecta (ARAI). So far, only one KLK4 and two MMP20 mutations have been reported. We have identified an ARAI-causing point mutation (c.l02G>A, g.l02G>A, and p.W34X) in exon 1 of MMP20 in a proband with autosomal recessive hypoplastic-hypomaturation amelogenesis imperfecta. The G to A transition changes the tryptophan (W) codon (TGG) at amino acid position 34 into a translation termination (X) codon (TGA). No disease-causing sequence variations were detected in KLK4. The affected enamel is thin, with mild spacing in the anterior dentition. The enamel layer is hypomineralized, does not contrast with dentin on radiographs, and tends to chip away from the underlying dentin. An intrinsic yellowish pigmentation is evident even during eruption. The phenotype supports current ideas concerning the function of enamelysin.
Keywords: Enamelysin, MMP20, Enamel, Tooth, Amelogenesis imperfecta
INTRODUCTION
Dental enamel forms in stages. During the secretory stage amelogenin, enamelin, and ameloblastin are secreted by ameloblasts. These proteins have been isolated from developing enamel and accumulate during the secretory stage, mainly as proteolytic cleavage products of the originally secreted proteins (Fincham et al., 1999). Early in the maturation stage, the accumulated cleavage products are degraded and reabsorbed into the overlying ameloblasts, and the rate of mineral deposition accelerates as the enamel crystallites thicken (Smith, 1998). Protease inhibition studies of secretory and maturation stage enamel extracts demonstrated that matrix metalloproteinase (MMP) activity predominates during the secretory stage, while serine protease activity is mainly observed during the maturation stage (Overall and Limeback, 1988).
Cloning of mRNA extracted from porcine developing teeth discovered two novel proteases: enamelysin (MMP-20) (Bartlett et al., 1996) and kallikrein 4 (KLK4) (Simmer et al., 1998). Enamelysin appears as a doublet at 45 and 41-kDa on casein zymograms (Fukae et al., 1998). Both bands represent the active protease, containing the catalytic and hemopexin domains, but lacking the propeptide (Yamada et al., 2003). KLK4 shows up as two smear bands between 30 and 36-kDa on gelatin zymograms (Tanabe, 1984). The expression of MMP-20 and KLK4 during tooth formation has been characterized by in situ hybridization (Hu et al., 2002; Simmer et al., 2004). Both enzymes are expressed by ameloblasts and odontoblasts. Ameloblasts express MMP-20 prior to the onset of mineralization (Begue-Kirn et al., 1998), and expression continues into early maturation. MMP-20 catalyzes the partial degradation, or processing, of enamel proteins during the secretory stage. Recombinant enamelysin cleaves recombinant amelogenin (Ryu et al., 1999) and recombinant ameloblastin (Iwata et al., 2007) at the same sites in vitro as are cleaved in vivo to generate the cleavage products that accumulate in the secretory stage matrix. Ameloblasts first express KLK4 in the transition between the secretory and maturation stages and KLK4 expression continues throughout the maturation stage (Hu et al., 2000). KLK4 degrades recombinant amelogenin in vitro, generating cleavage products that are not the same as those found in secretory stage enamel (Ryu et al., 2002).
The distinctive but partially overlapping patterns of MMP-20 and KLK4 expression, as well as the sharp differences in the way they individually cleave enamel proteins, suggests these enzymes play similar but separate roles in enamel formation. It is a general principle that disturbances of tooth development are manifested as dental malformations that are characteristic of the timing of the disturbance (Hu and Simmer, 2007). For instance, defects in genes expressed early in tooth development, such as PAX9, MSX1, and AXIN2, are associated with oligodontia or familial tooth agenesis (Lammi et al., 2004; Stockton et al., 2000; Vastardis et al., 1996). This principle also applies to the stages of amelogenesis. Amelogenin, enamelin and ameloblastin null mice all exhibit thin or absent enamel layers, which correlate with their critical expression during the secretory stage (Fukumoto et al., 2004; Gibson et al., 2001; Masuya et al., 2005). Humans with AMELX mutations in males (Kida et al., 2007) or ENAM mutations affecting both alleles (Ozdemir et al., 2005) have severely hypoplastic (thin) enamel.
The relationship between the normal timing of a gene’s expression and the character of the enamel malformations that occur in its absence appears to be more complex in the case of enamelysin. The enamelysin null mice deposit an enamel layer that is thinner than normal, tends to chip off from the underlying dentin, has disorganized prisms, is less highly mineralized, and retains more protein than normal (Bartlett et al., 2004; Caterina et al., 2002). The phenotype is indicative of disturbances occurring in both the secretory and maturations stages of amelogenesis. The enamel phenotype in two autosomal recessive amelogenesis imperfecta (ARAI) kindreds caused by MMP20 mutations have been described (Kim et al., 2005; Ozdemir et al., 2005). In both cases the enamel was pigmented, typically yellowish to brownish in color, but appeared to be normal in thickness. The enamel layer was less highly mineralized and did not contrast with dentin on radiographs. This phenotype is classic hypomaturation amelogenesis imperfecta, and suggests that in humans, MMP20 functions primarily in enamel maturation (Witkop Jr. and Sauk Jr., 1976). A classic hypomaturation phenotype was also observed in a kindred with ARAI caused by defects in KLK4 (Hart et al., 2004). In this report we ask the experimental questions: what is the range of phenotypes exhibited by kindreds with MMP20 defects, and how can these phenotypes be rationalized in terms of MMP-20 function? We describe the third MMP20 mutation in a kindred with ARAI, and the first displaying hypoplastic enamel as well as hypomaturation defects.
MATERIALS & METHODS
Protocol Approval
The study protocol and patient consents forms were reviewed and approved by the Institutional Review Boards at the University of Michigan and at Taipei Medical University Hospital.
Subject Recruitment and Examination
Proband is an 11-year-old Chinese girl. A thorough medical history review of the proband revealed no health concerns and no known drug or food allergies. The proband’s craniofacial examination, dental hygiene assessment, and dietary history were non-contributory. The nuclear family of four, including the proband, was interviewed and recruited. Due to their dispersed geographical locations, the recruitment of other relatives was not successful. All participating subjects were given oral and radiographic examinations to characterize their dental phenotype and to determine their affection status. The dental phenotype was only observed in the proband, and was consistent with a diagnosis of autosomal recessive hypoplastic hypomaturation amelogenesis imperfecta (Witkop, 1988).
Mutation Analysis
Based upon the clinical diagnosis and inheritance pattern, a list of target genes for mutational analyses was prioritized as KLK4 and MMP20, ENAM, AMBN, and AMELX. The strategy was to generate PCR amplification products that would allow DNA sequencing of each coding exon and about 50 basepairs of bordering intron. The PCR amplification primer sets used in this study were described previously (Kim et al., 2005; Kim et al., 2006). Genomic DNA was isolated from 5 mL of peripheral whole blood obtained from each family member, using the QIAamp® Maxi Blood Kits (Qiagen, Valencia, CA). PCR amplifications used the Platinum® DNA polymerase (Invitrogen, Carlsbad, CA). The reactions had a 5 min denaturation at 94 °C, followed by 40 cycles each with denaturation at 94 °C for 30 sec, primer annealing at 53–62 °C for 30–45 sec, and product extension at 72 °C for 30–90 sec. In the final cycle the 72 °C extension was for 7 min. PCR amplification products were run in a 1.2% agarose gel and photographed. PCR products were purified by QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and sent for direct sequencing analysis.
DNA Sequence Analysis
DNA sequencing was performed on an ABI Model 310 DNA sequencer (Applied Biosystems, Foster City, CA) at the DNA Sequencing Core, University of Michigan. The same oligonucleotide primers used to generate the PCR amplification products were used to prime the sequencing reactions. The amplifications and analyses were repeated to ensure accuracy. The sequencing results were checked against the human genome sequence and nucleotide variations were noted and checked against the dbSNPs database of NCBI, http://www.ncbi.nlm.nih.gov/sites/entrez?db=Gene and http://www.ncbi.nlm.nih.gov/SNP/
RESULTS
Subject evaluation
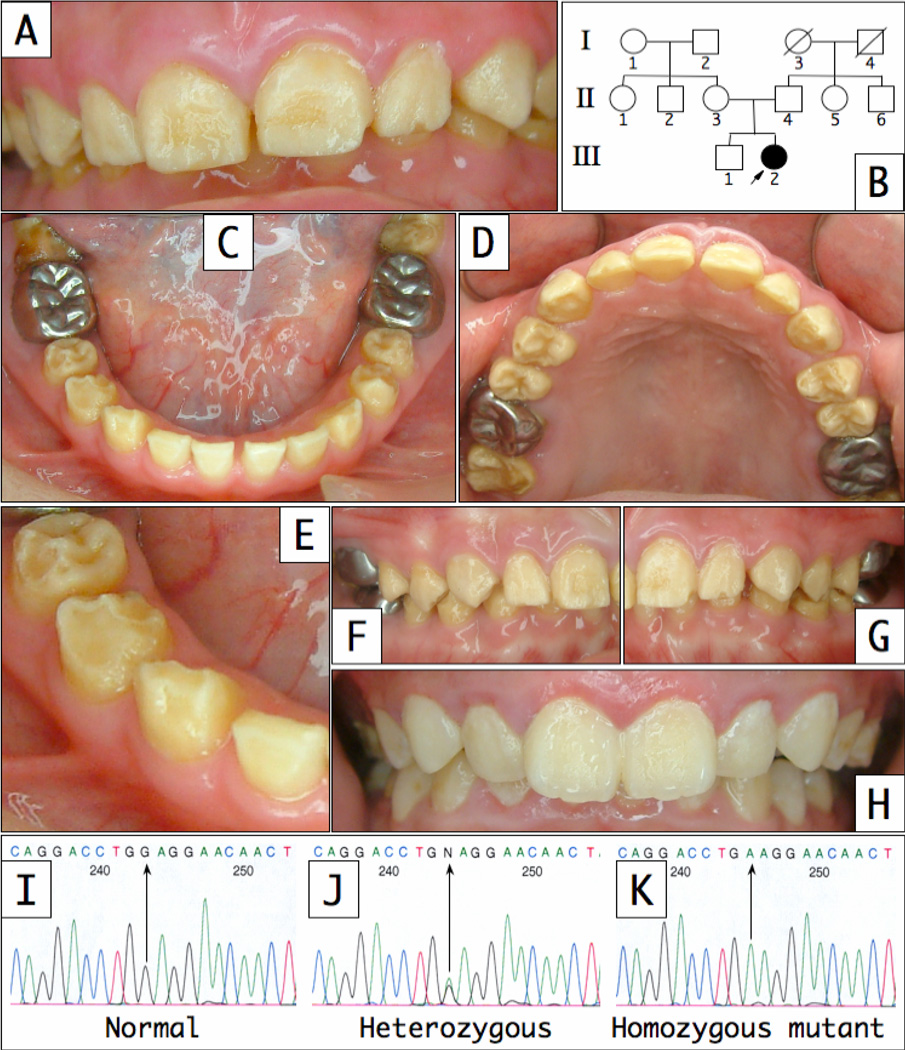
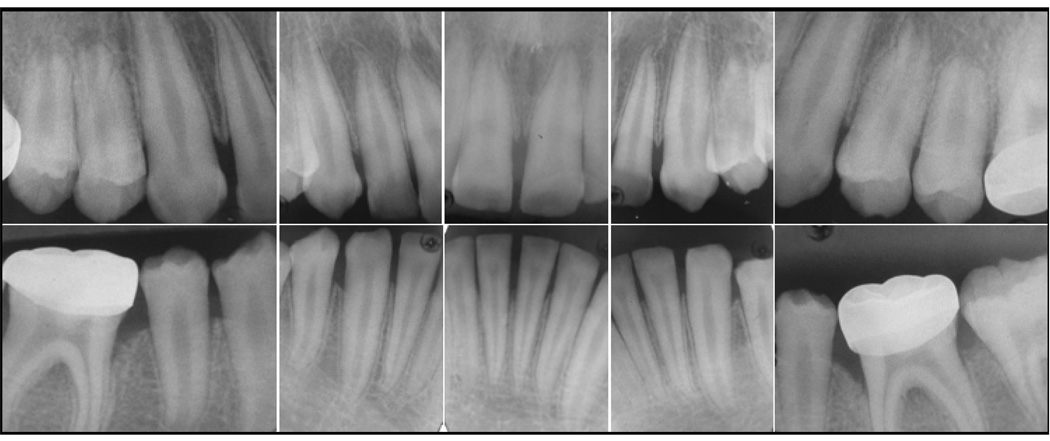
Clinically, the enamel layer of the proband was characterized by a generalized surface roughness and non-homogenous yellowish brown stain (Fig. 1). The erupting second permanent molars were severely discolored, suggesting that the discoloration was intrinsic in nature. Vertical shear fractures of the enamel on functional cusps exposed the underlying dentin on many teeth. Radiographically, tooth number and size were within normal limits, but the enamel layer was thin and showed little contrast with dentin, so the position of the DEJ could not be discerned (Fig. 2). No additional oral pathology was noted. Mild bone loss was observed around the first molars, and was associated with stainless steel crown restorations and inadequate dental hygiene. Following a thorough clinical and radiographic evaluation of the proband’s father, mother and older brother, it was concluded that no other members of the family were affected. Taken together, the clinical findings are consistent with a diagnosis of autosomal recessive hypoplastic-hypomaturation AI.
Figure 1.
Clinical photographs of proband (III-2) at age 11. (A) Frontal view; (B) Three generation pedigree; (C) Mandibular occlusal view; (D) Maxillary occlusal view; (E) Enamel shear fractures can be seen on the incisai edge of tooth #26 and the functional cusps of teeth #27, 28 and 29; (F and G) Right and left lateral views with the teeth in occlusion; (G) Frontal view of proband’s older brother; DNA sequence chromatograms showing the homozygous normal (I), heterozygous (J) and homozygous mutant (K) sequences. The (g.l02G>A) mutation sites are marked by arrows. The proband’s permanent dentition shows a generalized, intrinsic, yellow staining that is already evident during tooth eruption. The enamel surface is rough, and shows signs of previous chipping and wear. There is mild spacing related to the thinness of the enamel layer. A deep overbite and normal overjet are observed.
Figure 2.
Radiographs of proband at age 11. The affected enamel has reduced radiopacity and cannot be distinguished from the underlying dentin. Where the enamel can be distinguished, is appears thinner than normal. The posterior radiographs were enlarged relative to the anteriors.
Mutational analysis
DNA amplification of the KLK4 coding region was performed first. Single nucleotide changes were identified, but all of the variations are recognized single nucleotide polymorphisms in the databases and are considered not likely to be disease causing. Mutational analysis of MMP20 identified a nonsense mutation in the coding region of the exon 1 (Fig 1). This single nucleotide (G to A) substitution converts the tryptophan (W) codon (TGG) at amino acid position 34 into a translation termination (X) codon (TGA). The standard designations for this mutation in the cDNA, gene, and protein are c.l02G>A, g.l02G>A, and p.W34X, respectively. The distribution of this mutation in the four participating members of the kindred was determined. The proband is the only person with both MMP20 alleles affected and she is the only subject displaying an enamel phenotype.
DISCUSSION
The G>A transition in codon 34 of MMP20 was observed in both MMP alleles in the proband with autosomal recessive hypoplastic-hypomaturation amelogenesis imperfecta. This single nucleotide substitution introduces a premature translation termination codon in exon 1 and would likely cause the defective MMP20 transcripts to be degraded by the nonsense-mediated decay system (Wagner and Lykke-Andersen, 2002). If translated, the defective MMP20 mRNA transcripts could only have directed the synthesis of a 33 amino acid peptide (MKVLPASGLAVFLIMALKFSTAAPSLVAASPRT) containing the 22 residue signal peptide and 11 amino acids from the beginning of the propeptide domain (Llano et al., 1997). It is certain then, that no functional MMP-20 was expressed during tooth formation in the proband. Based upon the known importance of enamelysin in dental enamel formation, it is highly probable that the p.W34X mutation caused the enamel phenotype in the proband. It also seems safe to conclude that the enamel defects observed in the proband resulted from the MMP20 null condition, although a toxic effect from translation of the truncated protein cannot be ruled out.
There are now three defined MMP20 mutations reported in families with ARAI. The affected enamel phenotype in these families all showed stained, hypomineralized enamel that did not sharply contrast with dentin on radiographs and tended to chip away from the underlying dentin. The pigmentation is evident during eruption, suggesting the stain is intrinsic and not acquired, and is presumed to be due to the retention of enamel proteins. The enamel layer is clearly thin in our proband and caused a mild spacing in the anterior dentition, which contrasted with the tight contacts observed in her older brother. Although thin enamel was not evident in the two previously reported MMP20 phenotypes, a reduction in enamel thickness is hard to quantify because the DEJ is not plainly delineated on radiographs and because the formed enamel layer is altered by chipping and accelerated wear. Given the difficulties in assessing hypoplasia when associated with enamel hypomaturation, the enamel phenotypes observed in the three MMP20 kindreds are remarkably consistent and the minor variations can be attributed to differences in genetic background.
The enamel phenotype displayed in the MMP20 null condition provides insights into the function of enamelysin, although it must be kept in mind that the phenotype might reflect secondary events resulting from ameloblasts reacting pathologically to disturbances in the matrix caused by the absence of MMP-20 activity. The cleavage of enamel proteins by enamelysin may not be critical for crystal elongation, as the enamel crystals must lengthen significantly to achieve the observed thickness of hypoplastic enamel. The absence of secretory stage proteolytic activity resulting in enamel hypomaturation recalls earlier studies showing that significant amounts of enamel maturation occurs during the secretory stage, where enamel crystals increase in both width and thickness going from the enamel surface to the DEJ (Daculsi and Kerebel, 1978).
The accumulation of enamel matrix cleavage products during the secretory stage is a steady state process. Enamel proteins and MMP-20 are continually secreted by ameloblasts at the enamel surface. Enamelysin cleaves the secreted proteins into progressively smaller products. The abundance of any particular cleavage product is determined by its rate of generation (by proteolysis or secretion) and its rate of elimination (by further proteolysis) (Simmer and Hu, 2002). With increasing distance from the secretory front at the enamel surface there is a decline in the amount of organic matrix and the crystallites are able to increase in width and thickness. Failure to process and slowly degrade enamel proteins during the secretory stage may interfere with the progressive thickening of enamel crystals with depth. The resulting structural weaknesses may account for the observed chipping of enamel formed without enamelysin. The intrinsic staining suggests that KLK4 activity during the maturation stage is insufficient to overcome the increased enamel protein content and reduced crystal thickness in the deeper enamel that occurs in the absence of MMP-20 activity. It may also be that MMP-20 activity is necessary to activate KLK4. It is not known how the propeptide is removed from the KLK4 zymogen in vivo, but enamelysin is able to catalyze removal of the KLK4 propeptide in vitro (Ryu et al., 2002). We conclude that MMP-20 is necessary to gradually degrade enamel proteins during the secretory stage to remove the organic matrix between enamel crystallites and allow them to progressively thicken with depth. Without MMP-20 there may be insufficient maturation of the crystallites in the deeper enamel, which manifests itself clinically as a tendency for the enamel to shear.
ACKNOWLEDGMENTS
We thank the family for their participation and contributions. This investigation was supported by USPHS Research Grants DE15846 and DE016276 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 29892. All authors declare that there are no conflicting interests.
Contributor Information
P. Papagerakis, Email: petrosp@umich.edu.
H-K. Lin, Email: linhsikuei27@hotmail.com.
K. Y. Lee, Email: yunleone@yahoo.com.
Y. Hu, Email: yyhu@umich.edu.
J. P. Simmer, Email: jsimmer@umich.edu.
J. D. Bartlett, Email: JBartlett@forsyth.org.
J. C-C Hu, Email: janhu@umich.edu.
REFERENCES
- Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183:123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Beniash E, Lee DH, Smith CE. Decreased mineral content in MMP-20 null mouse enamel is prominent during the maturation stage. J Dent Res. 2004;83:909–913. doi: 10.1177/154405910408301204. [DOI] [PubMed] [Google Scholar]
- Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sei. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, et al. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- Daculsi G, Kerebel B. High-resolution electron microscope study of human enamel crystallites: size, shape, and growth. J Ultrastruct Res. 1978;65:163–172. doi: 10.1016/s0022-5320(78)90053-9. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- Fukae M, Tanabe T, Uchida T, Lee SK, Ryu OH, Murakami C, et al. Enamelysin (matrix metalloproteinase-20): localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis. J Dent Res. 1998;77:1580–1588. doi: 10.1177/00220345980770080501. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, et al. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, et al. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, Ryu OH, Chen JJ, Uchida T, Wakida K, Murakami C, et al. Localization of EMSP1 expression during tooth formation and cloning of mouse cDNA. J Dent Res. 2000;79:70–76. doi: 10.1177/00220345000790011301. [DOI] [PubMed] [Google Scholar]
- Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. Eur J Oral Sci. 2002;110:307–315. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- Hu JC, Simmer J. Developmental biology and genetics of dental malformations. Orthod Craniofac Res. 2007;10:45–52. doi: 10.1111/j.1601-6343.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, et al. Processing of ameloblastin by MMP-20. J Dent Res. 2007;86:153–157. doi: 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- Kida M, Sakiyama Y, Matsuda A, Takabayashi S, Ochi H, Sekiguchi H, et al. A novel missense mutation (p.P52R) in amelogenin gene causing X-linked amelogenesis imperfecta. J Dent Res. 2007;86:69–72. doi: 10.1177/154405910708600111. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, et al. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Lin BP, Seymen F, Bartlett JD, Hu JC. Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci. 2006;114(Suppl 1):3–12. doi: 10.1111/j.1600-0722.2006.00278.x. [DOI] [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano E, Pendas AM, Knauper V, Sorsa T, Salo T, Salido E, et al. Identification and structural and functional characterization of human enamelysin (MMP-20) Biochemistry. 1997;36:15101–15108. doi: 10.1021/bi972120y. [DOI] [PubMed] [Google Scholar]
- Masuya H, Shimizu K, Sezutsu H, Sakuraba Y, Nagano J, Shimizu A, et al. Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI) Hum Mol Genet. 2005;14:575–583. doi: 10.1093/hmg/ddi054. [DOI] [PubMed] [Google Scholar]
- Overall CM, Limeback H. Identification and characterization of enamel proteinases isolated from developing enamel. Amelogeninolytic serine proteinases are associated with enamel maturation in pig. Biochem J. 1988;256:965–972. doi: 10.1042/bj2560965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. Phenotype of ENAM Mutations is Dosage-dependent. J Dent Res. 2005;84:1036–1041. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, et al. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci. 2002;110:358–365. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, et al. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Fukae M, Tanabe T, Yamakoshi Y, Uchida T, Xue J, et al. Purification, characterization, and cloning of enamel matrix serine proteinase 1. J Dent Res. 1998;77:377–386. doi: 10.1177/00220345980770020601. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Hu JC. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43:441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Sun X, Yamada Y, Zhang CH, Bartlett JD, Hu JC-C. Enamelysin and kallikrein-4 expression in the mouse incisor. In: Kobayashi I, Ozawa H, editors. Biomineralization: formation, diversity, evolution and application Proceedings of the 8th International Symposium on Biomineralization; Sept 25–28, 2001; Niigata, Jpn. Hadano, Jpn: Tokai University Press; 2004. pp. 348–352. [Google Scholar]
- Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- Stockton DW, Das P, Goldenberg M, D’Souza RN, Patel PI. Mutation of PAX9 is associated with oligodontia. Nature Genet. 2000;24:18–19. doi: 10.1038/71634. [DOI] [PubMed] [Google Scholar]
- Tanabe T. Purification and characterization of proteolytic enzymes in porcine immature enamel. Tsurumi U Dent J. 1984;10:443–452. [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- Wagner E, Lykke-Andersen J. mRNA surveillance: the perfect persist. J Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- Witkop CJ., Jr Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol. 1988;17:547–553. doi: 10.1111/j.1600-0714.1988.tb01332.x. [DOI] [PubMed] [Google Scholar]
- Witkop CJ, Jr, Sauk JJ., Jr . Heritable defects of enamel. In: Stewart RE, Prescott GH, editors. Oral Facial Genetics. St. Louis: C.V. Mosby Co.; 1976. pp. 151–226. [Google Scholar]
- Yamada Y, Yamakoshi Y, Gerlach R, Hu C, Matsumoto K, Fukae M, et al. Purification and characterization of enamelysin from secretory stage pig enamel. Arch Comp Biol Tooth Enam. 2003;8:21–25. [Google Scholar]