Abstract
Ricin is a family member of the lethal ribosome-inactivating proteins (RIP) found in plants. Ricin toxin A-chain (RTA) from castor beans catalyzes the hydrolytic depurination of a single base from a GAGA tetraloop of eukaryotic ribosomal RNA to release a single adenine from the sarcin-ricin loop (SRL). Protein synthesis is inhibited by loss of elongation factor binding resulting in cell death. We report a sensitive coupled assay for the measurement of adenine released from ribosomes or small stem-loop RNAs by RTA catalysis. Adenine phosphoribosyl transferase (APRTase) and pyruvate orthophosphate dikinase (PPDK) convert adenine to ATP for quantitation by firefly luciferase. The resulting AMP is cycled to ATP to give sustained luminescence proportional to adenine concentration. Sub-picomole adenine quantitation permits the action of RTA on eukaryotic ribosomes to be followed in continuous, high-throughput assays. Facile analysis of RIP catalytic activity will have applications in plant toxin detection, inhibitor screens, mechanistic analysis of depurinating agents on oligonucleotides and intact ribosomes, and in cancer immunochemotherapy. Kinetic analysis of the catalytic action of RTA on rabbit reticulocyte 80S ribosomes establishes a catalytic efficiency of 2.6 × 108 M−1s−1, a diffusion limited reaction indicating catalytic perfection even with large reactants.
INTRODUCTION
Ricin from castor beans is among the most potent toxins and is a Category B bioterrorism threat. It is toxic by inhalation, oral, and intravenous exposure.1 The ricin type II RIP is comprised of a catalytic A-chain (RTA) and lectin B-chain linked by a single disulfide bond. Ricin entry into cells is mediated by lectin B-chain binding to cell surface galactose receptors.2 Following endocytosis, the toxin undergoes retrograde transport from the golgi to the endoplasmic reticulum where disulfide bond cleavage occurs and RTA is transferred to the cytosol in a chaperone-dependent process.3–5 In the cytosol, RTA binds the 28S portion of the 60S ribosomal subunit and catalyzes the hydrolytic depurination of A4324, the first adenosine of the conserved GAGA loop portion of the sarcin-ricin loop (SRL).6 Depurination of the SRL causes loss of elongation factor binding, inhibition of protein synthesis, and cellular death.
The sensitive detection of adenine is essential to establish the catalytic mechanism of RTA, screening inhibitor libraries against RTA action using intact ribosomes, and detection of ricin catalytic activity in unknown samples. Current methods of quantifying adenine from RIP depurination reactions include separation of adenine by HPLC (with or without fluorescent derivatization) and a continuous colorimetric enzyme coupled assay.7–9 These methods lack the sensitivity to measure the continuous rates of ribosome depurination by RIPs. Immunochemistry methods for ricin detect both the active and denatured enzyme.10, 11 A gel-resolved RNA fragment method using [32P]-radiolabeled ribosomes and aniline digestion at depurination sites is sensitive but cumbersome.12, 13
We report an enzymatically coupled assay with sufficient sensitivity to continuously measure a single adenine release from nanomolar concentrations of intact eukaryotic ribosomes. Femtomoles of ricin can be detected in minutes. The conversion of adenine to AMP by adenine phosphoribosyl transferase (APRTase, EC 2.4.2.7) has been reported as the first step in an adenine colorimetric assay for the detection of RIP activity with nanomole sensitivity for adenine.7 The measurement of AMP with sub-femtomole sensitivity employs the pyruvate orthophosphate dikinase (PPDK, EC 2.7.9.1) cycling reaction with firefly luciferase.14 Our assay combines APRTase and PPDK in either coupled or discontinuous reactions where free adenine is converted to AMP with APRTase and then to ATP with PPDK. The ATP generates light via firefly luciferase and regenerates AMP, which is rapidly converted to ATP by PPDK (Figure 1). Continuous measurements are accomplished in a 96-well plate format by combining the luciferase reagent with APRTase/PPDK (adenine to ATP) coupling enzymes.
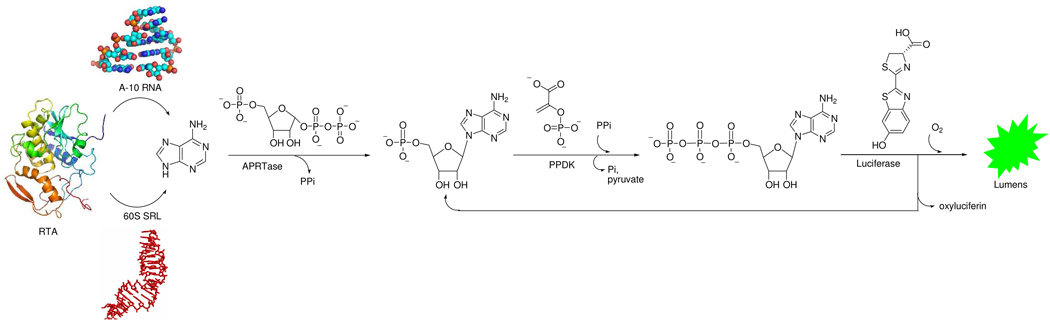
Figure 1.
Adenine released from the ricin A-chain (RTA) depurination of stem-loop (A-10) or ribosome (60S SRL) is converted to AMP with adenine phosphoribosyl transferase (APRTase) and then to ATP with pyruvate orthophosphate dikinase (PPDK). ATP drives the luciferase enzymatic reaction to produce luminescence and AMP to be recycled to ATP. At neutral pH all reactions can be combined to measure adenine continuously from ribosomes.
The depurination of 80S rabbit reticulocyte ribosomes and 60S yeast ribosomes by RTA was investigated using the adenine-luciferase coupled assay to establish the initial rate kinetic parameters benchmarked in both continuous and discontinuous assay formats. A-10, an RNA stem-loop (5’-CGCGAGAGCG-3’) mimic of the SRL was assayed at pH 4.0 with RTA to establish its kinetic parameters for comparison with kinetic results from an HPLC assay.8 A previously uncharacterized RNA/DNA hybrid stem-loop A-14 2-dA (5’-CGCGCGdAGAGCGCG-3’) was also assayed to characterize a high-efficiency synthetic substrate for routine detection and kinetic analysis of RIP activity.15
MATERIALS AND METHODS
Ricin toxin A-chain (RTA) was purchased from Sigma. Ricin (RCA60) from Vector Laboratories (Burlingame, CA) was a generous gift from Dr. Pamela Stanley (Albert Einstein College of Medicine). APRTase was the generous gift of Dr. Minkui Luo (Albert Einstein College of Medicine). The plasmid containing the PPDK clone from Clostridium symbiosum was a generous gift provided by Dr. Debra Dunaway-Mariano (University of New Mexico). 60S Yeast ribosomes were the generous gift from Dr. Uma Maitra (Albert Einstein College of Medicine). Rabbit reticulocyte lysate (untreated, $100) was purchased from Promega (Madison, WI) and is sufficient for ~100 assays as described below. Oligonucleotides A-10 and A-14 2-dA were purchased from Dharmacon (Lafayette,CO). Sufficient A-12 2-dA for 5,000 assays is currently $300. Firefly luciferase ATP assay kit (ATPlite) was purchased from Perkin Elmer (Waltham, Massachusetts). Phosphatase inhibitors (PhosSTOP) were purchased from Roche Applied Science (Indianapolis, IN). RNase inhibitor (SuperRNasin) was purchased from Ambion (Austin, TX). Buffers and enzyme preperations were checked for RNase activity using the RNaseAlert kit from Ambion (Austin, TX). DEPC treated water (0.1% DEPC stirred for 20 min followed by 30 min autoclave treatment) was used for all enzymatic reactions. All other reagents used were purchased in the highest purity available from Fisher Scientific (Pittsburgh, PA) or Aldrich Chemical Corp. (Ashland, MA). Spectrophotometric assays and concentrations of adenine, ribosomes, and oligonucleotides were measured using a Varian Cary 100 diode array spectrophotometer. Luminescence was measured on a GloMax 96-well luminometer from Promega (Madison, WI).
Adenine to ATP conversion buffer
50 mL of enzyme buffer was prepared with 100 mM Tris-acetate pH 7.7, 2 mM phosphoenolpyruvic acid, 2 mM sodium pyrophosphate, 2 mM 5-phospho-D-ribosyl-1-pyrophosphate (PRPP), 15 mM NH4SO4, and 15 mM (NH4)2MoO4 in DEPC treated water with phosphatase inhibitors (Roche PhosSTOP, 5 tablets). Charcoal/cellulose (1:5) powder was suspended in DEPC water and packed into a PD10 column with 100 mL of water at 2 mL/min. The enzyme buffer was passed through the column at 2 mL/min to remove adenine, AMP, and ATP contamination. The eluate was filtered (0.2 µm) and stored in 1 mL aliquots at −80 °C. Prior to use in adenine assays, 2× coupling buffer was prepared by adding 10 mM MgSO4, 4 units of APRTase , 8 units of PPDK , and 1 µL of SuperRNasin (Ambion) per 1 mL enzyme buffer. One unit of enzyme activity was defined as the amount which converts one µmole of substrate per minute at 20 °C. The enzymatic activity of PPDK was determined spectrophotometrically at 340 nM by coupling pyruvate formation to lactate dehydrogenase with 0.2 mM NADH and 50 µM AMP added to start the reaction at 20 °C in a buffer (500 µL 2× coupling buffer added to 500 µL H2O with 10 mM MgSO4 and 1 ul SuperRNasin (Ambion). APRTase activity was determined in similar conditions without AMP but with excess PPDK and 50 µM of adenine to start the reaction.
Expression and Purification of S. cerevisiae APRTase (scAPRTase)
scAPRTase (N-terminal 6 × His) was expressed and purified as described previously with some modification.16 The > 95% pure APRTase by SDS-PAGE was reconstituted with 10% glycerol (final volume ratio), concentrated and then purified on a Superdex75 (GE Science) in 50 mM Tris-HCl pH 7.4, 20 mM KCl, 5 mM MgCl2 containing 10% glycerol. The APRTase fractions were concentrated to ~10 mg/ml and flask frozen in dry ice/ethanol and stored at − 80 °C. The gel filtration purification step was needed to remove trace DNAase / RNAase activities in scAPRTase.
Expression and purification of Clostridium symbiosum PPDK
PPDK was expressed and purified as described previously with some modifications.17, 18 The plasmid encoding PPDK from Clostridium symbiosum was expressed in Escherichia coli JM101 grown in LB medium with 12.5 µM tetracycline to an A600 of 1.4. The cells were harvested by centrifugation at 2,000g for 30 minutes at 4°C. The cell pellet was suspended in buffer (20 mM imidazole, 2.5 mM Na2EDTA, 2 mM DTT, 75 mM KCl, pH 6.8 with complete protease inhibitor cocktail (Roche)) and the cells were disrupted by French press followed by centrifugation at 17,000g for 1 hour at 4 °C. Treatment with streptomycin sulfate, ammonium sulfate and DEAE cellulose column purification steps17, 18, PPDK was subsequently dialyzed in 20 mM imidazole, 2.5 mM Na2EDTA, 1 mM DTT, 88 mM KCl, pH 6.4 at 4 °C and loaded onto a Sephacryl S-200 column (GE Science) and eluted in dialysis buffer. Pure PPDK fractions were identified by SDS-PAGE. Glycerol to 10 % (final volume ratio) was added and aliquots were frozen in dry ice/ethanol and stored at −80°C.
Ribosomes
Rabbit reticulocyte 80S ribosomes were purified from untreated rabbit reticulocyte lysate. 500 µL of lystate was layered onto 250 µL of sucrose cushion (1 M sucrose, 20 mM Tris-HCl pH 7.5, 500 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, and 0.5 mM DTT) and centrifuged at 100,000 rpm in a TLA 120.2 rotor for 2 hours at 4 °C. The glassy pellet was washed twice on ice with ribosome storage buffer (20 mM Hepes-KOH pH 7.7, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 250 mM sucrose, and 1 mM DTT) and suspended in a minimal volume of the same buffer. The 80S ribosome was flash frozen in dry ice/ethanol and stored at − 80 °C. Ribosomes were stable while frozen, but multiple freeze-thaw cycles reduces their reactivity for the ricin assays. Once thawed, ribosome samples are stable on ice for the several hours needed to complete reactions.
Prior to use in catalytic assays, 60S and 80S ribosome were buffer exchanged into ribosome reaction buffer (20 mM Tris-HCl pH 7.4, 25 mM KCl, and 5 mM MgCl2) with a G-25 microcentrifuge desalting spin column at 4 °C (Pierce). Concentrations of 60S and 80S were calculated by absorbance at 260 nm.19 Active concentrations of 60S and 80S ribosome were determined by complete depurination by RTA in kinetic assays (see Methods). The contamination of AMP and ATP in isolated ribosomes were reduced by passing the ribosomes through a size exclusion spin column as described above.
Assaying samples for adenine
In the discontinuous assay format, 25 µL of 2× coupling buffer was added to 25 µL of sample and incubated at room temperature for several minutes. In a 96-well luminometer plate, 20 µL of the reaction was added to 80 µL of D-luciferin/luciferase reagent and was assayed for ATP with a luminometer as described by the manufacturer. Adenine content was calculated from a standard plot of Luminescence (RLU) versus varying pMoles of known adenine in identical sample buffer conditions.
For continuous assays of adenine, a 2× continuous assay buffer was prepared by adding 200 µl of D-luciferin/luciferase (ATPlite) to 1 mL of 2× coupling buffer. The 2× continuous assay buffer was equilibrated to room temperature (20 °C) for 10 minutes prior to assays. 25 µL of 2× continuous assay buffer was added to 25 µL of sample in a 96-well plate followed by 2 minutes of continuous luminescent (RLU) acquisition on a luminometer. Adenine content was calculated from a standard calibration curve in identical sample buffer conditions as described above.
In both assay formats, contamination of AMP and ATP was accessed in control reactions with sample by excluding APRTase or APRTase/PPDK respectively in the 2× coupling/continous assay buffers (Figure 1).
Discontinuous assay kinetics
Varying concentrations of substrate A-10 or A-14 2-dA were incubated at 37 °C for 5 minutes in acidic RTA buffer (10 mM potassium citrate-KOH and 1 mM EDTA at pH 4.0). Reactions were initiated by the addition of RTA at concentrations typically between 0.5–20 nM to a total reaction volume of 25 µL. The reactions were quenched to neutral pH at timed intervals with 25 µL of 2× coupling buffer to give < 15 % product formation. In parallel, five adenine standards and three controls (buffer, buffer + substrate, and RTA + buffer) were made in acidic RTA buffer diluted with 2× coupling buffer in volumes identical to the substrate assay condition. The quenched RTA reactions, standards, and control solutions were incubated at room temperature for at least 2 minutes (to convert adenine to ATP). 20 µL aliquots were assayed for pMoles of ATP as described. Luminescence (RLU) was converted to pMoles of adenine from the adenine standard curve fit (corrected for background from controls) and initial rate kinetics were fit to the Michaelis-Menten equation for the calculation of substrate kcat and Km.
For discontinuous ribosome assays, 60S or 80S ribosome were incubated for 5 minutes in pH 7.4 ribosome reaction buffer at 20 °C (a temperature identical to continuous assays). Reactions were initiated by the addition of RTA at concentrations between 30–70 pM in a total reaction volume of 25 µL. The reactions were quenched at timed intervals to give < 15 % product formation with 60 mM HCl , incubated on ice for 2 minutes, and brought back to neutrality with 60 mM KOH. An equal volume of 2× coupling buffer was added to the quenched reactions and incubated at room temperature for at least 2 minutes. Adenine standards and controls (as described for stem-loop assays) were prepared in ribosome reaction buffer and were quenched in an identical buffer condition as the ribosome RTA catalytic reactions. 20 µL aliquots were assayed for pMoles of ATP as described. Ribosome concentration was determined by depurinating (to completion) two stock concentrations with 500 nM RTA and comparing the final luminescence to the adenine standard curve fit. PicoMoles of adenine released during the assay and initial rate kinetics were calculated identically to stem-loop assays after appropriate background corrections were made from controls. Background degradation of any of the RNA substrates is not significant when standard RNA handling procedures are followed.
Continuous assay kinetics
In 96-well plate format, varying concentrations of ribosome were individually assayed in kinetic acquisition mode of the luminometer in a total reaction volume of 50 µL in 1× continuous assay buffer. RTA was prepared in ice cold ribosome reaction buffer with a single dilution from stock and equilibrated to room temperature before assays. A one minute blank reading for each ribosome concentration was made before the addition of RTA (30–70 pM) to ensure a stable luminescent signal. Luminescence was monitored for several minutes after RTA addition. Adenine standards in the concentration range of the assay reactions were preformed before and after continuous ribosome assays. Ribosome concentration was determined by depurinating (to completion) two stock concentrations with 500 nM RTA and comparing the final luminescence to the adenine standard curve fit. The initial rate of adenine formation from the catalytic reactions were calculated by converting luminescent rate (lumens / second) to enzymatic rate (pMoles of adenine / minute / pMoles enzyme) using the adenine standard curve fit and kinetic parameters kcat and Km were calculated by fitting initial rates to the Michaelis-Menten equation.
RESULTS AND DISCUSSION
Standard quantitation of adenine
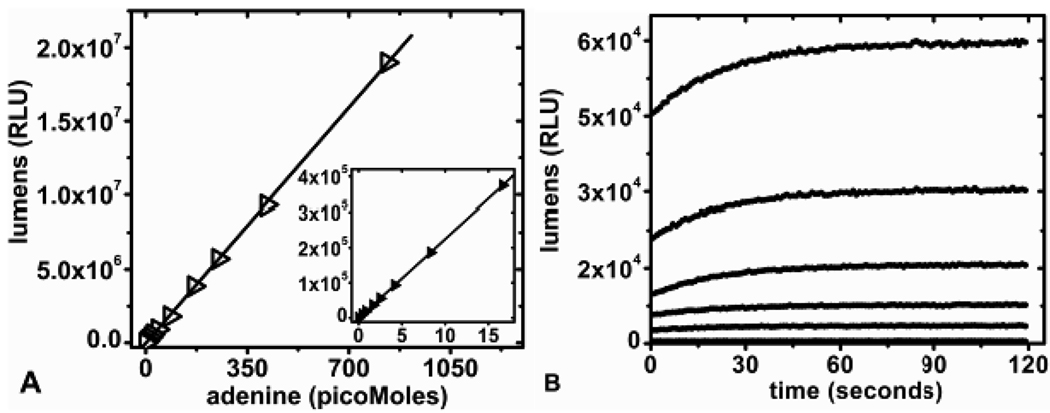
In the discontinuous assay format, adenine standards between 0.4 to 840 picomoles were used to establish a standard curve and gave a linear luminescence response when assayed with D-luciferin/luciferase reagent (Figure 2A). The conversion of adenine to ATP proceeds to chemical completion as determined by comparing luminescence signals from known quantities of adenine and ATP (data not shown). Both gave equivalent light signals under the conditions of Figure 2. In the continuous assay format, luminometer recordings of light output versus adenine concentration showed a stable luminescence signal after 2 min (Figure 2B). The Km values of adenine for APRTase and of AMP for PPDK are both 9 µM and several reactions of Figure 2A contained initial adenine concentrations lower than these Km values.16, 20 The coupling enzymes were in sufficient excess to reach full conversion to ATP in less than 2 min, even with sub-saturating adenine. Firefly luciferase has two Km values for ATP, 111 µM for the initial flash and 20 µM for the continuous but lower production of light.21 ATP concentrations of the aliquots assayed were below the initial flash and continuous light production Km values for the detection system. The linear response to adenine (ATP) in Figure 2A supports light production from the high Km process, resulting in a functionally linear response.
Figure 2.
Adenine standards: A) Plot of luminescence (RLU) versus adenine (0.4 – 840 pMoles) measured in the discontinuous assay format B) Continuous luminescent plots for increasing concentrations of adenine measured over a 2 min period. From bottom to top are blank, 0.5, 1, 2, 4, and 8 picomoles of adenine.
Discontinuous assay for adenine release from stem-loop RNA
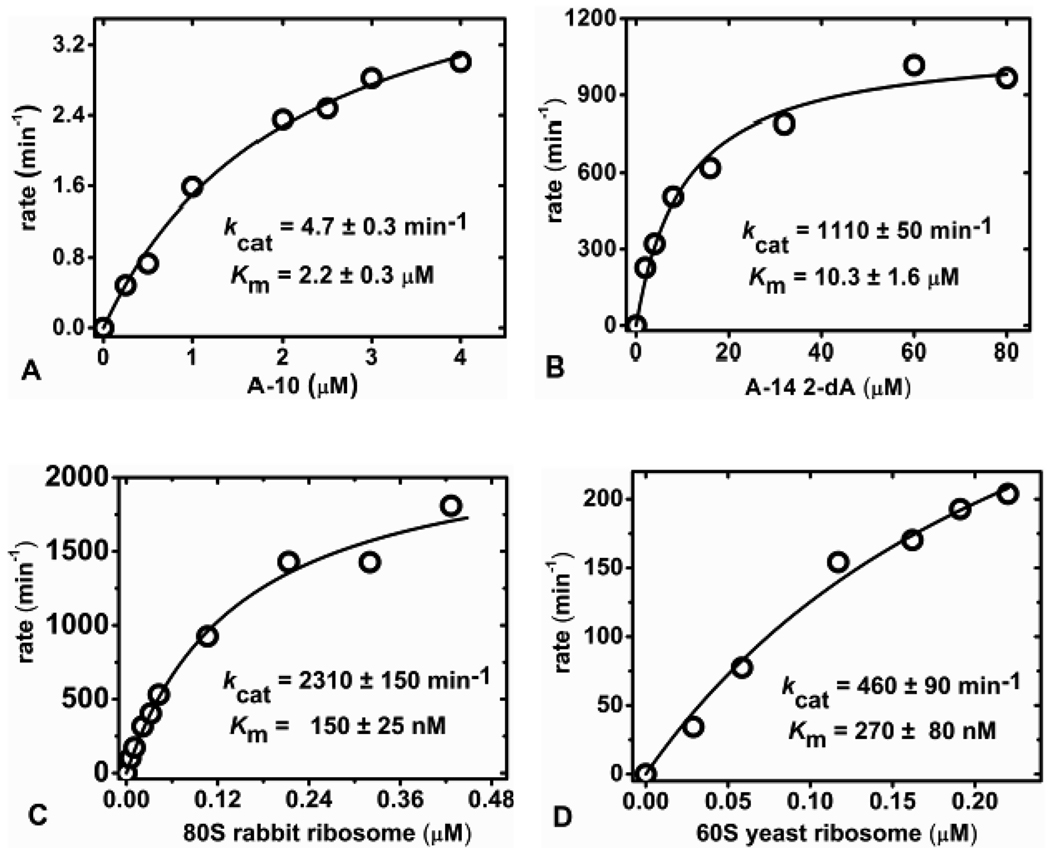
RTA depurinates an adenine from stem-loop RNA and RNA/DNA hybrid substrates at pH 4.0, but not at pH 7.8, 15, 22 Small RNA stem-loops were assayed discontinuously at pH 4.0 and the reaction stopped by the addition of 2× coupling buffer to provide a neutral pH. RTA initial rate kinetics for the catalysis of stem-loop substrate A-10 measured by the adenine-luciferase assay gave a saturating curve fit with a calculated kcat of 4.7 ± 0.3 min−1 and Km of 2.2 ± 0.3 µM (Figure 3A). Previously determined kinetics for RTA catalysis using HPLC analysis of adenine release in large scale reactions with A-10 are consistent with these values (kcat of 4.1 min−1 and Km of ~ 4.0 µM).8 With the adenine-luciferase assay, RTA catalysis of the A-14 2-dA stem-loop RNA/DNA hybrid was measured to give a kcat of 1110 ± 50 min−1 and a Km of 10.3 ± 1.6 µM at pH 4.0 (Figure 3B). The deoxyadenosine at the depurination site provides a better substrate compared to RNA A-14 (a reported kcat of 219 ± 14 min−1 and Km of 8.1 ± 0.7).8 Deoxyadenosine at the depurination site (GdAGA) of RNA stem-loop oligonucleotides increases the catalytic efficiency (kcat/Km) by RTA a factor of ~ 4 from catalysis.15 RTA is known to form a ribocation transition state and incorporation of the deoxyribosyl group specifically at the depurination site chemically destabilizes the ribosidic bond to the adenine and thereby increases the reaction rate.15, 23, 24
Figure 3.
Discontinuous adenine-luciferase assay format kinetic curve fits for RTA catalysis of stem-loop RNA A-10 (A) and A-14 2-dA (B) at pH 4.0. Kinetic curve fits for RTA catalysis of 80S rabbit (C) and 60S yeast (D) ribosome at pH 7.4 in 20 mM tris-HCl, 25 mM KCl, and 5 mM MgCl2.
RTA and other type I and II RIPs have a low pH catalytic optimum on nucleic acid substrates such as stem-loop RNA, poly(A) and herring sperm DNA, while the natural ribosome substrate is depurinated optimally at physiologic pH.9, 25 RTA catalysis of stem-loop substrates was not detectable above pH 6.5 with either A-10 or A-14 2-dA stem-loop oligonucleotides (up to 100 µM) with 2 µM RTA for 10 minutes. The pH 4.0 catalytic optimum of RTA on 6 to 18-mer stem-loop DNA and RNA 28S SRL mimic oligonucleotides is known to result from the protonation of two ionizable residues on substrate and/or RTA.8, 15, 22
Discontinuous assay of adenine release from ribosomes
Kinetic analysis of reactions using intact ribosomes as substrates where the Km for ribosomes is low and a single adenine is released from the macromolecular complex of approximately 5 million Daltons presents a challenge. AMP and ATP contamination in ribosome preparations were reduced with a size exclusion spin column (materials and methods). The initial rates of adenine release from ribosomes at pH 7.4 in a discontinuous assay involved quenching the reaction followed by adenine conversion to ATP. RTA reactions were quenched with HCl to inactivate RTA and subsequently neutralized to pH 7.0 with KOH. The depurination of ribosomes by RTA occurred with a stoichiometry of one adenine released per ribosome. This finding provides a unique method to quantify the chemical concentration of eukaryotic ribosomes. Using an enzyme concentration of 33 pM RTA, the initial rate kinetics for depurination of 80S rabbit reticulocyte ribosomes gave a saturation curve with a kcat of 2310 ± 150 min−1 and a Km of 150 ± 25 nM at 20 °C (Figure 3C). This reaction has a catalytic efficiency (kcat/Km) of 2.6 × 108 M−1s−1, at or near the diffusion limit for enzymatic catalysts with slowly diffusing substrates and a 32 kD enzyme.26 Initial rate kinetics for RTA depurination of 60S yeast ribosomes at pH 7.4 conformed to Michaelis-Menten kinetics with a kcat of 460 ± 90 min−1 and a Km of 270 ± 80 nM at 20 °C (Figure 3D).
Continuous assay for adenine release from ribosomes
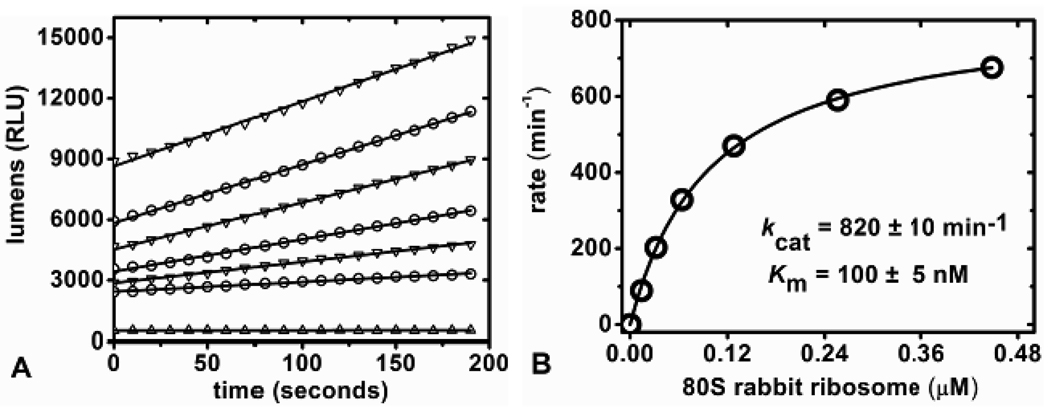
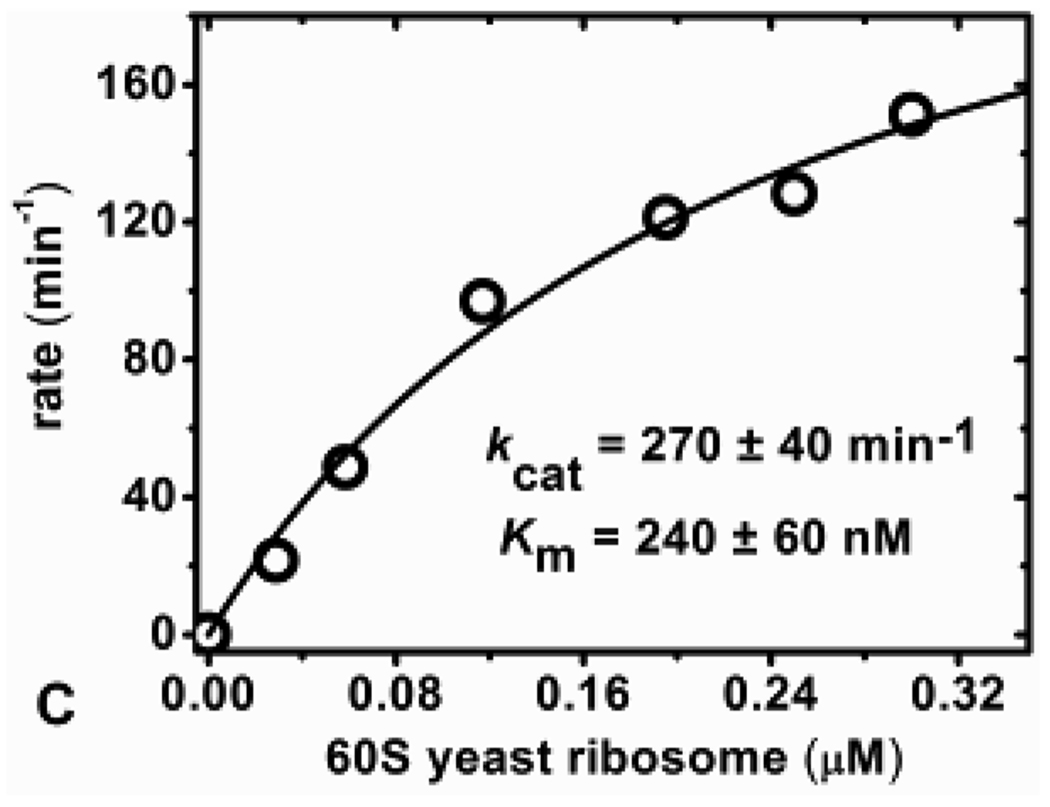
In continuous coupled assays for RTA, the conversion of adenine to ATP must be rapid relative to the release of adenine from the substrates. With fixed adenine concentrations, even at the highest concentrations, steady-state light production occurred within two minutes (Figure 2B). We considered it feasible to use continuous coupled assays for adenine release from intact ribosomes. RTA catalyzed release of adenine from 80S rabbit and 60S yeast ribosomes were measured continuously at pH 7.7 by monitoring a linear increase in luminescence. The concentration of RTA in continuous ribosome reaction assays was kept low to assure that production of adenine is rate limiting. In the continuous ribosome assays, the concentrations of RTA was 33 pM and 66 pM for 80S and 60S ribosomes respectively. The initial rates were dependent on ribosome concentration (Figure 4A). The initial rates of catalysis as a function of 80S ribosome concentration fit to the Michaelis-Menten equation with a kcat of 820 ± 10min−1 and Km of 100 ± 5nM (Figure 4B). Under the same conditions, the kcat and Km values for RTA catalysis of 60S yeast ribosomes were 270 ± 40 min−1 and 240 ± 60 nM respectively (Figure 4C). RTA action on ribosomes in continuous and discontinuous assays gave similar Km values but a ~ 2–3-fold difference in kcat (Table 1). This difference potentially results from varying conditions between the two assay formats but this result was not further investigated. 80S rabbit and 60S yeast ribosomes have Km values in the range of 100 to 270 nM with the tightest binding being 100 nM for 80S rabbit ribosomes under the conditions used for the continuous assay. Previous reports of the action of RTA on 80S rabbit reticulocyte ribosomes by aniline cleavage, [32P] end-labeling and gel separation gave a kcat of approximately 1500 min−1 and Km of 100 to 200 nM depending on conditions, completely comparable with the adenine-luciferase assays (Table 1).12 The ribosomes are depurinated at catalytic rates (kcat) comparable to that of deoxyadenosine (GdAGA) stem-loop oligonucleotides A-14 2-dA assayed at pH 4.0 (Table 1), but the Km value for the interaction with ribosomes indicates tighter binding.
Figure 4.
Continuous assay measurements. A) Initial rate slopes (lumens/second) of increasing concentrations of 80S rabbit ribosome with 33 pM RTA and B) kinetic curve fit of the calculated kinetic rate from (A) versus 80S ribosome concentration. C) Kinetic curve fit for RTA catalysis of 60S yeast ribosome measured in the continuous adenine-luciferase assay.
Table 1.
RTA kinetic parameters
| Substrate | Km (µM) | kcat (min−1) | kcat / Km (M−1s−1) |
|---|---|---|---|
| A-10 d* | 2.2 ± 0.3 | 4.7 ± 0.3 | 3.6 × 104 |
| A14 2-dA d* | 10.3 ± 1.6 | 1110 ± 50 | 1.8 × 106 |
| 60S yeast ribosome d | 0.270 ± 0.08 | 460 ± 90 | 2.8 × 107 |
| 60S yeast ribosome c | 0.240 ± 0.06 | 270 ± 40 | 1.9 × 107 |
| 80S rabbit ribosome d | 0.150 ± 0.025 | 2310 ± 150 | 2.6 × 108 |
| 80S rabbit ribosome c | 0.100 ± 0.005 | 820 ± 10 | 1.4 ×108 |
substrate assayed in discontinuous format
substrate assayed in continuous format
substrate assayed at 37 °C
We investigated if stem-loop catalysis of A-10 or A-14 2-dA occurs at neutral pH in the presence of ribosomes (rabbit or yeast), but only adenine released from ribosome depurination was observed with RTA. Mammalian ribosomal proteins L9 and L10e are adjacent to the RTA SRL binding site and are likely to provide recognition elements for RTA catalysis.27 Conformational changes in RTA upon ribosome binding have also been proposed.28 Thus, the pH-modulation effects occur only in the ribosome-RTA complex and are not maintained for RTA release and rebinding of small stem-loop RNAs under physiologic conditions with ribosomes.
Catalytic perfection in RTA depurination
Catalytic perfection in enzymatic reactions results when every diffusional collision between substrate and enzyme leads to product formation. In these cases, the catalytic rate is defined by the diffusion rate of the smaller reactant.29 Well characterized enzymes exhibiting catalytic perfection include triose phosphate isomerase (kcat/Km = 3.0 × 108 M−1s−1), acetylcholinesterase (kcat/Km = 1.4 × 108 M−1s−1), ketosteroid isomerase (kcat/Km = 1.3 × 108 M−1s−1) and β-lactamase (kcat/Km = 1.0 × 108 M−1s−1).26, 30 Remarkably, the catalytic efficiency (kcat/Km) for RTA on rabbit 80S ribosomes (2.6 × 108 M−1s−1 for the discontinuous assay conditions and 1.4 × 108 M−1s−1 for the continuous assay) are similar, even though diffusion for the RTA-ribosome system is limited by the relatively large molecular weight of RTA (Mr = 32 kDa). The finding that RTA has a kcat/Km value above the diffusion limit for uncharged reactants of this size indicates that the charge difference between RTA and the ribosome plays a significant role in its extraordinary catalytic efficiency.
Ricin/RTA detection
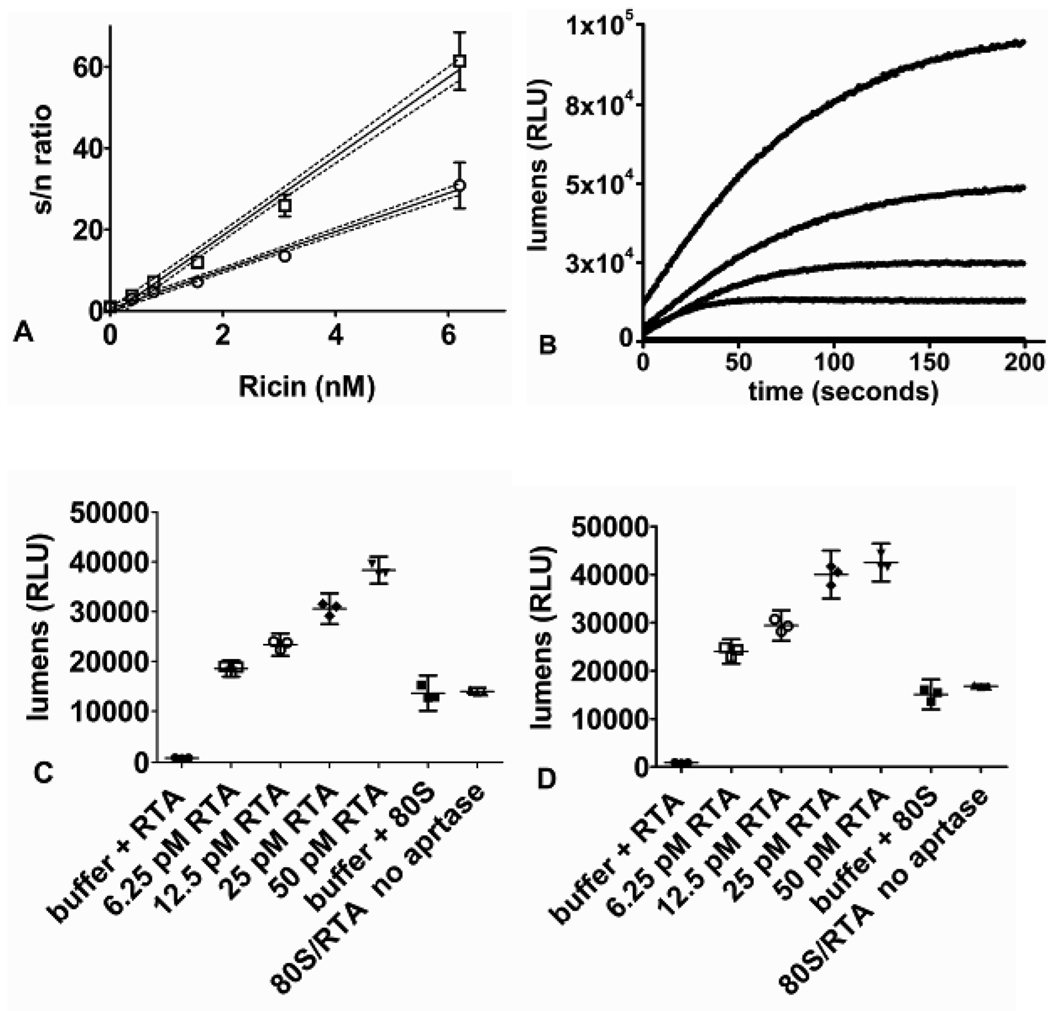
One application for a sensitive and rapid quantification of adenine is to permit the detection of RTA, ricin and related RIPs in samples using small stable nucleic acid constructs. Ricin holotoxin can depurinate synthetic substrates including A-14 2-dA without reductive pretreatment to release the catalytic A-chain (RTA).9 Truncated stem-loop substrates have a pH optimum of 4.0 for binding and catalysis, and the two-step discontinuous assay is optimal for this application. The RNA stem loop A-10 is hydrolyzed at 4.7 min−1 by RTA (Table 1). Based on the sensitivity of the assay here, a 50 µL assay containing 20 µM A-10 and 6.6 nM of RTA (~ 200 ng/mL) would provide a robust luminescent signal in 30 sec. The more active A-14 2-dA (kcat of ~ 1110 min−1) incorporates a labile adenine in an RNA/DNA hybrid with a 2'-deoxy ribose at the cleavage site and would be an ideal synthetic substrate for sub-nanomolar RTA/ricin detection (Table 1). In 25 µL reactions, depurination of 4 µM A-14 2-dA with ricin concentrations as low as 400 pM (25 ng/mL) were detected in an assay time of 20 min without reductive pretreatment with the detection limit defined as a signal to noise ratio (S/N) of 3 (Figure 5A). Ricin detection in environmental or serum samples would need to be desalted/buffer exchanged into acidic RTA buffer prior to detection assays to remove contaminating AMP/ATP and physiologic salts (inhibitory to the pH 4.0 RTA/ricin catalytic reaction). Sample RNase contamination is is analyzed in control reactions by omitting APRTase in the 2× coupling buffer and quantifying ATP formation from nuclease generated AMP (Figure 1, Figure 5C,D). Thus, samples containing interfering nuclease contamination would be identified and require applicable commercial RNase inhibitor treatment.
Figure 5.
A) Plot of luminescent signal to noise ratio (s/n ratio) versus concentrations of ricin. Ricin concentrations were from 0.380 to 6.2 nM in the discontinuous assay format. Reaction mixtures of 25 µL contained acidic RTA buffer + 0.005% triton X-100 with 4 µM A-14 2-dA at 37 °C for 10 min (open circles) and 20 min (open squares). Triplicate samples gave the mean and 95% confidence interval error bars. A linear fit to 95% confidence interval data points are shown (disconnected lines). B) Plot of lumens (RLU) versus time (seconds) for the detection of 140 pM (4.5 ng/mL) RTA with increasing concentrations of 80S rabbit ribosome in 50 µL reactions ( bottom to top: control, 20 nM , 40 nM , 80 nM, and 160 nM 80S ribosome). The continuous assay format has the background signal subtracted from the curves. C,D) Data points of lumens (RLU) versus controls: (buffer + RTA, buffer + 80S, 80S/RTA no APRTase) and samples containing: (RTA (6.25 to 50 pM) with 50 nM 80S ribosome) repeated in triplicate and measured for 1 second in the continuous assay format after 5 (C) and 10 (D) minutes of reaction respectively. Error bars were calculated as 95% confidence intervals.
80S ribosome is the most efficient RTA detection probe with the lowest Km and fastest kcat of substrates described herein (Table 1). 140 pM (4.5 ng/mL) of RTA in 50 µL reactions was detected within minutes using increasing concentrations of gel filtered 80S ribosome in the continuous adenine-luciferase assay format (Figure 5B). In the same format using 50 nM crude 80S ribosome substrate, the RTA detection limit (S/N ratio of 3) was 50 pM ( 1.6 ng/mL) in 5 minutes and 25 pM (0.8 ng/mL) in 10 minutes (Figure 5C,D). For the routine detection of ricin/RTA using 80S ribosomes, reductive pretreatment of the sample (ricin holotoxin), stringent nuclease control, and sensitive ribosome preparations make this method less practical than the discontinuous ricin detection assay using truncated stem-loop A-14 2-dA substrate (Figure 5A).
CONCLUSIONS
Coupling firefly luciferase to ATP formed from adenine permitted sensitive quantification of the RTA depurination reaction. Luminescence from adenine is produced from APRTase, PPDK, and D-luciferin/luciferase coupling enzymes (Figure 1). The assay can measure sub-picomole quantities of adenine in discontinuous and/or continuous assay formats (Figure 2). A luminescent signal stable for several minutes is observed with an optimized D-luciferin/luciferase formulation along with AMP cycling from PPDK and luciferase enzymes (Figure 2, Figure 1). We envision that the buffer conditions and D-luciferin/luciferase preparation of the adenine-luciferase assay buffers can be further optimized to adapt it to other applications where adenine detection is desired. For assaying larger quantities of adenine (mM), the adenine to ATP coupling buffer (2× coupling buffer) can be used spectrophotometrically by coupling the pyruvate generated by PPDK to a lactate dehygrogenase/NADH assay (Figure 1, materials and methods).
RTA kinetics were investigated on truncated sarcin-ricin loop (SRL) mimics A-10 and A-14 2-dA at acidic pH discontinuously where the adenine-luciferase assay provided a neutral pH quench and converted adenine to ATP (Figure 3 A,B). A-14 2-dA, a hybrid RNA/DNA stem-loop, was efficiently catalyzed (kcat/Km = 1.8 × 106 M−1s−1) by RTA and could serve as a rapid detection probe for the active toxin or access RTA immunoconjugate catalytic activity. Using A-14 2-dA substrate, untreated ricin was detected in 20 minutes with a limit of 400 pM (25 ng/mL) toxin (Figure 5A).
RTA kinetics on ribosomes demonstrated low nanomolar binding (Km = 100 and 270 nM) for 80S rabbit reticulocyte and 60S yeast respectively (Table 1). The catalysis of 80S ribosome by RTA approached the diffusion rate limit for enzymatic reactions with a catalytic efficiency (kcat/Km) of 2.6 × 108 M−1s−1 for the discontinuous assay conditions and 1.4 × 108 M−1s−1 for the continuous assay format. Catalytic perfection is achieved through the combined interactions of the polycationic nature of the RTA active site, anionic distribution of the substrate, and potential remote contacts of RTA with adjacent SRL ribosomal proteins. The 80S rabbit ribosome serves as a rapid continuous detection probe for active RTA. 140 pM (4.5 ng/mL) of RTA in 50 µL reactions was detected within minutes by directly measuring adenine release from 80S rabbit ribosome (Figure 5B). The RTA detection limit using 80S ribosome in continuous assays was 50 pM (1.6 ng/mL) in 5 minutes and 25 pM (0.8 ng/mL) in 10 minutes (Figure 5C,D). Ricin holotoxin detection via 80S ribosome catalysis would require reductive pretreatment of the sample. The adenine-luciferase assay can easily be adapted for high throughput to detect potential RTA inhibitors or the activity based detection of other RIP enzymes to aid in isolation and discovery.
Abbreviations
- RTA
ricin A-chain
- RIP
ribosome inactivating protein
- SRL
sarcin-ricin loop
- AMP
adenosine 5’-monophosphate
- APRTase
adenine phosphoribosyltransferase
- PPDK
pyruvate orthophosphate dikinase
- ATP
adenosine 5’-triphosphate
- HPLC
high performance liquid chromatography
- PAGE
polyacrylamide gel electrophoresis
- PEP
phosphoenolpyruvate
- PPi
inorganic pyrophosphate
- DEPC
diethylpyrocarbonate
- PRPP
5-phospho-α-D-ribosyl-1-pyrophosphate
- NADH
reduced nicotinamide adenine dinucleotide
- LB
Luria broth
- DTT
dithiothreitol
- SDS
sodium dodecylsulfate
- EDTA
ethylenediaminetetraacetic acid
- A-10
stem-loop RNA 5’-CGCGAGAGCG-3’
- A-14 2-dA
stem-loop RNA/DNA 5’-CGCGCGdAGAGCGCG-3’, where dA is 2’-deoxy-AMP in 3’,5’-linkage
Footnotes
Supported by NIH research grant CA072444 and NIH training grant GM07288
REFERENCES
- 1.Franz DR, Jaax NK. 1997;32:631–642. [Google Scholar]
- 2.Nicolson GL, Blaustein J, Etzler ME. Biochemistry. 1974;13:196–204. doi: 10.1021/bi00698a029. [DOI] [PubMed] [Google Scholar]
- 3.Sandvig K, vanDeurs B. Physiol. Rev. 1996;76:949–966. doi: 10.1152/physrev.1996.76.4.949. [DOI] [PubMed] [Google Scholar]
- 4.Wesche J, Rapak A, Olsnes S. J Biol Chem. 1999;274:34443–34449. doi: 10.1074/jbc.274.48.34443. [DOI] [PubMed] [Google Scholar]
- 5.Slominska-Wojewodzka M, Gregers TF, Wälchli S, Sandvig K. Mol. Biol. Cell. 2006;17:1664–1675. doi: 10.1091/mbc.E05-10-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo Y, Mitsui K, Motizuki M, Tsurugi K. J. Biol. Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 7.Heisler I, Keller J, Tauber R, Sutherland M, Fuchs H. Analytical Biochemistry. 2002;302:114–122. doi: 10.1006/abio.2001.5527. [DOI] [PubMed] [Google Scholar]
- 8.Chen XY, Link TM, Schramm VL. Biochemistry. 1998;37:11605–11613. doi: 10.1021/bi980990p. [DOI] [PubMed] [Google Scholar]
- 9.Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F. Nucleic Acids Research. 1997;25:518–522. doi: 10.1093/nar/25.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becher F, Duriez E, Volland H, Tabet JC, Ezan E. Anal. Chem. 2007;79:659–665. doi: 10.1021/ac061498b. [DOI] [PubMed] [Google Scholar]
- 11.Poli MA, Rivera VR, Hewetson JF, Merrill GA. Toxlcon. 1994;32:1371–1377. doi: 10.1016/0041-0101(94)90409-x. [DOI] [PubMed] [Google Scholar]
- 12.Olsnes S, Fernandez-Puentes C, Carrasco L, Vazquez D. Eur. J. Biochem. 1975;60:281–288. doi: 10.1111/j.1432-1033.1975.tb21001.x. [DOI] [PubMed] [Google Scholar]
- 13.Endo Y, Tsurugi K. The Journal of Biological chemistry. 1988;263:8735–8739. [PubMed] [Google Scholar]
- 14.Sakakibara T, Murakami S, Eisaki N, Nakajima M-o, Imai K. Analytical Biochemistry. 1999;268:94–101. doi: 10.1006/abio.1998.3028. [DOI] [PubMed] [Google Scholar]
- 15.Amukele TK, Schramm VL. Biochemistry. 2004;43:4913–4922. doi: 10.1021/bi0498508. [DOI] [PubMed] [Google Scholar]
- 16.Shi W, Tanaka KSE, Crother TR, Taylor MW, Almo SC, Schramm VL. Biochemistry. 2001;40:10800–10809. doi: 10.1021/bi010465h. [DOI] [PubMed] [Google Scholar]
- 17.Wang HC, Ciskanik L, Dunaway-Mariano D, von-der-Saal W, Villafranca JJ. Biochemistry. 1988;27:625–633. doi: 10.1021/bi00402a020. [DOI] [PubMed] [Google Scholar]
- 18.Goss NH, Evans CT, Wood HG. Biochemistry. 1980;19:5805–5809. doi: 10.1021/bi00566a022. [DOI] [PubMed] [Google Scholar]
- 19.Wool IG. Annual review in biochemistry. 1979;48:719–754. doi: 10.1146/annurev.bi.48.070179.003443. [DOI] [PubMed] [Google Scholar]
- 20.Ye D, Wei M, McGuire M, Huang K, Kapadia G, Herzberg O, Martin BM, Dunaway-Mariano D. J. Biol. Chem. 2001;276:37630–37639. doi: 10.1074/jbc.M105631200. [DOI] [PubMed] [Google Scholar]
- 21.DeLuca M, McElroy WD. Biochemical and Biophysical Research Communications. 1984;123:764–770. doi: 10.1016/0006-291x(84)90295-x. [DOI] [PubMed] [Google Scholar]
- 22.Amukele TK, Roday S, Schramm VL. Biochemistry. 2005;44:4416–4425. doi: 10.1021/bi0474362. [DOI] [PubMed] [Google Scholar]
- 23.Chen XY, Berti PJ, Schramm VL. J. Am. Chem. Soc. 2000;122:1609–1617. [Google Scholar]
- 24.Chen XY, Berti PJ, Schramm VL. J. Am. Chem. Soc. 2000;122:6527–6534. [Google Scholar]
- 25.Barbieri L, Ciani M, Girbés T, Liu W, VanDamme EJM, Peumans WJ, Stirpe F. FEBS Letters. 2004;563:219–222. doi: 10.1016/S0014-5793(04)00286-8. [DOI] [PubMed] [Google Scholar]
- 26.Stroppolo ME, Falconi M, Caccuri AM, Desideri A. Cell. Mol. Life Sci. 2001;58:1451–1460. doi: 10.1007/PL00000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vater CA, Bartle LM, Leszyk JD, Lambert JM, Goldmacher VS. Journal of biological chemistry. 1995;270:12933–12940. doi: 10.1074/jbc.270.21.12933. [DOI] [PubMed] [Google Scholar]
- 28.Argent RH, Parrott AM, Day PJ, Roberts LM, Stockley PG, Lord JM, Radford SE. J Biol Chem. 2000;275:9263–9269. doi: 10.1074/jbc.275.13.9263. [DOI] [PubMed] [Google Scholar]
- 29.Burbaum JJ, Raines RT, Albery WJ, Knowles JR. Biochemistry. 1989;28:9293–9305. doi: 10.1021/bi00450a009. [DOI] [PubMed] [Google Scholar]
- 30.Fersht A. W.H. Freeman and Co. New York. 1985. p. 475. [Google Scholar]