Abstract
Protein expression of p63 is used to differentiate prostate cancer from benign mimickers. Recent studies suggest that it may also distinguish aggressive prostate cancer with down-regulated expression occurring in men with more advanced disease. We conducted a prospective study among 298 men aged 51–84 years who were diagnosed with prostate cancer in the Physicians’ Health Study in 1983–2004 and whose tissue was available for immunohistochemical (IHC) staining. We used Cox proportional hazards regression to evaluate the association of p63 protein expression with fatal prostate cancer. We correlated p63 expression with tumor cell proliferation (ki67) and apoptosis (TUNEL staining). The predominant location of tumor p63 staining occurred in the cytoplasm, an uncommon departure from the strong nuclear staining usually observed in non-neoplastic basal cells. Increasing expression of cytoplasmic p63 (tertiles) was associated with prostate cancer mortality (n=19 deaths); the hazard ratios and 95% confidence intervals were: 1.0 [referent], 4.0 (0.9, 18.9) and 5.9 (1.3, 27.5), p for trend = .03). The positive trend remained significant (p=.047) after multivariable adjustment for age, year of diagnosis and Gleason score. Higher tertiles of cytoplasmic p63 were also associated with reduced levels of apoptosis (p for trend = 0.0408) and increased cellular proliferation (p for trend = 0.0026). We found aberrant expression of p63 in the cytoplasm to be associated with increased prostate cancer-specific mortality up to 20 years after diagnosis. The mislocalized expression was associated with reduced apoptosis and higher proliferative activity, and may suggest an oncogenic role in prostate cancer progression and survival.
Keywords: p63, prostate, cytoplasmic, mortality, apoptosis
INTRODUCTION
Expression of the p63 gene, a member of the p53 family (1) is down-regulated in adenocarcinoma of the prostate as compared with normal prostate and is used as a basal cell marker in the diagnosis of prostate cancer (2). Differences in p63 expression are associated with cancer progression or a poor prognosis for several cancer sites, including over-expression in the ovaries and oral squamous cell carcinoma (3, 4), down-regulated expression in the upper urinary tract and prostate (5–7) and aberrant cytoplasmic expression in lung adenocarcinoma (8).
The p63 gene is critical to embryonic development of the epidermis and its derivative structures including the prostate gland (2) (9–11). The p63 protein is normally expressed in basal cells of epithelial structures, including the prostate epithelium and is involved in epithelial differentiation and proliferation (2). The role of this transcription factor in carcinogenesis is complex as it encodes two classes of proteins with opposing tumor suppressor and oncogenic functions including transactivation, apoptosis and cell proliferation (2) (12–17). In adenocarcinomas, p63 tends to be under-expressed (18) and in prostate cancer specifically, negative immunohistochemical staining of p63 is a clinically useful tool for identifying benign mimickers (2). Recent studies have also identified p63 as important in signatures of advanced disease, with lower expression associated with disease progression and the development of lethal prostate cancer (6, 7). We undertook this study to further evaluate the role of p63 in distinguishing fatal disease in men with prostate cancer followed up to 20 years.
MATERIALS and METHODS
Study population
This study was nested within the Physicians’ Health Study (PHS) I and II randomized trials of aspirin and nutritional supplements for the primary prevention of cancer and cardiovascular disease among U.S. male physicians (described in detail elsewhere) (19, 20). Briefly, PHS I began in 1982 among 22,071 physicians aged 40–84 years without a history of cardiovascular disease or cancer at baseline and PHS II began in 1997, among 14,641 physicians aged 50 years and older. Follow-up information and mortality data are 97% complete on all participants.
Case identification
A prostate cancer diagnosis was based initially on self-report and then confirmed through a review of medical records and pathology reports by an End Point Committee of physicians (Dr. Stampfer is a member). Deaths were identified from the National Death Index, postal system and next of kin and medical records were reviewed to adjudicate causes of death, including prostate cancer.
Tissue microarrays
For this study, we obtained archival formalin-fixed, paraffin embedded tissue specimens for men who had radical prostatectomies or TURP’s between 1983 and 2004. Two study pathologists (MAR and SP) conducted a systematic re-review of all tissue specimens for standardized Gleason grading and to identify the dominant prostate cancer nodule with the highest Gleason score pattern from each specimen. A manual tissue arrayer was used to construct three high-density tissue microarrays (TMA) in which at least three tumor tissue cores from the targeted areas were transferred, arraying 0.6 mm cores per case.
Immunohistochemistry
p63
Five micron sections of each TMA were mounted on charged slides and were subjected to microwave treatment for antigen retrieval and incubated with a 1:600 dilution of the 4A4 mouse monoclonal antibody (Lab Vision Corporation, Santa Cruz), which binds to all isoforms of p63. A semi-automated image analysis system with high reproducibility (Chromavision) was used to measure protein expression of p63 (21). Using scanned digital images of the cores (22), the percent of positively stained area for p63 was scored on a scale of 0–100%. A study pathologist (MB) targeted those areas with histologically recognizable prostate cancer to focus on protein expression of p63 from tumor tissue only and was blinded to clinical outcomes.
Ki67
Five micron sections were used for the Ki67 antigen, which was utilized as a proliferation marker using a Rabbit polyclonal antibody (Vector, Burlingame, CA) diluted 1:1500 with a citrate-based antigen retrieval. The Ki67 score was assessed as the number of stained nuclei over the total number of tumor nuclei using the Ariol instrument SL-50 (Applied Imaging, Grand Rapids, MI) after selection of the tumor areas of each core for full quantitative image analysis.
Apoptosis
Based on five micron sections, the TUNEL assay was used to identify the percent of tumor cells undergoing apoptosis using the Apoptag peroxidase in situ Kit (Chemicon Internationak, Inc.) according to the instructions of the manufacturer. The Apoptag score was assessed as the number of positive cells out of the total number of tumor cells independently by two study pathologists (MF, AF). The whole area of each tumor core was evaluated for the score. Due to fixation and technical artifacts, the total number of specimens with reliable staining for apoptosis data was reduced to n=227 for analyses.
Clinical and demographic characteristics
We collected baseline questionnaire data on demographics such as age, height and weight in 1982 for PHS I participants and in 1997 on new PHS II participants. We collected information on clinical stage and PSA at diagnosis through medical records. Clinical stage of disease was determined through a review of medical records and pathology reports and categorized according to the tumor-node-metastasis (TNM) staging system (2002 AJCC). If data were missing from medical records, self-reported stage and diagnostic PSA were supplemented from information collected through follow-up questionnaires. Tumor grade was assigned by a study pathologist (MAR), who conducted a systematic re-review of all tissue specimens to determine a pathological Gleason score and grade for each patient. Assignment of tumor grade and stage was done while blinded to p63 levels and mortality information.
Statistical Analyses
The current study is based on 298 men with sufficient tumor tissue available for immunohistochemical analysis (n=270 from prostatectomy and n=28 from TURP). The percentage of area that stained positive for p63 was assessed as a continuous marker and in tertiles, based on the distribution in the study population. The means of continuous variables, such as age, year of diagnosis and PSA were compared across tertiles of p63 expression using analysis of variance and the F-test for trend for statistical significance. The percent of cells positive for ki67 and proportion of cells undergoing apoptosis were log-transformed and analyzed across tertiles of p63-positive area as linear variables using analysis of variance and the linear F-test for trend (p<0.05). The duration of survival was calculated as the interval from time of cancer diagnosis to either a prostate cancer death (time to event) or censored at time of death from other causes or at end of follow-up (March 31, 2007). Cox proportional hazards regression was conducted to compute the hazards ratio and 95% confidence interval (CI) for survival duration. Multivariate hazard ratios were adjusted for age (years), year of diagnosis, prostate-specific antigen (PSA) levels at the time of diagnosis (continuous, log-transformed) Gleason score (4–6, 7–10) and clinical stage (dichotomized - localized [T1–T2, NO/NX and M0/MX] vs. extra-prostatic disease [T3–T4 or N1 or M1]). We used the SAS program package, version 9.1 (SAS Institute, Cary, NC) to carry out statistical analyses with a significance level of .05. This study was approved by the Institutional Review Board at Partners Healthcare.
RESULTS
Men had a mean age at diagnosis of 66.3 years and a mean BMI of 24.5 kg/m2 (Table 1). Ten percent of the tissue samples were collected from TURP (n=30). Most had clinically localized disease (92%) but pathological staging at the time of surgery revealed 22% (n=66) with extra-prostatic disease, 6% (n=18) with positive seminal vesicles and 3% (n=9) with lymph node invasion (data not shown). Nearly a third of participants had low-grade disease (Gleason 4–6) and the mean PSA at diagnosis was 11.1 ng/mL.
Table 1.
Clinical characteristics by tertiles of p63 percent-positive area of men diagnosed with prostate cancer in PHS, 1983–2007
| Tertiles of p63 percent positivity |
||||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| (0 – 1.76) | (1.77 – 3.89) | (3.90 – 53.79) | ||
| Characteristics | 99 | 100 | 99 | |
| Mean (SE) or % | p-value | |||
| Median p63-% positive (IQR) | 1.1 (0.8, 1.4) | 2.5 (2.0, 3.0) | 6.6 (4.9, 9.1) | |
| Age at diagnosis (yrs) | 65.8 (0.6) | 67.2 (0.6) | 65.9 (0.6) | 0.83* |
| Body mass index (baseline, kg/m2) | 24.6 (0.3) | 24.4 (0.3) | 24.6 (0.3) | 0.99* |
| Follow-up time (yrs) | 10.0 (0.4) | 9.7 (0.4) | 8.0 (0.4) | <.001 * |
| PSA at diagnosis (ng/mL) | 11.2 (3.4) | 7.4 (3.6) | 14.8 (3.6) | 0.49* |
| missing (%) § | 7.1 | 17.0 | 17.2 | |
| Gleason score | ||||
| 4–6 | 29 (29.3) | 23 (23.0) | 38 (38.4) | 0.05† |
| 7 | 57 (57.6) | 61 (61.0) | 41 (41.4) | |
| 8–10 | 13 (13.1) | 16 (16.0) | 20 (20.2) | |
| Clinical stage | ||||
| T1, NX/NO | 45 (45.5) | 47 (47.0) | 58 (58.6) | 0.15† |
| T2, NX/NO | 47 (47.5) | 45 (45.0) | 33 (33.3) | |
| T3/T4 or N1/M1 | 4 (4.0) | 4 (4.0) | 1 (1.0) | |
| missing (%) | 3.0 | 4.0 | 7.1 | |
| Prostate cancer death | ||||
| Yes | 2 (2.0) | 8 (8.0) | 9 (9.1) | 0.04‡ |
| No | 97 (98.0) | 92 (92.0) | 90 (90.9) | |
F-test for trend
Global chi-squared test
Chi-square test for trend across tertiles
49% of men with missing PSA (n=20) were diagnosed in pre-PSA era
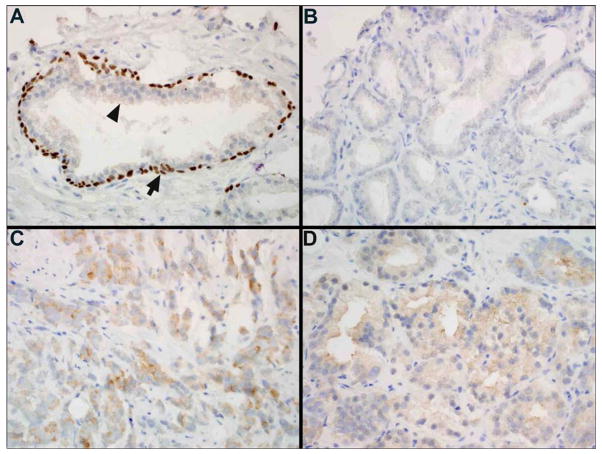
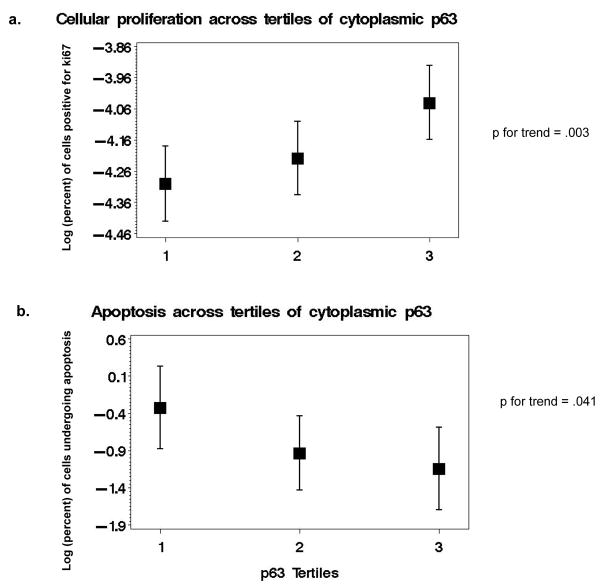
We observed predominantly cytoplasmic staining for p63-positive tumor cells (only two men had concomitant nuclear staining), which is a rare expression pattern for a protein that is normally absent in prostate adenocarcarcinoma and that usually exhibits strong nuclear staining in basal cells of benign prostate glands (Figure 1). P63-percent positivity reflects the proportion of examined area that stained positive for this protein and a mean of 4.3% of the area (median=2.5%) displayed p63 immunoreactivity (Table 1) with a maximum score of 54%. The low scores were expected given the under-expression of p63 in adenocarcinoma of the prostate and its use as a negative marker in the diagnostic work-up of prostate cancer. Although we observed no significant differences in age, PSA levels at diagnosis or pathological stage across tertiles of p63-percent positivity (Table 1), we found an increasing proportion of prostate cancer deaths in higher tertiles of cytoplasmic p63 expression (p for trend = .04) and a borderline significant difference in Gleason scores across p63 tertiles (p=.05). In the same histological specimens, higher levels of cytoplasmic p63 were associated with a significantly higher frequency of ki67-positive cells (p for trend = 0.0026, n=279) - a marker of cellular proliferation - and lower proportion of cells undergoing apoptosis (p for trend = 0.0408, n=227) (Figure 2).
Figure 1.
(A) Normal prostate gland showing p63 staining of basal cells (arrow) and lack of staining of luminal cells (arrowhead) (x40). (B) Prostate adenocarcinoma showing no cytoplasmic or nuclear staining for p63 (x40). (C & D) Prostate adenocarcinoma showing cytoplasmic staining for p63 (x40).
Figure 2.
Log (percent) of ki67-positive cells (a) and log (percent) of cells undergoing apoptosis (TUNEL assay) (b) across tertiles of cytoplasmic p63.
Over the 20-year follow-up period, we observed 19 prostate cancer deaths, which occurred an average of 9.3 years after diagnosis (SD=4.0; Table 1). In univariate analyses, we observed a significant positive association between the percent of area staining positive for p63 (as a continuous measurement) and fatal prostate cancer, with an approximate 7% increase in prostate cancer mortality for each additional percent of cytoplasmic p63-positivity (RR=1.07; 95 % CI: 1.04, 1.10, p <.0001). The association persisted after we excluded the two men with co-localized nuclear p63 (RR= 1.10; 95% CI: 1.05, 1.16, p=0.0001). We also observed a significant trend with increasing tertiles of p63 positivity (RR’s = 1.0 [T1-reference], 4.0; 95% CI: 0.9, 18.9 [T2] and 5.9; 95% CI: 1.3, 27.5 [T3], p for trend = .028; Table 2). As expected, univariate Cox proportional hazards models were significant for fatal prostate cancer for important clinical characteristics, such as age and PSA at diagnosis (Table 2). The increasing risk associated with higher tertiles of p63 cytoplasmic immunoreactivity (RR’s = 1.0 [T1-reference], 2.7; 95% CI: 0.6, 13.0[T2] and 4.8; 95% CI: 1.0, 22.9 [T3]) yielded a borderline significant trend (p for trend = 0.047) after multivariable adjustment for age, year of diagnosis and Gleason score. The trend persisted after further adjustment for clinical stage (p for trend =0.04, data not shown).
Table 2.
Univariate and Multivariate Cox Proportional Hazard Ratios for prostate cancer mortality in PHS, 1983–2007
| Univariate | Multivariate * | ||||||
|---|---|---|---|---|---|---|---|
| Clinical variable at diagnosis | N | Hazards Ratio | 95% CI | p-value | Hazards Ratio | 95% CI | p-value |
| Age at diagnosis, per year | 298 | 1.16 | (1.08, 1.26) | 0.0001 | 1.15 | (1.06, 1.24) | <.001 |
| Gleason score (4–6 [ref], 7–10) | 298 | 3.3 | (0.76, 14.29) | 0.11 | 2.3 | (0.5, 10.3) | 0.26 |
| Log(PSA at diagnosis), per 1 unit increase | 257 | 3.5 | (1.92, 6.22) | <.0001 | 2.9 | (1.46, 5.58) | 0.002 |
| p63 percent, per 1% increase | 298 | 1.07 | (1.04, 1.10) | <.0001 | 1.06 | (1.03, 1.09) | <.001 |
| p63 percent - tertiles | |||||||
| Low | 99 | 1.0 | ref | 1.0 | |||
| Medium | 100 | 4.0 | (0.9, 18.9) | 0.08 | 2.7 | (0.6, 13.0) | 0.21 |
| High | 99 | 5.9 | (1.3, 27.5) | 0.02 |
4.8 | (1.0, 22.9) |
0.05 |
| test for trend | 0.028 † | 0.047 † | |||||
DISCUSSION
We found a positive association between cytoplasmic expression of p63 in prostate tumor tissue at the time of diagnosis and fatal prostate cancer. This association remained significant after adjustment for age, year of diagnosis, Gleason score and stage. The low percent staining of p63 in our data is consistent with previous studies (23–26), however the majority of staining occurred in the cytoplasm rather than the nucleus, the usual location for p63 expression in benign and neoplastic epithelial cells. In our data, higher levels of cytoplasmic p63 were also significantly associated with increased proliferative activity (ki67) and lower rates of apoptosis. Similar to the localization shift of tumor suppressor gene proteins that induces cellular events leading to carcinogenesis (27, 28), the cytoplasmic staining of p63, a transcription factor involved in transactivation, apoptosis and proliferation that normally stains in the nucleus, may suggest an altered and potentially oncogenic function (8) for the mislocalized protein in prostate cancer progression and survival.
One of the strengths of our study is the use of a semi-automated, quantitative scoring system to determine objectively p63 expression on a continuous scale. We obtained sufficient tissue from the targeted areas, with 3 cores per patient (29) and evaluated differences in ranges below common threshold levels for p63-positivity (eg- <5%, <10%). We found no suggestion of degradation or altered p63 expression of tissue over time; for tumors of similar grade and stage, there was no overall trend in the variation of p63 percent staining by time since tissue collection (data not shown). Another strength of our study is the diversity of prostate cancers represented in pre- and post-PSA periods of diagnosis and a standardized Gleason scoring system. The cohort of men with available tissue did not significantly differ from the overall cohort of men treated by prostatectomy in PHS with respect to demographics such as age, BMI, smoking and physical activity, or clinical characteristics such as Gleason score or stage of disease (p>05; data not shown). One disadvantage of this cohort is that it was restricted to men who were surgically treated, which may limit the generalizability of findings and does not address whether biopsy specimens can also yield prognostic data. Our study was limited to 19 deaths, but we detected a significant association even after adjustment for important clinical covariates.
We observed low p63 expression in our population (median=2.5%), which is consistent with previous studies showing reduced levels of p63 in adenocarcinoma of the prostate (18, 23–26). Our findings are in contrast with two recent studies that reported an inverse association between p63 expression (as part of a genetic signature) and prostate cancer progression (6, 7). Bismar et al. generated a 12-gene signature for aggressive prostate cancer that included p63 based on its under-expression in metastatic cancer compared to benign tissue and localized disease, and the model was validated on a population of men followed for biochemical recurrence (6). In contrast to Bismar et al. who used PSA recurrence as an endpoint, our population was followed for the development of fatal disease, a more definitive outcome in terms of aggressive disease. Mucci et al. also reported lower levels of p63 staining with lethal prostate cancer in a Swedish watchful waiting cohort of men with T1a-T1b disease followed for up to 28 years (7). In our data, a low proportion of men presented with TURP-diagnosed T1a–T1b disease (10.4%), yet the findings in these men also showed a positive association between cytoplasmic p63 immunoreactivity and prostate cancer mortality (p = 0.06; data not shown). Our results may reflect a chance finding, a recently reported rare phenomenon in prostatectomy cases (30), or a real shift from the nucleus to the cytoplasm that has functional significance for prostate cancer progression.
The nuclear localization of p63 is essential for its role as a transcription factor. Similar to p53, alterations in nuclear-cytoplasmic shuttling may lead to cellular mislocalization, which disrupts regulation of cell cycle checkpoints and apoptosis, contributing to the initiation or progression of cancer (27, 28, 30–36). The cytoplasmic sequestration of p53 is associated with metastasis and poor long-term survival in patients with inflammatory breast carcinoma and colorectal carcinoma (32, 33, 37, 38) and similar aberrant immunoreactivity of p63 in the cytoplasm is associated with higher lung cancer mortality (8). The localization shift may arise from disruptions in the nuclear transport pathway (28), such as those mediated by the murine double minute-2 gene (Mdm2) (34, 39–42) where laboratory data show that p63-induced apoptosis is reduced when Mdm2 exports two isoforms of p63 (TAp63α and TAp63γ) from the nucleus to inhibit their transcription and pro-apoptotic activity (42). Our data linking higher levels of cytoplasmic p63 with reduced apoptosis and increased proliferation provide evidence for a potential mechanism of effect. The mislocalization and imbalance in p63 isoforms may alter p63 stability and function and thereby disrupt cell cycle arrest and apoptosis, which may have prognostic significance for cytoplasmic sequestration of p63 and the progression of prostate cancer.
Acknowledgments
Funding sources
The Physicians’ Health Study is supported by grants CA-34944, CA-40360, and CA-097193 from the National Cancer Institute and grants HL-26490 and HL-34595 from the National Heart, Lung, and Blood Institute, Bethesda, MD. This work was funded by US Army Prostate Cancer Program W81XWH-05-1-0562 and NIH 5R01 CA090598. Dr. Dhillon is supported by a Cancer Epidemiology Training Grant NCI T32 CA009001.
We are grateful to Haiyan Zhang and Jennifer Sinnott for programming assistance and to Michael Grady for his assistance with the figures. We are indebted to the men in the PHS Prostate Cancer Survivors Study. The Physicians’ Health Study is supported by grants CA-34944, CA-40360, and CA-097193 from the National Cancer Institute and grants HL-26490 and HL-34595 from the National Heart, Lung, and Blood Institute, Bethesda, MD. This work was funded by US Army Prostate Cancer Program W81XWH-05-1-0562 (PI Mucci) and NIH 5R01 CA090598 (PI Stampfer). Dr. Dhillon is supported by a Cancer Epidemiology Training Grant NCI T32 CA009001.
References
- 1.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998;4:747–8. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- 2.Signoretti S, Waltregny D, Dilks J, et al. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–75. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchini S, Marabese M, Marrazzo E, et al. DeltaNp63 expression is associated with poor survival in ovarian cancer. Ann Oncol. 2008;19:501–7. doi: 10.1093/annonc/mdm519. [DOI] [PubMed] [Google Scholar]
- 4.Lo Muzio L, Santarelli A, Caltabiano R, et al. p63 overexpression associates with poor prognosis in head and neck squamous cell carcinoma. Hum Pathol. 2005;36:187–94. doi: 10.1016/j.humpath.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Zigeuner R, Tsybrovskyy O, Ratschek M, Rehak P, Lipsky K, Langner C. Prognostic impact of p63 and p53 expression in upper urinary tract transitional cell carcinoma. Urology. 2004;63:1079–83. doi: 10.1016/j.urology.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Bismar TA, Demichelis F, Riva A, et al. Defining aggressive prostate cancer using a 12-gene model. Neoplasia. 2006;8:59–68. doi: 10.1593/neo.05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mucci LA, Pawitan Y, Demichelis F, et al. Testing a multigene signature of prostate cancer death in the Swedish Watchful Waiting Cohort. Cancer Epidemiol Biomarkers Prev. 2008;17:1682–8. doi: 10.1158/1055-9965.EPI-08-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narahashi T, Niki T, Wang T, et al. Cytoplasmic localization of p63 is associated with poor patient survival in lung adenocarcinoma. Histopathology. 2006;49:349–57. doi: 10.1111/j.1365-2559.2006.02507.x. [DOI] [PubMed] [Google Scholar]
- 9.Hall MC, Womack S, Sagalowsky AI, Carmody T, Erickstad MD, Roehrborn CG. Prognostic factors, recurrence, and survival in transitional cell carcinoma of the upper urinary tract: a 30-year experience in 252 patients. Urology. 1998;52:594–601. doi: 10.1016/s0090-4295(98)00295-7. [DOI] [PubMed] [Google Scholar]
- 10.Urist MJ, Di Como CJ, Lu ML, et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol. 2002;161:1199–206. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 12.De Laurenzi V, Costanzo A, Barcaroli D, et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–8. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores ER. The roles of p63 in cancer. Cell Cycle. 2007;6:300–4. doi: 10.4161/cc.6.3.3793. [DOI] [PubMed] [Google Scholar]
- 14.Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–99. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–5. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto K, Prives C. A role of cyclin G in the process of apoptosis. Oncogene. 1999;18:4606–15. doi: 10.1038/sj.onc.1202821. [DOI] [PubMed] [Google Scholar]
- 17.Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 18.Di Como CJ, Urist MJ, Babayan I, et al. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002;8:494–501. [PubMed] [Google Scholar]
- 19.Final report on the aspirin component of the ongoing Physicians’ Health Study. Steering Committee of the Physicians’ Health Study Research Group. N Engl J Med. 1989;321:129–35. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 20.Christen WG, Gaziano JM, Hennekens CH. Design of Physicians’ Health Study II--a randomized trial of beta-carotene, vitamins E and C, and multivitamins, in prevention of cancer, cardiovascular disease, and eye disease, and review of results of completed trials. Ann Epidemiol. 2000;10:125–34. doi: 10.1016/s1047-2797(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 21.Rubin MA, Bismar TA, Andren O, et al. Decreased alpha-methylacyl CoA racemase expression in localized prostate cancer is associated with an increased rate of biochemical recurrence and cancer-specific death. Cancer Epidemiol Biomarkers Prev. 2005;14:1424–32. doi: 10.1158/1055-9965.EPI-04-0801. [DOI] [PubMed] [Google Scholar]
- 22.Kim R, Demichelis F, Tang J, et al. Internet-based Profiler system as integrative framework to support translational research. BMC Bioinformatics. 2005;6:304. doi: 10.1186/1471-2105-6-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molinie V, Herve JM, Lugagne PM, Lebret T, Botto H. Diagnostic utility of a p63/alpha-methyl coenzyme A racemase (p504s) cocktail in ambiguous lesions of the prostate upon needle biopsy. BJU Int. 2006;97:1109–15. doi: 10.1111/j.1464-410X.2006.06069.x. [DOI] [PubMed] [Google Scholar]
- 24.Oliai BR, Kahane H, Epstein JI. Can basal cells be seen in adenocarcinoma of the prostate?: an immunohistochemical study using high molecular weight cytokeratin (clone 34betaE12) antibody. Am J Surg Pathol. 2002;26:1151–60. doi: 10.1097/00000478-200209000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Parsons JK, Gage WR, Nelson WG, De Marzo AM. p63 protein expression is rare in prostate adenocarcinoma: implications for cancer diagnosis and carcinogenesis. Urology. 2001;58:619–24. doi: 10.1016/s0090-4295(01)01311-5. [DOI] [PubMed] [Google Scholar]
- 26.Shah RB, Zhou M, LeBlanc M, Snyder M, Rubin MA. Comparison of the basal cell-specific markers, 34betaE12 and p63, in the diagnosis of prostate cancer. Am J Surg Pathol. 2002;26:1161–8. doi: 10.1097/00000478-200209000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res. 2003;282:59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 28.Hood JK, Silver PA. Diverse nuclear transport pathways regulate cell proliferation and oncogenesis. Biochim Biophys Acta. 2000;1471:M31–41. doi: 10.1016/s0304-419x(00)00018-4. [DOI] [PubMed] [Google Scholar]
- 29.Rubin MA, Dunn R, Strawderman M, Pienta KJ. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol. 2002;26:312–9. doi: 10.1097/00000478-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Osunkoya AO, Hansel DE, Sun X, Netto GJ, Epstein JI. Aberrant diffuse expression of p63 in adenocarcinoma of the prostate on needle biopsy and radical prostatectomy: report of 21 cases. Am J Surg Pathol. 2008;32:461–7. doi: 10.1097/PAS.0b013e318157020e. [DOI] [PubMed] [Google Scholar]
- 31.Inoue T, Stuart J, Leno R, Maki CG. Nuclear import and export signals in control of the p53-related protein p73. J Biol Chem. 2002;277:15053–60. doi: 10.1074/jbc.M200248200. [DOI] [PubMed] [Google Scholar]
- 32.Moll UM, Ostermeyer AG, Haladay R, Winkfield B, Frazier M, Zambetti G. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol. 1996;16:1126–37. doi: 10.1128/mcb.16.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci U S A. 1992;89:7262–6. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie L, Sasaki M, Maki CG. Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. J Biol Chem. 2007;282:14616–25. doi: 10.1074/jbc.M610515200. [DOI] [PubMed] [Google Scholar]
- 35.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–64. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci U S A. 1999;96:3077–80. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosari S, Viale G, Roncalli M, et al. p53 gene mutations, p53 protein accumulation and compartmentalization in colorectal adenocarcinoma. Am J Pathol. 1995;147:790–8. [PMC free article] [PubMed] [Google Scholar]
- 38.Sun XF, Carstensen JM, Stal O, et al. Prognostic significance of p53 expression in relation to DNA ploidy in colorectal adenocarcinoma. Virchows Arch A Pathol Anat Histopathol. 1993;423:443–8. doi: 10.1007/BF01606533. [DOI] [PubMed] [Google Scholar]
- 39.Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–73. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 40.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 41.Inoue T, Geyer RK, Howard D, Yu ZK, Maki CG. MDM2 can promote the ubiquitination, nuclear export, and degradation of p53 in the absence of direct binding. J Biol Chem. 2001;276:45255–60. doi: 10.1074/jbc.M107477200. [DOI] [PubMed] [Google Scholar]
- 42.Kadakia M, Slader C, Berberich SJ. Regulation of p63 function by Mdm2 and MdmX. DNA Cell Biol. 2001;20:321–30. doi: 10.1089/10445490152122433. [DOI] [PubMed] [Google Scholar]