Abstract
Proteins participate in various biological processes and can be harnessed to probe and control biological events selectively and reproducibly, but the genetic code limits the building block to 20 common amino acids for protein manipulation in living cells. The genetic encoding of unnatural amino acids will remove this restriction and enable new chemical and physical properties to be precisely introduced into proteins. Here we present new strategies for generating orthogonal tRNA-synthetase pairs, which made possible the genetic encoding of diverse unnatural amino acids in different mammalian cells and primary neurons. Using this new methodology, we incorporated unnatural amino acids with extended side chains into the K+ channel Kv1.4, and found that the bulkiness of residues in the inactivation peptide is essential for fast channel inactivation, a finding that had not been possible using conventional mutagenesis. This technique will stimulate and facilitate new molecular studies using tailored unnatural amino acids for cell biology and neurobiology.
The use of proteins as experimental tools has led to significant advances in neurobiology. These proteins can be selectively targeted to cells of interest or engineered, enabling them to make precise measurements or to perturb signaling1,2. For instance, some genetically encoded biosensors have provided new ways to `see' the activity of ion channels3. Proteins derived from green fluorescent protein (GFP), such as cameleon4 and synapto-pHluorin5, can report on changes in calcium flux or neurotransmitter release. In addition, channelrhodopsin-2, a light-gated cation channel, allows action potentials to be noninvasively evoked using light, facilitating the study and modulation of behavior in intact animals6. However, a protein-based sensor that can report a single action potential is not available, and there are no ideal protein tools for the in vivo study of other neurobiological events such as protein misfolding, aggregation, trafficking and interactions. In addition to known proteins with unique properties, further advances would be expected if it were possible to make changes in neuronal proteins that generate new properties. Currently, we are limited to the naturally occurring 20 common amino acids as building blocks.
One solution to this limitation is to use unnatural amino acids that possess unique side chains. Incorporation of unnatural amino acids has provided a powerful method for studying the physical, chemical and biological properties of proteins7. Unnatural amino acids have been incorporated into ion channels and receptors by microinjecting the chemically acylated tRNA and UAG-containing mutant mRNA into Xenopus oocytes8, and have greatly contributed to our understanding of receptor functions9-11. Unfortunately, application of this method in neurobiology is restricted. The requirement for microinjection limits the technique mainly to large Xenopus oocytes, and it is not suitable for studies that require large numbers of cells. A more significant obstacle is that the tRNA is chemically acylated with the unnatural amino acid in vitro, and the acylated tRNA is consumed as a stoichio-metric reagent during translation and cannot be regenerated. Therefore, yields of mutant proteins are low and data cannot be collected over extended periods.
The genetic encoding of unnatural amino acids would overcome such restrictions and enable proteins and related biological events to be investigated directly in living cells and organisms. Previously, bacterial Eschericia coli have been used to expand the genetic code to include unnatural amino acids12. This method involves the generation of a new tRNA-aminoacyl-tRNA-synthetase pair that is specific for the unnatural amino acid and decodes a blank codon that is unused by a common amino acid (such as a stop codon or an extended codon with four or more bases). The tRNA-synthetase pair needs to be compatible with the protein biosynthesis machinery of the host cell, and crosstalk with endogenous tRNA-synthetase pairs must be avoided. Such orthogonal tRNA-synthetase pairs for use in E. coli have been evolved from large RNA and protein libraries (>109 members) through high-throughput selection7. However, mammalian cells and E. coli differ significantly in tRNA transcription, processing and transportation, leading to inefficient biosynthesis of orthogonal tRNAs in mammalian cells. Moreover, primary neurons have low transfection efficiency and cannot divide. It will therefore be impractical, if not impossible, to devise and implement the high-throughput selections necessary for engineering orthogonal tRNAs and unnatural-amino-acid-specific synthetases in neurons.
Here we report new strategies that enable unnatural amino acids to be efficiently encoded in various types of mammalian cell, including primary neurons. We used a type-3 pol III promoter to efficiently express and process bacterial tRNAs, and unnatural-amino-acid-specific synthetases generated in yeast were successfully transferred for use in mammalian cells and neurons. To show that unnatural amino acids can alter the function of a neuronal protein, we chose to study the fast, N-type inactivation of voltage-gated K+ channels13. Upon membrane depolarization, such as during an action potential, some voltage-gated K+ channels open and then quickly enter a non-conducting state; this process is known as inactivation. Structural studies14 and muta-genesis studies15-17 have suggested that fast inactivation occurs when the amino-terminal domain of the channel feeds through a small portal in the cytoplasmic domain and binds to the inner channel cavity, preventing the efflux of K+ ions. Here, we used unnatural amino-acid mutagenesis to investigate whether the bulkiness of an amino acid in the N-terminal inactivation peptide affects the fast inactivation of the neuronal K+ channel Kv1.4. Our data support a model in which the overall diameter of the inactivation peptide is crucial for determining the rate of channel inactivation. We discuss these results as well as the potential impact of unnatural amino-acid mutagenesis in neurons on neurobiology.
RESULTS
Efficient expression of orthogonal tRNA in mammalian cells
An efficient way of generating an orthogonal tRNA-synthetase pair is to import a tRNA-synthetase pair from species in a different kingdom18,19, because the cross-aminoacylation between different species is often low. The tRNATyr-TyrRS pair from the archaebacterium Methanococcus jannaschii was successfully used in E. coli. Similarly, the E. coli. tRNATyr-TyrRs pair has been used in yeast, and should be orthogonal in mammalian cells. To test and use E. coli tRNAs, one challenge lies in the expression of functionalE. coli tRNAs in mammalian cells. E. coli and mammalian cells differ significantly in tRNA transcription and processing. E. coli tRNAs are transcribed by the sole RNA polymerase through promoters upstream of the tRNA structural gene. However, the transcription of mammalian tRNA genes depends principally on promoter elements within the tRNA known as the A and B box sequences, which are recognized by RNA polymerase III (pol III) and its associated factors20. Whereas all E. coli tRNA genes encode full tRNA sequences, mammalian tRNAs have the 3′-CCA sequence added enzymatically by the tRNA nucleotidyltransferase after transcription. In addition, the 5′- and 3′-flanking sequences, the removal of introns, and the export from nucleus to cytoplasm also affect mammalian tRNA expression and function. Owing to these differences, E. coli tRNAs, especially those that diverge from the preserved eukaryotic A and B box sequences, are not efficiently biosynthesized or correctly processed in mammalian cells.
We reasoned that a pol III promoter lacking any requirement for intragenic elements could efficiently transcribe tRNAs without the preserved internal A and B boxes in mammalian cells. The H1 promoter21 was chosen for the following reasons: (i) it drives the expression of human H1RNA and thus is of mammalian origin; (ii) it is a type-3 pol III promoter that has no downstream transcriptional elements; (iii) the transcription initiation site of the H1 promoter is well defined, and can be used to generate the 5′ end of the tRNA without further post-transcriptional processing; and (iv) the H1 promoter has been successfully used to express short interfering RNAs in mammalian cells.
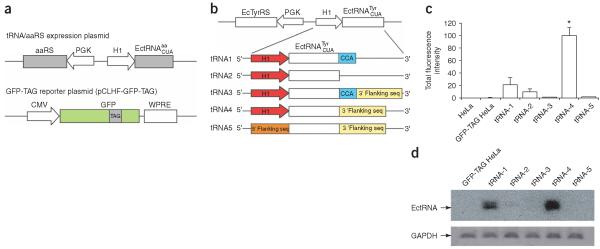
We developed a fluorescence-based functional assay in mammalian cells to identify those expression elements that can efficiently drive the transcription of E. coli tRNAs to generate functional tRNAs in mammalian cells (Fig. 1a). The gene for the candidate E. coli amber suppressor tRNA (, whose anticodon is changed to CUA to decode the amber stop codon TAG) is co-expressed with its cognate synthetase (aaRS). A TAG stop codon is introduced at a permissive site of the GFP gene, and this mutant GFP gene is co-expressed with the -aaRS pair in mammalian cells. If the is expressed and correctly processed to a functional tRNA, the synthetase will aminoacylate this tRNA with the cognate amino acid. The acylated will suppress the TAG codon in the GFP gene, producing full-length GFP and rendering cells fluorescent. By comparing the fluorescence intensities of cells, this method can also serve as a sensitive in vivo assay for the orthogonality of the to endogenous synthetases of host cells when the cognate E. coli synthetase is not expressed, and for the activity of the orthogonal towards an unnatural-amino-acid-specific mutant synthetase when the mutant synthetase is expressed in place of the cognate synthetase.
Figure 1.
Efficient expression of bacterial tRNA in mammalian cells using the H1 promoter. (a) A fluorescence-based assay for the expression of functional tRNA in mammalian cells. The candidate amber suppressor tRNA and its cognate synthetase were expressed using the tRNA-aaRS expression plasmid. A reporter plasmid was used to express GFP with an amber stop codon at a permissive site. (b) Schematic illustration of tRNA-aaRS expression plasmids using different elements to drive the tRNA transcription and processing. (c) Total fluorescence intensity of the fluorescent GFP-TAG HeLa cells after transfection with constructs shown in b. The intensities were normalized to those of cells transfected with tRNA-4. The values (± s.e.m.) were: GFP-TAG HeLa 0.3 ± 0.1, tRNA-1 21 ± 11, tRNA-2 10 ± 4.7, tRNA-3 1.3 ± 0.7, tRNA-4 100 ± 12, tRNA-5 1.4 ± 0.5. For all samples, n = 5. (d) Northern blot analysis of the amount of transcribed in HeLa cells. Transcript of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize the total amount of RNA in different samples.
The E. coli tyrosyl amber suppressor tRNA () was chosen as the candidate orthogonal tRNA because it is orthogonal to yeast synthetases and suppresses the amber stop codon efficiently in yeast when coexpressed with E. coli TyrRS22. In vitro aminoacylation assays indicate that E. coli TyrRS does not charge eukaryotic tRNAs23. Mammalian and yeast cells are eukaryotic, so we reasoned that the -E. coli TyrRS pair should be orthogonal in mammalian cells. For 3′-end processing of the , we used the 3′-flanking sequence of the human tRNAfMet. The 5′- and 3′-flanking sequences of the human tRNAfMet can drive the functional expression of E. coli (which has the A box and B box) in mammalian cells24. To test the importance of the 3′-CCA trinucleotide, these nucleotides were included or removed in the tRNA gene, resulting in four expression cassettes (tRNA-1 to tRNA-4; Fig. 1b). For comparison, we made a control plasmid (tRNA-5) in which the was placed downstream of the 5′-flanking sequence of the human tRNATyr.
To compare the ability of different expression cassettes to generate functional tRNAs, we established a clonal stable HeLa cell line expressing the GFP gene with a TAG stop codon introduced at the permissive site 182 (GFP-TAG HeLa). The tRNA-aaRS expression plasmid was transfected into the stable GFP-TAG HeLa cell line, and cells were analyzed with flow cytometry after 48 h. The total fluorescence intensity of the green fluorescent cells indicates the amount of GFP produced (Fig. 1c). When -TyrRS was not expressed, the fluorescence intensity of the GFP-TAG HeLa cell line was similar to that of HeLa cells, indicating that the background readthrough of the TAG codon in GFP is negligible. Using the 5′-flanking sequence of human tRNATyr in tRNA-5, only weak amber suppression was detected, confirming that bacterial tRNAs without the preserved A and B boxes could not be functionally expressed in mammalian cells. The highest fluorescence intensity was found in cells transfected with tRNA-4, which was 71-fold higher than that of tRNA-5, indicating that the H1 promoter can drive the functional biosynthesis of much more efficiently than the 5′-flanking sequence of the human tRNATyr. This also indicates that the H1 promoter can generate the correct 5′-end of the tRNA directly from the transcription initiation site without the post-transcriptional processing that is necessary for endogenously expressed tRNAs. The fluorescence intensity of cells transfected with tRNA-2 was 10% of that of cells transfected with tRNA-4, indicating that the 3′-flanking sequence of the human tRNAfMet is also needed for the efficient expression of the . It is intriguing to find that functional tRNA was produced in mammalian cells transfected with tRNA-1 (21% of tRNA-4), in which the CCA trinucleotide but no 3′-flanking sequence is included, as mammalian cells do not encode the CCA in the tRNA gene. However, when both the CCA trinucleotide and the 3′-flanking sequence were included in tRNA-3, the fluorescence intensity dropped markedly to 1.3%.
We used Northern blotting to measure the transcription levels of the expressed by different constructs in GFP-TAG HeLa cells (Fig. 1d). Very low levels of could be detected using an -specific probe in samples transfected with tRNA-5, tRNA-3 or tRNA-2. By contrast, in cells transfected with tRNA-4 and tRNA-1, the amounts of were about 93-fold and 19-fold higher, respectively, than in cells transfected with tRNA-5. The northern blotting data confirmed that the was transcribed in HeLa cells, and the increase in tRNA transcription is consistent with the increase in fluorescence intensity measured by cytometry in different samples.
To test the orthogonality of the to endogenous synthetases in HeLa cells, we removed the E. coli TyrRS in tRNA-4 to express the only. Transfection of the resultant plasmid into the GFP-TAG HeLa cell line did not change the fluorescence intensity of the cells, showing that the is not aminoacylated by any synthetases in HeLa cells.
To test whether the H1 promoter, together with the 3′-flanking sequence, can be used to express other E. coli tRNAs, we replaced the in construct tRNA-4 with the E. coli leucyl amber suppressor tRNA () and the TyrRS with the cognate leucyl-tRNA synthetase (LeuRS). When only the was expressed, we saw no fluorescence change in the GFP-TAG HeLa cells, indicating that the is also orthogonal in HeLa cells. By contrast, when the -LeuRS pair was expressed, the GFP-TAG HeLa cells became very bright. The total fluorescence intensity was 104% of that of cells transfected with the -TyrRS pair. The does not have a fully matched A box, whereas the has no fully matched A or B box sequences. Taken together, these results show that the H1 promoter can efficiently drive the expression of E. coli tRNAs, regardless of the internal promoter elements, in mammalian cells, and that the transcribed tRNAs are functional for amber suppression.
Unnatural-amino-acid-specific aaRS for mammalian cells
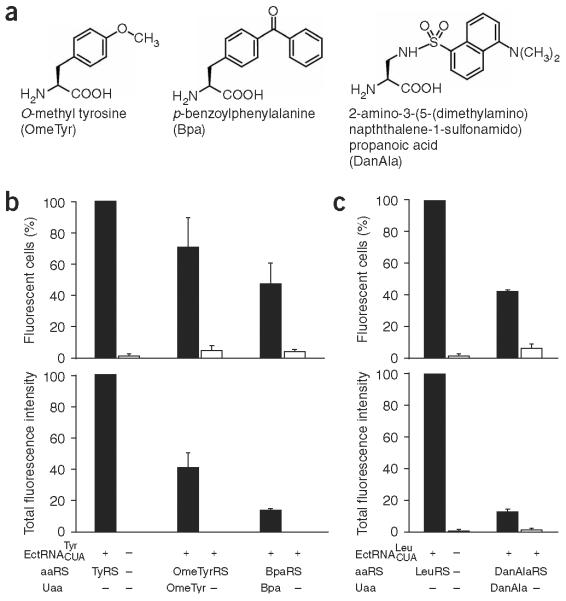
Synthetases that are specific for a variety of unnatural amino acids have been evolved in E. coli and later in yeast from large mutant synthetase libraries consisting of >109 members7. Similar strategies cannot be practically employed in mammalian cells and neurons because the transfection efficiencies of these cells are lower by several orders of magnitude than those of E. coli and yeast. As both yeast and mammalian cells are of eukaryotic origin, we decided to transfer the mutant synthetases that had been evolved in yeast to mammalian cells. To test the feasibility of this approach, the E. coli TyrRS gene in the tRNA-aaRS expression plasmid (Fig. 1a) was replaced with the gene for OmeTyrRS, a synthetase specific for the unnatural amino acid o-methyl-L-tyrosine (OmeTyr; Fig. 2a). The resultant plasmid was transfected into the GFP-TAG HeLa cell line, and cells were grown in the presence and absence of OmeTyr. As shown in Figure 2b, in the absence of OmeTyr, these cells were virtually nonfluorescent and similar to the GFP-TAG HeLa cells, indicating that the expression of the -OmeTyrRS pair does not suppress amber codons efficiently. When OmeTyr was added, 71% of cells (normalized to the total number of fluorescent cells transfected with the and wild-type TyrRS) became fluorescent, indicating that OmeTyr was incorporated into the GFP. The incorporation efficiency is about 41% by comparing the total fluorescence intensity of these cells to the intensity of cells transfected with the -TyrRS pair.
Figure 2.
Unnatural-amino-acid-specific synthetases evolved in yeast are functional in mammalian cells. (a) Structures of unnatural amino acids used in this study. (b) Incorporation of OmeTyr and Bpa into GFP in the GFP-TAG HeLa cells using the and corresponding synthetases evolved from E. coli TyrRS in yeast. All data were normalized to those obtained from GFP-TAG HeLa cells transfected with the and wild-type E. coli TyrRS. Percentages (± s.e.m.) of fluorescent cells were 71 ± 19 (+ OmeTyr, n = 3); 4.8 ± 3.4 (- OmeTyr, n = 3); 47 ± 14 (+ Bpa, n = 3); and 4.2 ± 1.5 (- Bpa, n = 3). The total fluorescence intensities (± s.e.m.) were 41 ± 9.5 (+ OmeTyr, n = 3); 0.17 ± 0.02 (- OmeTyr, n = 3); 13 ± 1.4 (+ Bpa, n = 3); and 0.11 ± 0.06 (- Bpa, n = 3). (c) Incorporation of DanAla into GFP in the GFP-TAG HeLa cells using the and a DanAla-specific synthetase evolved from E. coli LeuRS. Data were normalized as in b. Percentages (± s.e.m) of fluorescent cells were 42 ± 1.3 (+ DanAla, n = 3) and 5.9 ± 2.6 (- DanAla, n = 3). The total fluorescence intensities (± s.e.m.) were 13 ± 2.1 (+ DanAla, n = 3) and 1.4 ± 1.0 (- DanAla, n = 3).
To determine whether the transfer strategy could be generally applied to other synthetases evolved in yeast, we next tested the BpaRS, a synthetase that is specific for p-benzoylphenylalanine (Bpa; Fig. 2a). As expected, when the BpaRS was coexpressed with the in the GFP-TAG HeLa cell line, 47% of cells were fluorescent in the presence of Bpa, and virtually no fluorescent cells (≤4%) were detected in the absence of Bpa. The incorporation efficiency of this unnatural amino acid is about 13% (Fig. 2b).
In addition to tRNA-aaRS pairs derived from the E. coli tRNATyr-TyrRS, we also tested a tRNA-aaRS pair derived from E. coli tRNALeu-LeuRS. The and a mutant synthetase specific for a fluorescent unnatural amino acid 2-amino-3-(5-(dimethylamino)-naphthalene-1-sulfonamido)propanoic acid (DanAla)25 (Fig. 2a) were expressed in the GFP-TAG HeLa cell line. DanAla was incorporated with 13% efficiency, and 42% of the cells became fluorescent (Fig. 2c).
These results confirm that unnatural-amino-acid-specific synthetases evolved in yeast can generally be transferred for use in mammalian cells. The tested unnatural amino acids were incorporated less efficiently than Tyr by the wild-type TyrRS, indicating that the activities of the evolved synthetases are not optimal. More rounds of directed evolution of these synthetases in yeast might further improve their incorporation efficiencies.
Genetically encoding unnatural amino acids in neurons
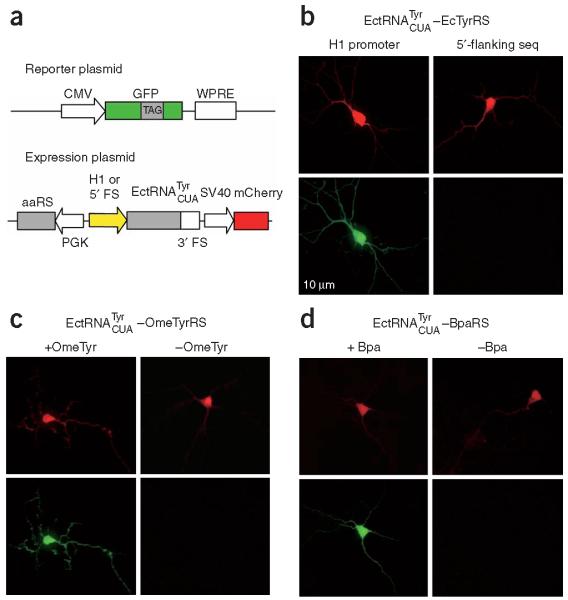
We first investigated whether the H1 promoter and the 3′-flanking sequence identified in HeLa cells could also generate functional amber suppressor tRNAs in neurons. Mouse hippocampal neurons were transfected with two plasmids simultaneously (Fig. 3a): the reporter plasmid pCLHF-GFP-TAG encoding a mutant GFP (182TAG), and the expression plasmid encoding the E. coli TyrRS, the driven by either the H1 promoter or the 5′ flanking sequence of human tRNATyr, and a red fluorescent protein, mCherry, as an internal marker for transfection. Fluorescence microscopy was used to look for red transfected cells, and then to image their green fluorescence. The presence of green fluorescence in transfected cells indicates that functional is biosynthesized to incorporate Tyr at the 182TAG position of the GFP gene. As shown in Figure 3b, neurons transfected with the expression plasmid in which the was driven by the H1 promoter showed intense green fluorescence, whereas no green fluorescence could be detected in neurons in which the was driven by the 5′-flanking sequence of the human tRNATyr.
Figure 3.
Genetically encoding unnatural amino acids in neurons. (a) Schematic illustration of the reporter plasmid expressing the GFP mutant gene with a TAG stop codon at site 182 and the expression plasmid encoding the , the synthetase and an internal transfection marker (mCherry). (b) Fluorescence images of neurons transfected with the reporter plasmid, the , and wild-type E. coli TyrRS. tRNA expression was driven by the H1 promoter in the left panels, and by the 5′-flanking sequence of the human tRNATyr in the right panels. (c) Fluorescence images of neurons transfected with the reporter plasmid, the , and the OmeTyrRS in the presence (left panels) and absence (right panels) of OmeTyr. (d) Fluorescence images of neurons transfected with the reporter plasmid, the , and the BpaRS in the presence (left panels) and absence (right panels) of Bpa.
Next we tested whether unnatural amino acids could be genetically encoded in neurons using the and mutant synthetases that are specific for different unnatural amino acids. Synthetases that had been evolved in yeast and proven to be functional in HeLa cells were used. When the OmeTyrRS was coexpressed with the , transfected neurons showed no green fluorescence in the absence of the corresponding unnatural amino acid OmeTyr (Fig. 3c), indicating that the is orthogonal to endogenous synthetases in neurons. Transfected neurons showed bright green fluorescence only when OmeTyr was fed to the growth medium. These results indicate that OmeTyr, but no common amino acid, is incorporated into GFP at the 182TAG position. The same results were obtained for the unnatural amino acid Bpa when the BpaRS was coexpressed with the (Fig. 3d). Using this approach, OmeTyr and Bpa were also genetically encoded in hippocampal and cortical neurons isolated from rats (data not shown).
Probing the inactivation mechanism of K+ channel Kv1.4
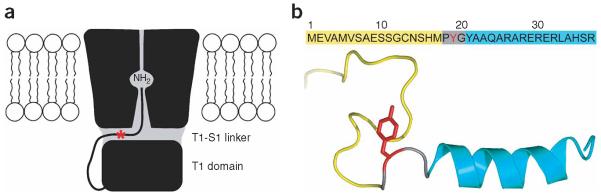
The fast inactivation of voltage-dependent K+ (Kv) channels involves ~20 amino acids on the channel's intracellular N terminus, or on the N terminus of an associated β-unit. The first ~10 amino acids are predominantly hydrophobic, and the other ~10 amino acids are hydrophilic and positively charged26. Recent crystal structure and mutation studies of Kv1.2 suggest that the inactivation peptide might snake through a side portal in the fully extended conformation, and allow the hydrophobic terminus to reach and plug the inner pore for inactivation14-17 (Fig. 4a). Through unnatural amino-acid mutagenesis of the inactivation peptide of Kv1.4 in live mammalian cells, we aimed to experimentally test this inactivation model by determining whether the bulkiness of the inactivation peptide would influence channel inactivation.
Figure 4.
A model for the N-type inactivation of Kv channels. (a) The model proposes that the N-terminal inactivation peptide snakes through the side portal, and then enters and plugs the inner pore of the Kv channel14,15. The side portals are formed by the transmembrane domain, the T1 domain and the T1-S1 linker. The hydrophobic region of the inactivation peptide reaches into the inner pore, and the positively charged residues in the hydrophilic region of the inactivation peptide make electrostatic interactions with negatively charged surfaces of the T1 domain and the T1-S1 linker. (b) The amino-acid sequences of the N-terminal inactivation peptide of Kv1.4, and its structure in aqueous solution determined by NMR28. Only one conformer of the random N-terminal Met1-Met17 is shown in yellow. The β-turn and α-helix are colored gray and cyan, respectively. Residue Tyr19 is colored red, and its approximate position is indicated with a red star in a.
The N-terminal domain of Kv1.4 possesses two inactivation domains, consisting of amino acids Met1-Ala40 for the primary and Ala40-Gly60 for the secondary inactivation domains27. The structure of the primary inactivation domain in aqueous solution has been analyzed using NMR spectroscopy28,29. The N-terminal Met1-Met17 is disordered, followed by a β-turn (Pro18-Tyr21) and a well-defined α-helix (Tyr21-Ala36) (Fig. 4b). In the Kv1.2 structure, a negatively charged patch of residues lies at the entrance to the side portal, and is proposed to interact with the positively charged amino acids on the inactivation peptide14. According to this hypothesis, several arginines (Arg26, Arg28, Arg30 and Arg32) in the α-helix of Kv1.4 will interact with the negatively charged residues at the side portal entrance, which will place Tyr19 inside the side portal (Fig. 4a). In addition, Tyr19 resides at the beginning of the α-helical portion of the inactivation domain (Fig. 4b). We hypothesized that increasing the bulkiness of the amino-acid side chain at Tyr19 would alter the channel inactivation owing to the size restriction of the side portal.
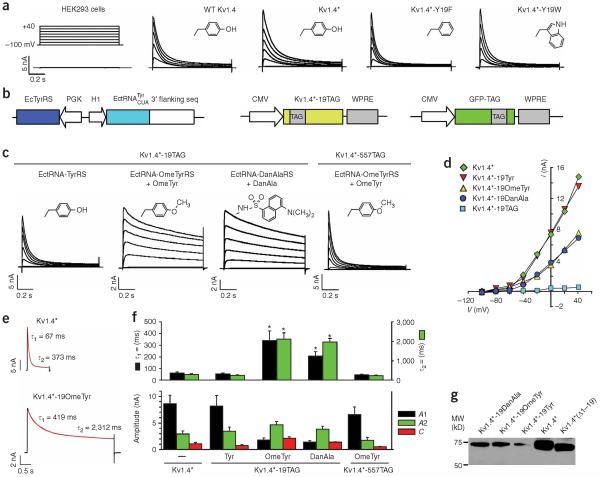
We first investigated the inactivation properties of wild-type Kv1.4 channels expressed heterologously in human embryonic kidney 293 (HEK293) cells using whole-cell patch-clamp recordings (Fig. 5a). A series of 1-s voltage steps from -100 mV to +40 mV elicited a rapidly inactivating macroscopic K+ current. To avoid undesired transcriptional initiation at the downstream Met109 site and thus the generation of truncated Kv1.4, the methionine at 109 was mutated to leucine (M109L; see Methods). Kv1.4-M109L (Kv1.4⋆) channels displayed fast inactivation similar to wild-type Kv1.4 channels (Fig. 5a). To investigate the role of bulkiness in determining fast inactivation, we first mutated Tyr19 of Kv1.4⋆ to Phe or Trp, using conventional site-directed mutagenesis. Compared to the native Tyr, Phe is smaller (lacking the hydroxyl) whereas Trp is considerably larger. The macroscopic K+ currents recorded from HEK293 cells expressing Kv1.4⋆-Y19F or Kv1.4⋆-Y19W channels were similar to those of Kv1.4⋆ channels (Fig. 5a). Therefore, the Y19F or Y19W mutation did not greatly alter the inactivation properties.
Figure 5.
Probing N-type inactivation using unnatural amino acid mutagenesis with Kv1.4 channels expressed in mammalian cells. (a) Comparison of macroscopic currents recorded from HEK293 cells expressing wild-type Kv1.4, Kv1.4⋆ (Kv1.4-M109L), Kv1.4⋆-Y19F or Kv1.4⋆-Y19W channels. Untransfected HEK293 cells displayed a small, background current (0.50 ± 0.03 nA, n = 5). Inserts in each panel show the side-chain structures of amino acids at position 19. (b) Constructs for expressing Kv1.4 through amber suppression. The GFP-TAG reporter plasmid was used as an indicator for transfection. (c) Potassium currents recorded from HEK293 cells expressing Kv1.4⋆-TAG channels through amber suppression using the indicated orthogonal tRNA-synthetase pair. Only background currents were recorded for Kv1.4⋆-19TAG alone (0.40 ± 0.05 nA, n = 5) or Kv1.4⋆-TAG with the EctRNA-OmeTyrRS pair in the absence of OmeTyr (0.42 ± 0.07 nA, n = 4). The Kv1.4⋆-19OmeTyr and Kv1.4⋆-19DanAla mutants exhibited significantly slower inactivation than Kv1.4⋆ and Kv1.4⋆-19Tyr. Inserts in each panel are the side chain structures. (d) Peak current plotted as a function of voltage for the indicated Kv1.4 channels. (e) Current (black trace) elicited by a 1-s (Kv1.4⋆) or 4-s (Kv1.4⋆-19OmeTyr) voltage step to +20 mV were fit with a sum of two exponentials (red line) relaxing to a small steady-state current with the indicated time constants. (f) Quantitative analysis of inactivation kinetics. Bar graphs show the average time constants (τ1, τ2) for the fast and slow inactivation components, current amplitudes (A1, A2) for the exponentials and steady-state current (C). Specific values are in Table 1. Asterisks indicate statistical difference from Kv1.4⋆ (P < 0.05). (g) Western blot analysis of Kv1.4⋆-19OmeTyr and Kv1.4⋆-19DanAla. The full-length Kv1.4⋆, Kv1.4⋆-19Tyr, and a truncated Kv1.4⋆ with the first 19 amino acid residues deleted were run as controls.
Next, we tested whether a functional Kv1.4 channel could be expressed in HEK293 cells through amber suppression using the orthogonal -TyrRs pair to incorporate the native Tyr. An amber stop codon (TAG) was introduced at Tyr19, and the mutant gene (Kv1.4⋆-19TAG) was cotransfected into HEK293 cells with the -TyrRs pair and the GFP-TAG reporter gene (Fig. 5b). Transfected cells fluoresced green owing to the suppression of the amber codon in the GFP-TAG gene by the -TyrRs pair. GFP-positive cells also showed large voltage-dependent K+ currents that inactivated with large depolarizations (Fig. 5c,d). To investigate possible differences in the rates of fast inactivation, we fit the decay of current elicited by the 1-s voltage-step to +20 mV with a sum of two exponentials relaxing to a small steady-state current (C). Neither the τ1 or τ2 time constants nor the amplitudes of current were significantly different between Kv1.4⋆ and Kv1.4⋆-19Tyr channels (Fig. 5e,f; Table 1), indicating that the incorporation of amino acids through amber suppression did not adversely affect the cell or the properties of the Kv1.4 channel. Importantly, no current could be detected in the absence of the -TyrRs pair, confirming that functional Kv1.4 is produced through amber suppression and that the tRNA-synthetase pair is required for the incorporation of Tyr.
Table 1.
Kinetics of current inactivation of Kv1.4 channels
| Time constant (ms) |
Amplitude (nA) |
Steady-state current (nA) |
Cells |
|||
|---|---|---|---|---|---|---|
| Kv1.4 channel | τ 1 | τ 2 | A1 | A2 | C | n |
| Kv1.4* | 63.7 ± 5.8 | 300 ± 31 | 8.67 ± 1.53 | 2.99 ± 0.52 | 1.09 ± 0.23 | 9 |
| Kv1.4*-19Tyr | 54.7 ± 6.4 | 250 ± 3.4 | 8.14 ± 2.04 | 3.42 ± 0.81 | 0.79 ± 0.16 | 7 |
| Kv1.4*-19OmeTyr | 340 ± 80 | 2,115 ± 323 | 1.81 ± 0.38 | 4.69 ± 0.56 | 2.19 ± 0.18 | 11 |
| Kv1.4*-19DanAla | 205 ± 39 | 1,978 ± 174 | 1.35 ± 0.35 | 3.83 ± 0.57 | 1.37 ± 0.10 | 11 |
| Kv1.4*-557OmeTyr | 47.6 ± 3.3 | 234 ± 18 | 6.58 ± 1.44 | 1.77 ± 0.52 | 0.53 ± 0.08 | 8 |
All values are mean ± s.e.m.
We then tested the effect of introducing a large bulky unnatural amino acid at position 19. OmeTyr was incorporated at the 19TAG position of Kv1.4⋆ using the -OmeTyrRS pair. When no OmeTyr was added to the growth medium, only small background levels of current could be detected in transfected cells (identified by cotransfected enhanced GFP in this case). By contrast, large voltage-gated K+ currents were seen when OmeTyr was added to the culture medium (Fig. 5c,d). Interestingly, the rate and extent of inactivation were markedly slower than with the wild-type channel — note the large amount of current at the end of the 1-s voltage step (Fig. 5c). To quantify this change in inactivation more accurately, we measured the macroscopic current elicited by a 4-s voltage step to +20 mV and fit the decay of current with the sum of two exponentials, as described above (Fig. 5e). The time constants of the fast (τ1) and the slow inactivation component (τ2) for the OmeTyr mutant increased 5- and 7-fold, respectively, compared to those of the wild-type Kv1.4⋆ channel. In addition, the amplitude of the fast component (A1) decreased to ~20%, whereas the amplitude of the slow component (A2) increased by ~1.5-fold for the OmeTyr mutant (Fig. 5f, Table 1). We then tested the effect of incorporating an amino acid with an even larger side chain, DanAla, at Tyr19 (Fig. 2a). As when OmeTyr was inserted, slowly inactivating, voltage-gated K+ currents were seen in HEK293 cells transfected with Kv1.4⋆-19TAG, and the -TyrRs pair, along with DanAla in the medium (Fig. 5c,d). Quantification of the rate of inactivation showed that DanAla increased the fast and slow time constants for inactivation by 3.2-fold and 7-fold, respectively (Fig. 5f, Table 1). Similarly, the amplitude of the slow component was larger than that of the fast component of inactivation, in contrast to the wild-type channel. Thus, both OmeTyr and DanAla can be functionally incorporated into Kv1.4 channels, leading to markedly slower inactivation.
We also carried out several important controls. First, only small background currents were evident in HEK cells when the unnatural amino acid was omitted from the medium (Fig. 5). Thus, there was no inappropriate `read-through' of the amber mutation. To exclude the possibility that OmeTyr is toxic to cells or to the Kv channel, OmeTyr was also incorporated into Kv1.4⋆ at 557TAG. Residue 557 is on the transmembrane helix S6 and lies at the inner pore, which is between the selectivity filter and intracellular solution. Like Kv1.4⋆, the inactivation rate of Kv1.4⋆-557OmeTyr was similar to that of wild-type Kv1.4 (Fig. 5c,f; Table 1). These experiments show that the abolition of fast inactivation of Kv1.4 by OmeTyr is site-dependent. To address whether the introduction of OmeTyr or DanAla rendered the N-terminal inactivation peptide subject to proteolytic cleavage, we added a FLAG tag to the C terminus of Kv1.4⋆-19TAG, and used western blotting to analyze channel proteins containing OmeTyr or DanAla at residue 19 expressed in HEK293 cells (Fig. 5g). In both samples, one band corresponding to the full-length Kv1.4⋆ channel was detected using FLAG-specific antibodies, indicating that there was no proteolytic cleavage accompanying the unnatural amino-acid incorporation.
DISCUSSION
Unnatural amino-acid mutagenesis in mammalian cells
Genetically encoding unnatural amino acids in mammalian cells, such as neurons, has been greatly hampered by two main challenges: inefficient biosynthesis of orthogonal bacterial tRNAs and difficulty in generating unnatural-amino-acid-specific synthetases in these cells. The transcription of mammalian tRNAs relies on conserved promoter elements within the tRNA gene, which are lacking in many bacterial tRNAs. Early attempts to create such internal promoters in the through mutation resulted in nonfunctional tRNAs30. As for bacterial tRNAs containing the internal promoter elements, expression of them using 5′-flanking sequences of mammalian tRNAs is inefficient even when multiple copies of the bacterial tRNA gene are used30. Here we developed a general strategy for efficient expression of bacterial tRNA in mammalian cells, regardless of the internal promoter elements, by using a type-3 pol III promoter, the H1 promoter. This promoter does not require downstream transcriptional elements, and has a well-defined transcription initiation site for generating the correct 5′-end of tRNA. We showed that the H1 promoter can drive the expression of different tRNAs ( and ) in various cell types (HeLa, HEK293, mouse and rat primary neurons) for the incorporation of diverse natural or unnatural amino acids. Other members of the type-3 class of pol III promoter, such as the promoter for U6 snRNA (ref. 31), 7SK (ref. 32) and MRP/7-2 (ref. 33) should also work in a similar manner. To evolve a synthetase specific for a desired unnatural amino acid, mutant synthetase libraries containing more than 109 members have been made and selected in E. coli and later in yeast. Owing to the low transfection efficiency, it is impractical to generate such huge libraries in mammalian cells. We show here that synthetases evolved in yeast are functionally compatible with mammalian HEK293 cells, HeLa cells and neurons. This transfer strategy will facilitate the future incorporation of diverse unnatural amino acids tailored for mammalian and neuronal studies. Using these strategies, we were able to genetically encode unnatural amino acids in different mammalian cells, and in primary neurons.
A key advance in our study was that the orthogonal tRNA and synthetase are genetically expressed in mammalian cells. Genetically encoding unnatural amino acids overcomes restrictions imposed by in vitro semisynthetic and biosynthetic unnatural-amino-acid incorporation methods on protein type, size, quantity and location34,35. Importantly, genetic expression enables studies of structure and function to be carried out in living cells and possibly organisms. For studies with cultured neurons or brain slices, the cDNA for tRNA-synthetase need only be introduced via transfection or virus into the cells. Many unnatural amino acids can be added directly to the growth medium and taken up by the cell through nonspecific amino-acid and amine transporters. Highly polar amino acids can be derivatized with enzymatically labile groups (for example, esterification or acylation) to facilitate cellular uptake. A second level of specificity could be achieved by engineering cell-specific promoters that would drive the expression of the mutant protein and the orthogonal tRNA-synthetase in a subset of neurons1. One consideration for future studies is that the combination of transfection efficiency and the efficiency of incorporating unnatural amino acids in neurons must not be too low. The incorporation efficiency of unnatural amino acids used in this study (relative to Tyr incorporation by the wild-type TyrRS) is in the range of 13% to 41%. Cytoplasmic GFP containing unnatural amino acids was expressed in sufficient amounts in neurons to be easily detected using fluorescence microscopy, and the integral membrane protein Kv1.4 was expressed in HEK293 cells sufficiently for patch clamping. Factors that could determine the incorporation efficiency of unnatural amino acids include the activity of the evolved synthetase, the gene context of the amber stop codon, the ability of cells to take up the unnatural amino acid, and the stability of the target mRNA and tRNA-synthetase pair. The overall efficiency should benefit from improvements in any of these aspects. For instance, a viral expression system might prove to be advantageous for expressing the tRNA-synthetase pair and the target gene. Genetically encoding unnatural amino acids in a multicellular organism will be more challenging. It is possible to engineer a transgenic mouse to express the mutant gene and tRNA-synthetase pair in a specific cell. The relevant unnatural amino acid could be added to the food or drinking water, or injected directly into the brain ventricles. The compatibility of this method with living systems is especially valuable for proteins whose function requires native complex cellular environments such as integral membrane proteins and proteins involved in signaling. Genetic stability and inheritance are well-suited for researching long-term biological processes such as developmental and evolutionary studies. In addition, this technology does not require special expertise, and is easily transferable to the scientific community in the form of plasmid DNA, stable cell lines or transgenic animals.
In the future, we anticipate that unnatural amino acids will be tailor designed and encoded to probe, and more excitingly, to control proteins and protein-related biological processes. For instance, fluorescent unnatural amino acids could be used to sense local environmental changes and serve as reporters for enzyme activity, membrane potential or neurotransmitter release; unnatural amino acids bearing photocrosslinking agents could be applied to identify protein-protein and protein-nucleic acid interactions in cells; and photocaged and photoisomerizable amino acids could be designed to switch on and off signal initiation and transduction noninvasively. Many of these unnatural amino acids have been encoded in E. coli and in yeast36. The method reported here will enable the genetic encoding of such novel amino acids in mammalian cells and neurons, and stimulate more precise and profound molecular studies of cell biology and neurobiology.
Investigating the fast inactivation of Kv1.4 channels
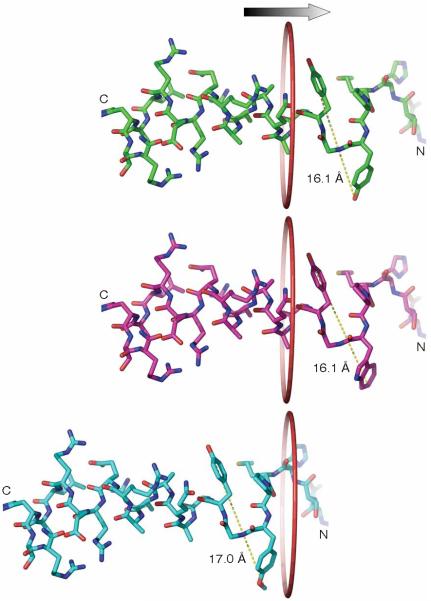
Neuronal Kv1.4 channels exhibit rapid inactivation (N-type) upon depolarization, and are important for regulating the firing rate of neurons13. Using the genetic expression of unnatural amino acids in mammalian cells, we were able to probe the role of a single amino acid in determining the rate of inactivation for Kv1.4 channels. We focused on the Tyr19 amino acid because of its unique position in the N-terminal domain—just downstream of the unfolded inactivation peptide and upstream of a structured α-helix (Fig. 4b). One model of fast inactivation proposes that the N-terminal inactivation domain is threaded through a small portal on the cytoplasmic domain, where the unfolded region extends into the channel's inner cavity. We hypothesized that the portal could exert steric constraints on the size of amino acids that could move through it. The incorporation of OmeTyr or DanAla at the Tyr19 position generated voltage-gated K+ channels with markedly slowed inactivation. Interestingly, the similarity in inactivation between Y19F and wild-type channels indicates that the hydroxyl group of Tyr is not required for fast inactivation. Thus it seems unlikely that OmeTyr alters fast inactivation by blocking hydrogen bonding. Furthermore, the current response of the Y19W mutant indicates that the increase in the lateral bulkiness of the side chain also does not affect channel inactivation. All three Phe, Tyr and Trp natural aromatic amino acids have a rigid phenyl ring. Incorporating the unnatural amino acids OmeTyr and DanAla enabled us to lengthen the side chain in the direction orthogonal to the peptide backbone and thus effectively to increase the diameter of the peptide cross section. The NMR structure of the inactivation peptide indicates that residues 19-21 are in a β-turn conformation28. The distal distance between Tyr19 and Tyr21 is ~19.9 Å (Fig. 6). We modeled Trp and OmeTyr at the Tyr19 site. The distal distance remained 19.9 Å for Trp, but increased to 20.8 Å for OmeTyr. In the crystal structure of Kv1.2, the side portal has a diameter of 15-20 Å (ref. 14). Changing Tyr19 to OmeTyr extends the length of the side chain by 0.9 Å. According to the model in which the inactivation peptide snakes through a side portal14,15, the change in fast inactivation could occur because the larger diameter of the activation peptide relative to that of the side portal interferes with the ability of the peptide to extend into the channel pore. Such effects would not be observable through conventional site-directed mutagenesis, as no natural amino acids can further extend the side-chain length rigidly.
Figure 6.
Diameter of the inactivation peptide affects channel inactivation owing to the size restriction of the side portal. Upper panel, NMR structure of the inactivation peptide from Asn14 to Arg37 of Kv1.4. The central distance between the oxygen atoms of the hydroxyl groups of Tyr19 and Tyr21 is 16.1 Å. The surface distance of these two residues is thus ~19.9 Å by including the Van der Waals radius of the hydrogen atom at both ends. Trp (middle panel) and OmeTyr (bottom panel) were modeled in the structure and the corresponding central distances were measured. A ring with a diameter of 20 Å is drawn to illustrate the side portal entrance. The change of Tyr19 to OmeTyr increases the distal distance by ~0.9 Å, which can impede the entry of residue 19 and following residues of the inactivation peptide into the portal, effectively abolishing the fast inactivation.
Another possibility is that the introduction of unnatural amino acids alters the interaction between the N-terminal inactivation peptide and other proteins, slowing its entry into the inactivation gate. Other proteins that are known to interact with the N terminus of Kv1.4 channels, such as KChIPs, DPPs and Kv- β subunits, were not coex-pressed with the channel cDNA in our experiments. If the presence of an unnatural amino acid leads to unexpected interactions between the inactivation peptide and unknown proteins, such interactions should vary with the nature of the unnatural amino acid. OmeTyr and DanAla differ significantly in structure and chemical properties. Nonetheless, the inactivation kinetics were similar for channels containing OmeTyr and channels containing DanAla, indicating that such interactions probably did not occur. In addition, proteolysis of the N-terminal domain can be ruled out because we did not observe any significant degradation of the Kv1.4⋆-OmeTyr and Kv1.4⋆-DanAla channel protein (Fig. 5f). Finally, incorporation of OmeTyr or DanAla at Tyr19 could completely disrupt fast N-type inactivation, leaving only C-type inactivation. However, deletion of the N-terminal domain, which eliminates N-type inactivation, produces channels with inactivation kinetics even slower than those containing OmeTyr or DanAla27.Thus, we favor the conclusion that the bulky unnatural amino acids OmeTyr and DanAla prevent the N-terminal peptide from fully extending into the channel pore cavity. Future studies will explore the effects of other unnatural amino acids substituted at Tyr19 and neighboring sites.
In summary, genetically encoding unnatural amino acids with orthogonal tRNA-synthetase pairs provides neuroscientists and other researchers with new tools for probing the function of proteins in mammalian cells.
METHODS
Chemicals
OmeTyr and Bpa were purchased from Chem-Impex. DanAla was synthesized as described25. All other chemicals were purchased from Sigma-Aldrich.
Constructs
All constructs were assembled by standard cloning methods and confirmed by DNA sequencing. Plasmid pCLHF is a derivative of pCLNCX (Imgenex), and has a hygromycin resistance gene instead of a neomycin resistance gene. The amber stop codon TAG was introduced into the enhancedGFP (EGFP) gene at position 182 through site-directed mutagenesis. The woodchuck hepatitis virus post-transcriptional regulatory element (WPRE)37 was added to the 3′-end of the GFP-TAG mutant gene. The GFP-TAG-WPRE gene fragment was ligated into the HindIII and ClaI sites of pCLHF to afford the plasmid pCLHF-GFP-TAG.
The E. coli TyrRS gene was amplified from E. coli genomic DNA using the primers 5′-CCACCATGGAACTCGAGATTTTGATGGCAAGCAGTAACTT GATTAAAC-3′ and 5′-ACAAGATCTGCTAGCTTATTTCCAGCAAATCAGAC AGTAATTC-3′. Genes for OmeTyrRS (Y37T, D182T and F183M) and Bpa-TyrRS (Y37G, D182G and L186A)38 were made from the E. coli TyrRS gene by site-directed mutagenesis using overlapping PCR. The gene for in construct tRNA2 was made using the primers 5′-GTGGGATCCCCGGT GGGGTTCCCGAGCGGCCAAAGGGAG CAGACTCTAAATCTGCCGTCATC GACTTCG-3′ and 5′-GATAAGCTTTTCCAAAAATGGTGGTGGGGGAAGG ATTCGAACCTTCGAAGTCGATGACGGCAGATTTAG-3′ through Klenow extension. Other tRNA constructs were made by PCR using tRNA2 as the template. Genes for and the mutant synthetase specific for DanAla were amplified from plasmid pLeuRSB8T252A25 using PCR. The E. coli LeuRS gene was amplified from E. coli genomic DNA using primers 5′-GCCTCGAGAAGAGCAATACCGCCCGG-3′ and 5′-CGCTAGCTTAGCC AACGACCAGATTGAGGAG-3′. The H1 promoter was amplified from plasmid pSUPER (OligoEngine).
To make the tRNA-aaRS expression plasmid pEYCUA-YRS, we used pBlue-script II KS (Stratagene) as the backbone for construction. The PGK promoter and the SV40 polyA signal were inserted between EcoRI and NotI sites. The E. coliTyrRS gene was inserted between the PGK and SV40 polyA sequences using the introduced XhoI and NdeI sites. The H1 promoter containing the BglII and HindIII sites at the 3′-end was cloned into the EcoRI and ClaI sites. The was then inserted between the BglII and HindIII sites. Finally, a gene cassette containing the SV40 promoter followed by the neomycin resistance gene and the SV40 poly A signal was amplified from pCDNA3 (Invitrogen) and inserted into the ClaI and KpnI sites. Other tRNA-synthetase plasmids were modified from plasmid pEYCUA-YRS by swapping the synthetase gene or the tRNA gene, or by inserting various 3′-flanking sequences after the tRNA.
We used rat Kv1.4 cDNA, which matched NM_012971 except for one difference (L42A). Mutations M109L, 19TAG, 557TAG, Y19F and Y19W were created using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). When Kv1.4-19TAG was expressed in HEK293 cells, voltage-dependent K+currents could be detected in the absence of the orthogonal EctRNA-synthetase pair. We discovered that this was due to undesired transcription initiation at residue Met109, which generates a truncated Kv1.4 channel. To solve this problem, we mutated Met109 to Leu to make Kv1.4⋆. No truncated Kv1.4 channel was expressed from Kv1.4⋆-19TAG alone, and only background currents were detected.
Cell culture and transfection
HeLa, HEK293T and HEK293 cells were cultured and maintained with Dulbecco's modified Eagle's medium (DMEM, Mediatech) supplemented with 10% FBS.
To establish the GFP-TAG HeLa stable cell line, we transfected 293T cells with the retroviral vector pCLHF-GFP-TAG and the packaging vector pCL-Ampho (Imgenex) using FuGENE 6 transfection reagent (Roche). Viruses were harvested after 48 h and used to infect HeLa cells grown in 50% conditioned medium in the presence of 8 ng/ml hexadimethrine bromide (Sigma). From the next day on, cells were split to a very low confluence. Stably infected cells were selected with 200 ng/ml hygromycin (Invitrogen). Hygromycin (50 ng/ml) was always present in subsequent cell culture to ascertain plasmid DNA maintenance.
For electrophysiological recordings, HEK293 cells were transiently transfected using Lipofectamine 2000 (Invitrogen). After 8 h, cells were reseeded onto 12-mm glass cover slips (Fisher) coated with poly-L-lysine (10 μg/ml; Sigma, MW30,000) in 24-well plates. OmeTyr or DanAla (1 mM) was added to the culture medium, and cells were incubated for 24 h at 37 °C before recording.
Hippocampi of postnatal day 0 Sprague-Dawley rats or mice were removed and treated with 2.5% trypsin (Invitrogen) for 15 min at 37 °C. The digestion was stopped with 10 ml of DMEM containing 10% heat-inactivated FBS. The tissue was triturated in a small volume of this solution with a fire-polished Pasteur pipette, and ~100,000 cells in 0.5 ml neuronal culture medium were plated per coverslip in 24-well plates. Glass coverslips were prewashed overnight in HCl followed by several rinses with 100% ethanol and flame sterilization. They were subsequently coated overnight at 37 °C with poly-L-lysine. Cells were plated and grown in Neurobasal-A (Invitrogen) containing 2% B-27 (Life Technologies), 1.8% HEPES, and 2 mM glutamine (Life Technologies). Half of the medium was replaced the next day. For imaging, cells cultured for 3 days were transfected with Lipofectamine 2000, changed into fresh medium with 1 mM OmeTyr or Bpa after 5 h, and cultured for another 24 h before the experiment.
Northern blot and western blot analysis
RNA was prepared from the GFP-TAG HeLa cells transfected with different tRNA-aaRS constructs using PureLink miRNA Kit (Invitrogen). The RNA was denatured, electrophoresed on 15% PAGE gel, blotted onto Hybond-N (Amersham) membrane, and crosslinked by ultraviolet fixation. 32P-labeled DNA probes specific for the were made using Klenow extension with primer 5′-AACCTTCG AAGTCGATGACGGCAGATTTACAGTCTGC-3′ and primer 5′-CCGTCTAAA TGTCAGACGAGGGAAACCGGCGAG-3′. After pre-hybridization for 4 h in the hybridization buffer (5× sodium chloride-sodium citrate buffer, 40 mM Na2HPO4 (pH 7.2), 7% sodium dodecylsulfate (SDS), 2× Denhardt's), membranes were hybridized with 32P-labeled cDNA probes (0.5-2 × 107 c.p.m./ml) in the same buffer plus 50 μg/ml salmon sperm DNA at 58 °C overnight. Hybridized membrane was sequentially washed with high stringency buffer (40 mM Na2HPO4, 1 mM EDTA, 1% SDS, 58 °C) twice and exposed to an X-ray film (Kodak) for 48 h. To control the total RNA amount loaded in each lane, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal standard.
A FLAG tag was added to the C termini of genes for Kv1.4⋆, Kv1.4⋆ with the first 19 amino acid residues deleted, and Kv1.4⋆-19TAG, respectively. Full-length and truncated (Δ1-19) Kv1.4⋆ were expressed in HEK293 cells. Kv1.4⋆-19TAG was co-expressed with -TyrRS, with -OmeTyrRS supplied with OmeTyr (1 mM), or with -DanAlaRS supplied with DanAla (1 mM) in HEK293 cells. Cells were lysed with 1% Nonidet P-40 (Calbiochem) in PBS at 4 °C for 30 min. After centrifugation, supernatants containing membrane proteins were loaded and separated by SDS-PAGE. A monoclonal antiflag antibody (Sigma) was used to detect the FLAG-containing proteins.
Flow cytometry
GFP-TAG HeLa cells were transfected with plasmid DNA by lipofection 2000 according to the protocol of the vendor (Invitrogen). Unnatural amino acids (1 mM) were added to the medium immediately after transfection. Cells were collected after 48 h, washed twice, and resuspended in 1 ml of PBS containing 0.05 μg/ml propidium iodide. Samples were analyzed with a FACScan (Becton & Dickinson).
Electrophysiology
Whole-cell patch clamp was used to record macroscopic currents from HEK293 cells. Borosilicate glass (Warner) electrodes had resistance of 3-8 MΩ. Membrane currents were recorded with an Axopatch 200B amplifier (Axon Instruments), adjusted electronically for cell capacitance and serial resistance (80-100%), filtered at 1 kHz, and digitized at 10 kHz. The intracellular solution consisted of 150 mM KCl, 5.46 mM MgCl2, 10 mM HEPES (pH7.4 with 3.3 mM Na+), 5 mM EGTA, 2.56 mM K2ATP and 0.3 mM Li2GTP. The external bath solution contained 155 mM NaCl, 5 mM KCl, 2 mM MgCl2, 0.5 mM CaCl2 and 10 mM HEPES (pH7.4). The osmolarity was 310-330 mOsm.
Macroscopic currents were elicited with 1-s voltage steps from -100 mV to +40 mV in 20-mV increments. The time interval between voltage steps was 10 s. For analysis, the current elicited by a 1-s voltage step to +20 mV was fit with a sum of two exponentials (A1exp(-t/τ1) + A2exp(-t/τ2) and a constant (C). A 4-s voltage step to +20 mV was used for OmeTyr and DanAla mutant channels. All recordings were performed at room temperature. Clampex8 was used for recording and data were analyzed using Clampfit 8 (Axon). All values shown are mean ± s.e.m. Statistical significance (P < 0.05) was assessed using Kruskal-Wallis one-way ANOVA on Ranks, followed by Dunn's post-hoc test for significance (SigmaStat 3.0).
Fluorescence microscopy
Fluorescence images were acquired on an Olympus X81 inverted microscope using a 20× objective. For the GFP channel, filters were 480/30 nm for excitation and 535/40 nm for emission. For the mCherry channel, filters were 580/20 nm for excitation and 675/130 nm for emission.
ACKNOWLEDGMENTS
We thank B.R. Martin and R.Y. Tsien for plasmid pCLHF, E. Cooper for Kv1.4 cDNA, and D. Summerer and P.G. Schultz for plasmid pLeuRSB8T252A before publication. We thank A.R. Parrish, K. Adams, J. Xu, F.D. Winter, R. Nassirpour, Z. Chen for technical support, and P. Aryal for helpful discussions. P.A. Slesinger acknowledges grant support from US National Institutes of Health. L. Wang acknowledges the support of the Searle Scholar Program and the Beckman Young Investigator Program.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
References
- 1.Callaway EM. A molecular and genetic arsenal for systems neuroscience. Trends Neurosci. 2005;28:196–201. doi: 10.1016/j.tins.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi CS, Isacoff EY. Shedding light on membrane proteins. Trends Neurosci. 2005;28:472–479. doi: 10.1016/j.tins.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 5.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 6.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Schultz PG. Expanding the genetic code. Angew. Chem. Int. Edn. Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 8.Nowak MW, et al. Nicotinic receptor binding site probed with unnatural amino acid incorporation in intact cells. Science. 1995;268:439–442. doi: 10.1126/science.7716551. [DOI] [PubMed] [Google Scholar]
- 9.Lummis SC, et al. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 10.Lummis SC, Beene DL, Harrison NJ, Lester HA, Dougherty DA. A cation-pi binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem. Biol. 2005;12:993–997. doi: 10.1016/j.chembiol.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Beene DL, Price KL, Lester HA, Dougherty DA, Lummis SC. Tyrosine residues that control binding and gating in the 5-hydroxytryptamine3 receptor revealed by unnatural amino acid mutagenesis. J. Neurosci. 2004;24:9097–9104. doi: 10.1523/JNEUROSCI.2429-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 13.Johnston D, et al. Dendritic potassium channels in hippocampal pyramidal neurons. J. Physiol. (Lond.) 2000;525:75–81. doi: 10.1111/j.1469-7793.2000.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 15.Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 16.Demo SD, Yellen G. The inactivation gate of the Shaker K+ channel behaves like an open-channel blocker. Neuron. 1991;7:743–753. doi: 10.1016/0896-6273(91)90277-7. [DOI] [PubMed] [Google Scholar]
- 17.Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Magliery TJ, Liu DR, Schultz PG. A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J. Am. Chem. Soc. 2000;122:5010–5011. [Google Scholar]
- 19.Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem. Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 20.Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- 21.Myslinski E, Ame JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–2509. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards H, Schimmel P. A bacterial amber suppressor in Saccharomyces cerevisiae is selectively recognized by a bacterial aminoacyl-tRNA synthetase. Mol. Cell. Biol. 1990;10:1633–1641. doi: 10.1128/mcb.10.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doctor BP, Mudd JA. Species specificity of amino acid accepter ribonucleic acid and aminoacyl soluble ribonucleic acid synthetases. J. Biol. Chem. 1963;238:3677–3681. [PubMed] [Google Scholar]
- 24.Drabkin HJ, Park HJ, Rajbhandary UL. Amber suppression in mammalian cells dependent upon expression of an Escherichia coli aminoacyl-tRNA synthetase gene. Mol. Cell. Biol. 1996;16:907–913. doi: 10.1128/mcb.16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summerer D, et al. A genetically encoded fluorescent amino acid. Proc. Natl. Acad. Sci. USA. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murrell-Lagnado RD, Aldrich RW. Interactions of amino terminal domains of Shaker K channels with a pore blocking site studied with synthetic peptides. J. Gen. Physiol. 1993;102:949–975. doi: 10.1085/jgp.102.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondoh S, Ishii K, Nakamura Y, Taira N. A mammalian transient type K+ channel, rat Kv1.4, has two potential domains that could produce rapid inactivation. J. Biol. Chem. 1997;272:19333–19338. doi: 10.1074/jbc.272.31.19333. [DOI] [PubMed] [Google Scholar]
- 28.Antz C, et al. NMR structure of inactivation gates from mammalian voltage-dependent potassium channels. Nature. 1997;385:272–275. doi: 10.1038/385272a0. [DOI] [PubMed] [Google Scholar]
- 29.Wissmann R, et al. Solution structure and function of the “tandem inactivation domain” of the neuronal A-type potassium channel Kv1.4. J. Biol. Chem. 2003;278:16142–16150. doi: 10.1074/jbc.M210191200. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy S, Di Liegro C, Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987;51:81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- 33.Yuan Y, Reddy R. 5′ flanking sequences of human MRP/7-2 RNA gene are required and sufficient for the transcription by RNA polymerase III. Biochim. Biophys. Acta. 1991;1089:33–39. doi: 10.1016/0167-4781(91)90081-v. [DOI] [PubMed] [Google Scholar]
- 34.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 35.Cornish VW, Mendel D, Schultz PG. Probing protein structure and function with an expanded genetic code. Angew. Chem. Int. Edn. Engl. 1995;34:621–633. [Google Scholar]
- 36.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 37.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus post-transcriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]