Abstract
Theorists have long speculated on the mechanisms driving directed and spontaneous cell polarization. Recently, experimentalists have uncovered many of the mechanisms underlying polarization, enabling these models to be directly tested. In the process, they have demonstrated the explanatory and predictive value of these models and, at the same time, uncovered additional complexities not currently explained by them. In this review, we discuss some of main theories regarding cell polarization and highlight how the intersection of mathematical and experimental biology has yielded new insights into these mechanisms in the case of budding yeast and eukaryotic chemotaxis.
Introduction
Cells are not static entities but rather dynamically reorganize in response to internal and external cues. The ability to spontaneously form specialized domains of regulatory and structural elements is critical to the function of many cellular processes including differentiation, communication, and directed migration [1]. While cell polarization has been well documented, the driving mechanism has proved challenging to understand. Namely, how does a cell transition from homogeneous state to a heterogeneous, asymmetric one? And, as one author elegantly put it “how are heads made different from tails and everything in between?” [2]. Theorists have long puzzled over this question and proposed a number of potential models to address it. In the past decade, substantial progress has been made towards understanding the mechanisms involved in different polarization processes. These results have enabled various mathematical models to be tested and also uncovered new phenomena lacking in them. The aim of this review is to briefly highlight some of these theories and illustrate how the intersection between mathematical modeling and experimentation has led to new insights into the mechanisms behind cell polarization.
Theoretical Foundations
Theorists employ at least two approaches when constructing models of biological processes. In the bottom-up approach, modeling has been used to test whether a proposed set of biochemical reactions is capable of generating a specific response, such as polarization; if not, then this approach can be used to explore what reactions are possibly missing. Alternatively, in a top-down approach, a general mechanism is proposed and then various molecules and reactions are assigned roles within this mechanistic framework. In the past, this top-down approach was the one modelers most often employed, as little was known about the underlying biology. The resulting top-down models made specific predictions about the mechanisms generating these responses; specific in the sense that fundamental feature of the reaction networks were identified, such as positive/negative feedback and mutual inhibition, but not so specific as to establish which proteins were involved. As more has became known about the underlying biology, modelers have increasingly employed a bottom-up approach. Both approaches are not mutually exclusive and many models employ a combination of the two. In addition, both provide a common framework for integrating experimental data and generating testable hypotheses.
We begin by briefly discussing some common models used to explain how polarization is generated, many of which were developed before the underlying biology was known (and thus are examples of a top-down approach). Nearly all of these models treat polarization as an induced transition from a homogeneous state to an inhomogeneous one (Figure 1). Two additional assumptions are typically employed in developing this framework. The first is that the homogenous state is stable to uniform perturbations by not to some spatially non-homogeneous ones. In other words, a cell is happy to remain in an unpolarized state until it is coaxed into transitioning to polarized one, where the coaxing arises typically from exogenous factors such as chemical gradients or, alternatively, from intrinsic random fluctuations that generate small spatial asymmetries. The second assumption is that the transition is irreversible. Once an asymmetry develops, the cell will polarizes and then remain in the polarized state. Based on these two postulates, a number of related mechanisms have been proposed. Far and away the most influential is the concept of a diffusion-induced instability, proposed by its namesake, Alan Turing, over fifty years ago [3] and subsequently refined in the context of cell polarization by Gierer and Meinhardt [4–6]. The basic idea is that polarization results from two competing processes with different spatial characteristics, one local and the other global (Figure 2). This model assumes that polarization is induced by a small fluctuation or some external cue that is then amplified by the local process. Amplification is usually achieved by a self-reinforcing or autocatalytic mechanism. For example, a protein may randomly associate to the membrane. Once membrane bound, it then recruits other proteins to membrane. Left unchecked, this process would continue until the entire membrane is bound with protein, assuming there are sufficient reservoirs of cytosolic protein. A second process, therefore, is needed to restrict protein to a single cluster and prevent it from binding the entire surface of the membrane. To accomplish this, the local activating process is assumed to induce a global, inhibitory process that prevents these clusters from growing too large in size.
Figure 1. Polarization involves a transition from a symmetric, homogenous state to an asymmetric, inhomogeneous one.
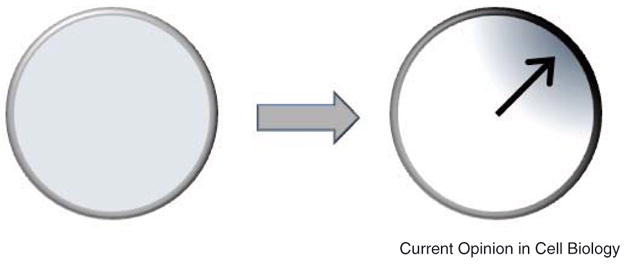
A key question concerns the mechanisms that enable cells to transition between from one state to the other.
Figure 2. Activator-inhibitor model for cell polarization.
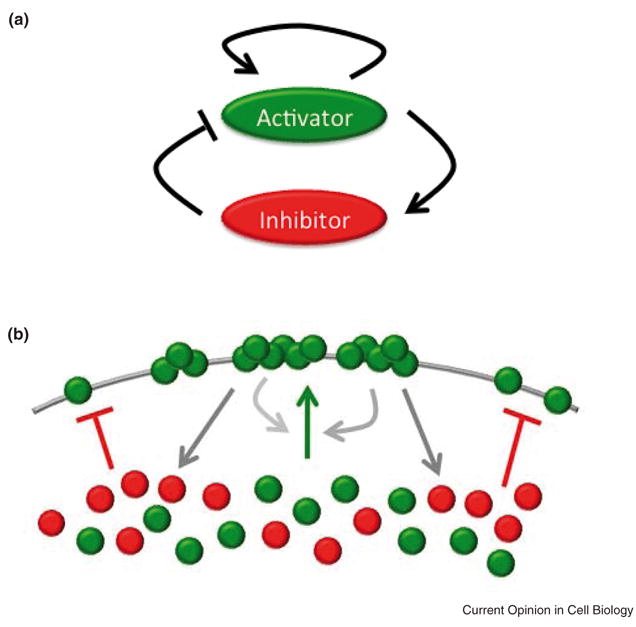
A. In this model, polarization is assumed to arise from the interplay between a local activator, capable of catalyzing its own production, and a global inhibitor. B. In the case of the formation of polarized clusters on the cell surface, membrane-bound activator (green) recruits other activator molecules to proximal regions of the membrane via a positive feedback mechanism. In addition, membrane-bound activator is assumed to recruit inhibitor molecules to the membrane (red). The inhibitor molecules prevent the activators from binding to the membrane. Unlike the activator, the inhibitor has a global reach. Competition between the activator and inhibitor limits the size and number of the clusters.
This inhibitory process is typically assumed to arise from the induction of some slow-acting, fast-diffusing molecule that counteracts the self-reinforcing mechanism involved in the local activation process. Slow action ensures that inhibition occurs after activation and fast diffusion ensures that the inhibitor has a rapid global reach within the cell, thereby limiting propagation of the local activating process. In terms of mechanisms, the fast-diffusing inhibitor is typically assumed to be a small, cytosolic molecule, whereas the local activation process is typically assumed to involve large, slowly-diffusing, membrane-bound molecules such as proteins.
As an alternative to the diffusion-induced mechanism described above, a number of researchers have proposed that substrate supply may limit the size of the cluster [5,7,8]. For example, in the case of proteins localizing to the membrane via an autocatalytic process, this process will continue until all of the available binding sites on the membrane are occupied or the cytosolic supply of protein is depleted. However, if protein supply is limiting, then membrane-associating proteins will not be able to saturate all of the binding sites. Moreover, as the local activation process is autocatalytic, the proteins will tend to associate in a few clusters.
Relative to a mechanism involving a global inhibitor, one involving substrate limitation is far simpler. No additional mechanisms involving global inhibitors are needed and, as a consequence, it is quite easy to convince oneself that any polarization process involving positive feedback involves substrate limitation. However, the substrate limitation mechanism is quite sensitive to parameters values. If the activator molecules or some co-factor is not limiting, then the transition will be from one homogenous state, where for example all of the activator is in the cytoplasm, to another, where all of it is bound uniformly to the membrane. Mechanisms involving diffusing inhibitors are often more robust to parameter variations, however they require additional mechanisms that, in many cases, have yet to be identified. We do note that there is some overlap from a mathematical perspective between these mechanisms, as depletion of a limiting substrate during the activation process can be viewed to play an equivalent role as a global inhibitor [6]. While in some cases these mechanisms are mathematically equivalent (or at least very similar), we note that in many other cases they are quite distinct [8]. Of course, other mechanisms and variations are equally possible. The basic formulation and key issues, nonetheless, are still the same: How is the cell able to robustly amplify a small fluctuation or signal on the surface of the cell yet limit activation to distinct regions of the cell?
Polarization in Budding Yeast
A classic model for polarization is the budding yeast Saccharomyces cerevisiae [9]. During the cell cycle, yeast transition from uniform to polarized growth in order to build a bud. A key step in initiating polarization involves the clustering of active, GTP-bound Cdc42 to the membrane [10]. Cdc42 then directs the nucleation of actin cables, which serve as conduits for delivering the necessary components for bud formation [11,12]. The transition to and maintenance of the polarized state is facilitated by both F-actin-dependent and independent positive feedback loops [11,13–17].
In an elegant series of papers involving modeling and experimentation, Altschuler, Li, Wu, and colleagues used budding yeast as a model for understanding the mechanism of spontaneous polarization [7,11,15]. Working with a constitutively active variant of Cdc42 that stably associates with the membrane in yeast arrested in the G1 phase (so that polarization is independent of any preexisting cues), they developed a simple experimental system that spontaneously polarizes in random directions when expression of Cdc42 is induced [11]. Using this system, they established that spontaneous polarization involves a positive feedback loop consisting of Cdc42-dependent actin polymerization and F-actin-dependent recruitment of Cdc42 to the membrane. These results suggest a mechanism where Cdc42 initially accumulates along the entire surface of membrane. Small fluctuations in the relative amount of Cdc42 are then amplified by the positive feedback loop, leading to the accumulation of Cdc42 at distinct clusters, known as caps, on the membrane. Based on this simple mechanism, they developed a mathematical model for polarization. A key prediction of their model was that as the initial amount of Cdc42 on the membrane increases the number of caps also increases; a result they experimentally validated. This result is easily interpreted with the model: induction of the positive feedback loop necessary for cluster formation requires that the proximal amount of Cdc42 exceed some threshold, in the sense that there is sufficient Cdc42 at a particular site on the membrane to effectively compete for co-factors necessary for activating the positive feedback loop. At low concentrations, the probability of exceeding this threshold is low and likely will occur at only one site. However, as the concentration increases, so does the probability that multiple sites will exceed this threshold.
One key issue that was not directly addressed in their initial studies was a mechanism for limiting cap growth. As emphasized in the previous section, some mechanism is needed to prevent a cluster from growing and eventually encompassing the entire membrane. In their initial model, Altschuler and colleagues implicitly assumed substrate limitation, both in terms of Cdc42 and the factors involved in forming the actin cables. In a subsequent study [7], they investigated polarization under conditions where actin polymerization was inhibited in order to eliminate F-actin-dependent positive feedback, leaving only the F-actin-independent one. By eliminating the F-actin-dependent loop, they could focus directly on how the supply of Cdc42 affects polarization independent of changes in the cytoskeleton. Consistent with their model predictions specific for this system, they found that increasing the supply of Cdc42 decreased the probability that a cell would polarize. In particular, as the supply of Cdc42 increases, there is a greater tendency for the molecule to associate along the entire membrane rather than cluster in distinct locations, an expected result from a mechanism involving substrate limitation.
Note that these two models make different predictions. In the first, where the actin-dependent feedback loop is present, increasing Cdc42 leads to the formation of multiple clusters while in the second, where this actin-dependent loop is absent, increasing Cdc42 decreases the probability that a cluster will form (and increases the probability that Cdc42 will be uniformly distributed on the membrane). These two predictions regarding the responses to increasing Cdc42 concentrations result from how the positive feedback was modeled. In case where actin-dependent feedback is present, the actin cables were assumed to provide a stable conduit for recruiting Cdc42 to the membrane. In other words, once the cables form (where the probability of nucleation is proportional to the proximal density of membrane-bound Cdc42), they stay formed and continue to deliver Cdc42 to the membrane at the site of nucleation, irrespective of the proximal concentration of Cdc42. In the model where only the actin-independent feedback loop is present, the process is transient as the kinetics are second order; the instantaneous activity of the positive feedback loop is directly proportional to the proximal concentration of membrane-bound Cdc42, whereas in former case the activity is independent of proximal concentration once the cables form. These differences illustrate how differences in the underlying kinetics can lead to different results in otherwise equivalent mechanisms.
In another remarkable study involving budding yeast, Ozbudak and colleagues investigated spontaneous polarization in the absence of the Rsr1 (Bud1) landmark protein [18]. Unlike wild-type cells where Rsr1 provides an internal cue directing polarization, in cells lacking Rsr1 polarization will occur at random locations [13,19]. Moreover, Ozbudak and colleagues found that the cap will move around the cell when Rsr1 is missing whereas it will remain fixed when present. In addition, they found that motion of cap was F-actin dependent. Based on their results, they constructed a mathematical model that predicted that polarization must involve an F-actin-dependent negative feedback loop that removes Cdc42 from the membrane. When Rsr1 is present, the F-actin-dependent positive feedback loop dominates the negative one and stably fixes the cap at one location. However, in the absence of Rsr1, the negative feedback loop dominates and the cap migrates as a traveling wave around the cell. One immediate question is whether this negative feedback loop has a role in regulating polarization when Rsr1 is present and, if so, what that role is. One possibility is that this putative negative feedback loop serves to control the dynamics of Cdc42 recruitment to the membrane and subsequent accumulation within the cap, in effect fine-tuning the polarization process.
These models are all examples of where a top-down approach was employed. In particular, these models investigated how positive and negative feedback can control different aspects of polarization without regard to many mechanistic details. One notable example of a bottoms-up approach to modeling yeast polarization is the work of Goryachev and Pokhilko [20]. Here, the authors constructed a mechanistic model of spontaneous yeast polarization in the absence of landmark proteins and actin polymerization. Key elements of their model include the cycling of Cdc42 between the active GTP-bound and the inactive GDP-bound states and the role of guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs), and GDP dissociation inhibitors (GDIs) in controlling Cdc42 recruitment and persistence on the membrane. Using their model, they were able to show that these biochemical reactions were sufficient for inducing polarization. Furthermore, to understand the general mechanism involved, Goryachev and Pokhilko were able to simplify their model and demonstrate that polarization results from a Turing-like instability where the cytoplasmic depletion of Cdc24-Bem1 GEF complex plays the role of the global inhibitor, a mechanism somewhat different than the one proposed in the models previously described where Cdc42 was assumed to be the limiting component. This kind of analysis is an elegant example of how a bottoms-up approach can be coupled with a top-down one. More often than not, detailed mechanistic models are difficult to analyze, as the complexity of governing reaction networks often obscures the key underlying processes and general mechanisms. By simplifying their model, they were able to merge these two model approaches, in the process gaining the insight offered by top-down models while simultaneously capturing much of the biological detail afforded by the bottoms-up approach.
Polarization and Gradient Sensing
Many kinds of cells are able to migrate in response to external cues [21,22]. For example, neutrophils and the slime mold Dictyostelium discoideum are able to detect shallow gradients of chemoattractants and crawl towards the source of these chemicals [23]. Prior to stimulation with chemoattractant, these cells exist in an unpolarized, non-motile state. When stimulated, they migrate by extending pseudopods at their front and contracting at their rear. This transition, from an unpolarized state to a polarized one, involves the selective recruitment of a number of proteins and lipids, initially distributed uniformly on either the membrane or in the cytosol, to either the front or back of the cell. These molecules align themselves with the external gradient and thus are thought to serve as a compass for the migrating cell [24,25]. With regards to modeling polarization, chemotaxis has attracted the most attention from theoreticians [26,27]. While a complete survey of these models is beyond the scope of this review, we wish to highlight a few key ideas and issues to emerge from them.
Both neutrophils and Dictyostelium discoideum sense chemoattractants using G protein coupled receptors (GPCR), which are uniformly distributed on their surface [28]. These cells sense gradients by detecting the number of ligand-bound receptors on their surface and then migrate in the direction where their number is greatest [29]. As receptors are activated along the entire surface of the cell, some mechanism is needed to discriminate between the front and back of the cell.
Perhaps the simplest and most elegant models proposed for gradient sensing, championed by Devreotes, Iglesias, and coworkers [30,31], involve local excitation and global inhibition (LEGI) (Figure 3). In the LEGI models, ligand-bound receptors recruit and/or produce both activator and inhibitor molecules on their membrane in their vicinity. The activator is produced locally, and its concentration is proportional to the number of ligand-bound receptors in its proximal vicinity. The inhibitor, on the other hand, has a global reach, as it is assumed to be rapidly diffusing and, therefore, does not accumulate in any particular region of the membrane. The net result is that more activator is present at the front of the cell than the rear in relation to the chemoattractant gradient, whereas the inhibitor is more uniformly distributed throughout the cell. Differences in the local concentration of activator and inhibitor are then hypothesized to establish the front and back of the cell.
Figure 3. Local excitiaton, global Inhibition (LEGI) mechanism for gradient sensing.
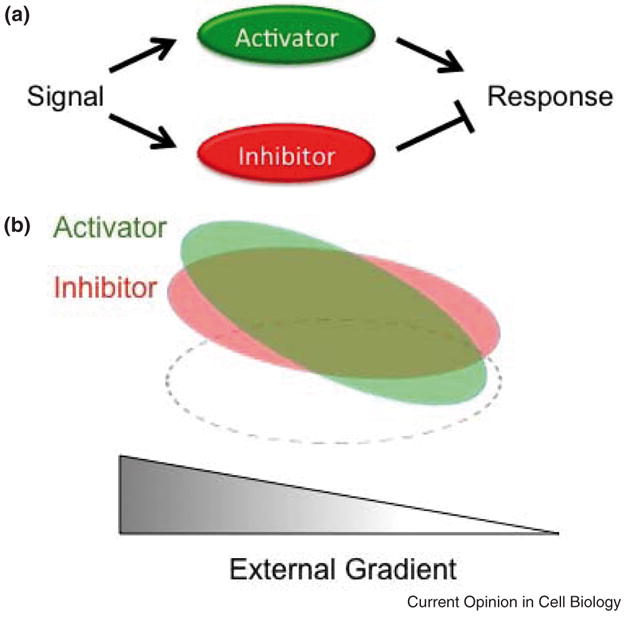
A. An external signal (eg, ligand-bound receptor), is assumed to activate two pathways, one involving a local activator and the other a global inhibitor of the downstream pathway used to establish the front of the polarized cell. B. As the activator (green) is more sensitive to the local strength of the external signal, more of it will accumulate in front of the cell than in the rear, mimicking the external gradient. The inhibitor (green) is less sensitive to the local strength of the external signal as it is assumed to be rapidly diffusing and, therefore, will be more homogeneously distributed throughout the cell. Local differences in the concentration of the activator and inhibitor are thought to establish to front and back of the cell. The relative timing of induction of the activator and inhibitor can also provide a mechanism for sensory adaptation [31]. Note the dashed ellipse is used to denote the cell boundary.
While the LEGI mechanism provides both a simple and robust mechanism for polarization in response to a gradient, experimental corroboration is still lacking. Whereas a number of candidates exist for the local activator, such as phosphatidylinositol-3-kinase (PI3K) or its product, phosphatidyl-3-4-5-triphosphate (PIP3) [25], no viable candidate for the global inhibitor has yet been identified. One additional limitation of the simple LEGI model as described above is that it fails to provide a mechanism for polarization in the absence of a gradient. Both neutrophils and Dictyostelium discoideum will randomly polarize and then migrate when stimulated with a uniform field of chemoattractant [32].
This discrepancy between model and experiments led to development of several new models for gradient sensing and polarization, each with select strengths and weaknesses [26]. Some have extended the LEGI mechanism to include, for example, random polarization in uniform chemoattractant fields by assuming that positive feedback enables a Turing-like instability to occur [33]. Others have employed a bottom-up approach to test if the known biological pathways are sufficient to cause spontaneous polarization [34–36].
As one example, we recently proposed a mathematical model for gradient sensing and polarization in neutrophils using a bottom-up approach [37]. Our model was motivated by recent work from Bourne and colleagues that suggested ligand-bound receptors activate two antagonizing pathways, one for establishing the front of the cell and the other the rear (Figure 4) [38]. Through modeling, we set out to explain how these two antagonizing pathways could possibly enable gradient sensing and polarization. In order to capture the general chemotactic behavior along with key mutant data, we first thought constructively about the underlying mechanism. One obvious fact was that the front pathway included a number of positive feedback loops. Another was that there was no reasonable candidate for a global inhibitor, at least based on the data at hand. Given these constraints, we naturally concluded that a mechanism involving substrate limitation must be involved. In our case, we assumed that PI3K was the limiting molecule though other candidates such as Ras had an equivalent effect. While sufficient for recruiting the components of two pathways to distinct and separate regions of the membrane in response either to a gradient or a uniform field of chemoattractant, this mechanism still did not explain why one pathway always localizes to the front of the cell and the other to the rear in response to gradient. To achieve selective localization of the two pathways, we needed to find a mechanism where the front pathway is more sensitive to the local concentration of ligand-bound receptors than the rear pathway. Based on the data available, we assumed that localization of the front pathway required the coincident activation of Ras and PI3K by ligand-bound receptors [39]. Coincident activation yielded a simple mechanism for robustly positioning the two pathways in response to an external gradient of chemoattractant. We note that a number of other models have also been proposed using a similar line of reasoning and that ours is in no way unique [35,40].
Figure 4. Schematic of proposed pathway regulating polarization and gradient sensing in neutrophils.
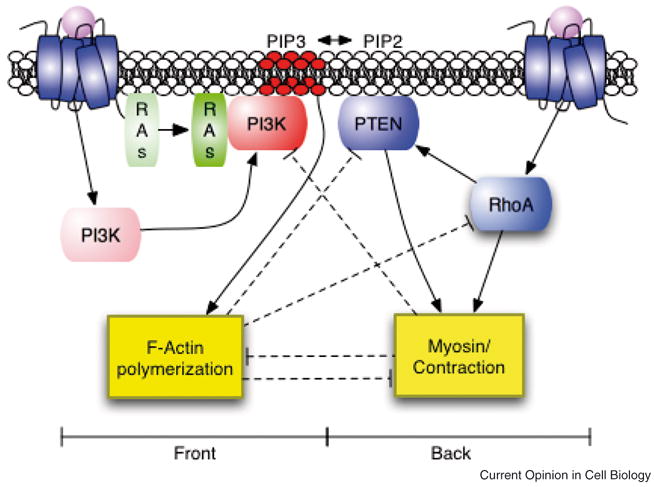
Ligand-bound receptors activate two antagonizing pathways that establish the front and back of the cell [38]. The “front” pathway activates F-actin polymerization and the “back” pathway activates myosin contractions. The two pathways are proposed to inhibit one another at five points (four are denoted by the dashed lines and the other results from the reciprocal action of PI3K and PTEN on PIP3 formation). From [37].
The example above describes a scenario where modeling was used to establish how the parallel activation of two antagonizing pathways, based on known molecular biology, may give to gradient sensing and polarization. While providing a sufficient mechanism for some aspects of chemotaxis, the model fails to account for others. For example, as positive feedback plays a key role in inducing polarization, the response to a chemoattractant is switch-like, and not dose dependent as observed experimentally [41]. In fact, this issue associated with models involving substrate limitation provides, arguably, the strongest case of a LEGI mechanism, which does not have this problem [42]. Of course, other issues associated with LEGI models provide reciprocal support other mechanisms, such as those involving substrate limitation. More significantly, subsequent experimental investigations have questioned the central role of PI3K signaling in chemotaxis [43,44] and found that multiple, redundant pathways control this process [45,46], indicating that we have just scratched the surface with regard to understanding the mechanisms driving chemotaxis.
Conclusions
We are now in a position to experimentally test many competing models of directed and spontaneous cell polarization. For theorists, the results so far have been very encouraging, as they have validated many key predictions from their models. At the same time, these results have uncovered new behaviors and mechanisms, requiring new concepts and models that will keep theorists busy for the foreseeable future. For experimentalists, mathematical models offer a systematic framework for quantitatively exploring the ingredients necessary for cell polarization and also analyzing the nonintuitive consequences that arise from the interplay of coupled processes such as diffusion and positive feedback. We have briefly covered only a few examples of how modeling can aid in this understanding; many more exciting examples exist that we were unfortunately unable to cover due to space limitations. However, we believe the examples cited demonstrate the explanatory and predictive power of mathematical modeling coupled with experimental endeavors, and that this approach can accelerate our understanding of complex biological processes.
Acknowledgments
We thank members of the Merrimack Pharmaceuticals and the Rao lab for helpful comments. C.V.R is supported is National Institutes of Health grant GM083601.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellettieri J, Seydoux G. Anterior-posterior polarity in C. elegans and Drosophila--PARallels and differences. Science. 2002;298:1946–1950. doi: 10.1126/science.1072162. [DOI] [PubMed] [Google Scholar]
- 3.Turing AM. The chemical basis of morphogenesis. Phil Trans Roy Soc B. 1952;237:37–72. [Google Scholar]
- 4.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 5.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci. 1999;112 (Pt 17):2867–2874. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]
- 6.Meinhardt H, Gierer A. Applications of a theory of biological pattern formation based on lateral inhibition. J Cell Sci. 1974;15:321–346. doi: 10.1242/jcs.15.2.321. [DOI] [PubMed] [Google Scholar]
- 7.Altschuler SJ, Angenent SB, Wang Y, Wu LF. On the spontaneous emergence of cell polarity. Nature. 2008;454:886–889. doi: 10.1038/nature07119. **This study employs a combination of experimental analysis and mathematical modeling to study a simplified mechanism for cell polarization. In the process, they are able to relate how the degree of polarization depends on the supply of the Cdc42 substrate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori Y, Jilkine A, Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys J. 2008;94:3684–3697. doi: 10.1529/biophysj.107.120824. *This paper develops the theory behind polarization involving substrate limitation. It also demonstrates that this mechanism is not mathematically equivalent to a Turing-like instability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- 10.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wedlich-Soldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 12.Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113 (Pt 4):571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 13.Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- 14.Wedlich-Soldner R, Wai SC, Schmidt T, Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–422. doi: 10.1016/j.cell.2007.02.043. *Using a combination of experimental analysis and mathematical modeling, the authors explore the role of endocytosis in established polarity in budding yeast expressing activated Cdc42. The authors conclude from their analysis that endocytosis rates are optimized in order to maximize both the height and sharpness of the polarized cap. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- 17.Butty AC, Perrinjaquet N, Petit A, Jaquenoud M, Segall JE, Hofmann K, Zwahlen C, Peter M. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. Embo J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozbudak EM, Becskei A, van Oudenaarden A. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev Cell. 2005;9:565–571. doi: 10.1016/j.devcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- 20.Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. **This paper develops a mathematical model of yeast polarization using a bottoms-up approach. It also provides an elegant example of how a detailed mechanistic model can be used to derive a simplified, top-down model. [DOI] [PubMed] [Google Scholar]
- 21.Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 22.Stephens L, Milne L, Hawkins P. Moving towards a better understanding of chemotaxis. Curr Biol. 2008;18:R485–494. doi: 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 23.Kay RR, Langridge P, Traynor D, Hoeller O. Changing directions in the study of chemotaxis. Nat Rev Mol Cell Biol. 2008;9:455–463. doi: 10.1038/nrm2419. [DOI] [PubMed] [Google Scholar]
- 24.Bourne HR, Weiner O. A chemical compass. Nature. 2002;419:21. doi: 10.1038/419021a. [DOI] [PubMed] [Google Scholar]
- 25.Rickert P, Weiner OD, Wang F, Bourne HR, Servant G. Leukocytes navigate by compass: roles of PI3Kgamma and its lipid products. Trends Cell Biol. 2000;10:466–473. doi: 10.1016/s0962-8924(00)01841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iglesias PA, Devreotes PN. Navigating through models of chemotaxis. Curr Opin Cell Biol. 2008;20:35–40. doi: 10.1016/j.ceb.2007.11.011. **This paper provides an insightful review of many of key issues and questions involved in modeling eukaryotic chemotaxis. [DOI] [PubMed] [Google Scholar]
- 27.Iglesias PA, Levchenko A. Modeling the cell’s guidance system. Sci STKE. 2002;2002:RE12. doi: 10.1126/stke.2002.148.re12. [DOI] [PubMed] [Google Scholar]
- 28.Servant G, Weiner OD, Neptune ER, Sedat JW, Bourne HR. Dynamics of a chemoattractant receptor in living neutrophils during chemotaxis. Mol Biol Cell. 1999;10:1163–1178. doi: 10.1091/mbc.10.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herzmark P, Campbell K, Wang F, Wong K, El-Samad H, Groisman A, Bourne HR. Bound attractant at the leading vs. the trailing edge determines chemotactic prowess. Proc Natl Acad Sci U S A. 2007;104:13349–13354. doi: 10.1073/pnas.0705889104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 32.Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: a focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 33.Narang A. Spontaneous polarization in eukaryotic gradient sensing: a mathematical model based on mutual inhibition of frontness and backness pathways. J Theor Biol. 2006;240:538–553. doi: 10.1016/j.jtbi.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 34.Meier-Schellersheim M, Xu X, Angermann B, Kunkel EJ, Jin T, Germain RN. Key role of local regulation in chemosensing revealed by a new molecular interaction-based modeling method. PLoS Comput Biol. 2006;2:e82. doi: 10.1371/journal.pcbi.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otsuji M, Ishihara S, Co C, Kaibuchi K, Mochizuki A, Kuroda S. A mass conserved reaction-diffusion system captures properties of cell polarity. PLoS Comput Biol. 2007;3:e108. doi: 10.1371/journal.pcbi.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skupsky R, Losert W, Nossal RJ. Distinguishing modes of eukaryotic gradient sensing. Biophys J. 2005;89:2806–2823. doi: 10.1529/biophysj.105.061564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onsum M, Rao CV. A mathematical model for neutrophil gradient sensing and polarization. PLoS Comput Biol. 2007;3:e36. doi: 10.1371/journal.pcbi.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, Kelly K, Takuwa Y, Sugimoto N, Mitchison T, Bourne HR. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 39.Suire S, Condliffe AM, Ferguson GJ, Ellson CD, Guillou H, Davidson K, Welch H, Coadwell J, Turner M, Chilvers ER, et al. Gbetagammas and the Ras binding domain of p110gamma are both important regulators of PI(3)Kgamma signalling in neutrophils. Nat Cell Biol. 2006;8:1303–1309. doi: 10.1038/ncb1494. [DOI] [PubMed] [Google Scholar]
- 40.Dawes AT, Edelstein-Keshet L. Phosphoinositides and Rho proteins spatially regulate actin polymerization to initiate and maintain directed movement in a one-dimensional model of a motile cell. Biophys J. 2007;92:744–768. doi: 10.1529/biophysj.106.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zigmond SH. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci U S A. 2004;101:8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Iijima M, Tang M, Landree MA, Huang YE, Xiong Y, Iglesias PA, Devreotes PN. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev Cell. 2007;12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN. PIP3-independent activation of TorC2 and PKB at the cell’s leading edge mediates chemotaxis. Curr Biol. 2008;18:1034–1043. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 46.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]


