Abstract
Human Pin1 is a key regulator of cell-cycle progression and plays growth-promoting roles in human cancers. High-affinity inhibitors of Pin1 may provide a unique opportunity for disrupting oncogenic pathways. Here we report two high-resolution X-ray crystal structures of human Pin1 bound to non-natural peptide inhibitors. The structures of the bound high-affinity peptides identify a type-I β-turn conformation for Pin1 prolyl peptide isomerase domain–peptide binding and an extensive molecular interface for high-affinity recognition. Moreover, these structures suggest chemical elements that may further improve the affinity and pharmacological properties of future peptide-based Pin inhibitors. Finally, an intramolecular hydrogen bond observed in both peptide complexes mimics the cyclic conformation of FK506 and rapamycin. Both FK506 and rapamycin are clinically important inhibitors of other peptidyl-prolyl cis-trans isomerases. This comparative discovery suggests that a cyclic peptide polyketide bridge, like that found in FK506 and rapamycin or a similar linkage, may significantly improve the binding affinity of structure-based Pin1 inhibitors.
Prolyl peptide isomerases (PPIases) catalyze the rapid equilibration of cis–trans conformers of proline-containing peptide bonds. These backbone conformational changes play a pivotal role in protein folding (1). Moreover, the polypeptide backbone structural change centered around proline-containing peptide bonds can result in a kink in the polypeptide chain and, therefore, conformationally regulate a wide range of biological activities (2), including some activities resulting in disease (3, 4). To date, three families of highly conserved PPIases have been identified, namely, cyclophilins (Cyp) (5), FK506 binding proteins (FKBPs) (6) and parvulins (7). The Cyp and FKBP family members are also collectively termed immunophilins because of their role as binding partners for cyclosporine A (8, 9) and FK506 (6, 10), both of which serve as immunosuppressants. The most widely studied member of the parvulin family, human Pin1, distinguishes itself from Cyp and FKBP through its unique substrate specificity for the phosphorylated Ser/Thr-Pro (P.Ser/P.Thr-Pro) motif (11, 12).
The identification and characterization of Pin1 led to the discovery of a novel post-phosphorylation regulatory mechanism in proteins possessing –P.Ser/P. Thr-Pro– motifs (11, 12). Cyclin-dependent protein kinase, glycogen synthase kinase 3β, and mitogen-activated protein kinases, as well as some of the protein phosphatases such as PP2A, are proline-directed enzymes that recognize Ser/Thr-Pro segments in their respective substrates (13). These enzymes act catalytically in a conformationally specific manner, only catalyzing phosphorylation or dephosphorylation of the trans conformation of proline-containing peptide bonds in their protein substrates. The intrinsic rate of peptide bond isomerization of these dipeptide segments is especially slow in phosphorylated proteins because of the steric hindrance presented by the phosphate group. Therefore, the PPIase activity of Pin1 will greatly accelerate the reestablishment of the trans-to-cis equilibria of these dipeptide segments, ultimately affecting the underlying biological pathways (recently reviewed in ref 14).
Over the last decade, >30 proteins have been identified as Pin1 targets, most of which are involved in cell cycle regulation and growth promotion, including cyclin D1 (15), c-Jun (16), c-Myc (17), β-catenin (18), and p53 (19, 20). Through a P.Ser/P.Thr-Pro-specific N-terminal WW domain binding module (21, 22) and a P.Ser/P.Thr-Pro-specific recognition motif on Pin1’s C-terminal PPIase catalytic domain (12, 23), Pin1 can both bind to and modify the function of phosphorylated protein targets. By altering the cis–trans state of P.Ser/P.Thr-Pro peptide bonds, Pin1 catalyzes peptide backbone conformation changes that can then alter the catalytic activity, stability, subcellular localization, and the rate and extent of dephosphorylation of phospho-protein targets (24, 25).
Pin1 can amplify oncogenic signals (17, 26) and is highly expressed in several human tumors (27), including breast (16), colorectal (28), prostate (29), and thyroid cancers (30). Studies have shown a direct correlation between Pin1 protein levels, Pin1 catalytic activity, and tumor growth (14). Furthermore, Pin1 appears to be a highly predictive and early diagnostic marker for prostate cancer prognosis (29). Loss of Pin1 function triggers apoptosis (31), suppresses the transformed phenotype in some cell lines (31, 32), and can even prevent oncogenic transformation (32, 33). In contrast, Pin1 overexpression induces cancer development in vitro and in vivo (34).
Given Pin1’s role in cell cycle regulation and oncogenesis, Pin1 has been used as a unique target for anti-neoplastic agents. Here, we designed and synthesized two non-natural peptide inhibitors that can bind to the PPIase domain of Pin1 with nanomolar affinity. The structural analyses of these high-affinity peptides bound to human Pin1 provide additional insight into the structure–affinity relationships of these peptide-based inhibitors. More importantly, the high-resolution structures clarify the chemical features underlying high-affinity Pin1-peptide recognition and shed light on the peptide bond isomerization mechanism.
RESULTS AND DISCUSSION
Peptide Inhibitor Design
By examining the structure of Pin1 bound to a low-affinity Ala-Pro dipeptide (Protein Data Bank (PDB) code 1PIN), we speculated that the hydrophobicity and excess volume of the proline-binding pocket in Pin1 might serve as a prominent target for enhancing inhibitor binding to Pin1’s PPIase domain. Indeed, more bulky proline mimics like pipe-colic acid (Pip) are found in PPIase inhibitors such as the natural products FK506 and rapamycin, and this amino acid is used in other synthetic pharmacophores as a surrogate for proline. In addition, based upon previous results obtained with a Pin1 substrate library (23), the inclusion of aromatic residues surrounding the core P.Ser/P.Thr-Pro motif may provide additional hydrophobic contacts around the central proline-binding pocket.
Using a cellulose-bound combinatorial peptide library containing non-natural amino acids, we qualitatively identified Ac-Phe1-L-(or D-)P.Thr2-Pip3-Nal4 (naphthylalanine)-Gln5-NH2 (called L-PEPTIDE and D-PEPTIDE, respectively) (Figure 1, panel a) as high-affinity, Pin1-interacting peptides. Each peptide was then synthesized in solution to quantitatively assess peptide-based inhibiton of Pin1’s PPIase activity (35). Both the L- and D-PEPTIDE exhibited a high degree of inhibitory activity with Ki values of 507 ± 37 and 20.4 ± 4.3 nM (Supplementary Table 1), respectively. Furthermore, the metabolically stable D-PEPTIDE was shown to block cell cycle progression in HeLa cells in a dose-dependent manner (35).
Figure 1.
D- and L-PEPTIDE peptide-based PPIase inhibitors of Pin1. a) Chemical structure of the peptides used in crystallization experiments. P.Thr2-Pip3 mimicking the substrate recognition motif is colored blue. The peptide bond analogous to the one subject to catalytic isomerization in peptide/protein substrates is colored red. b) Ribbon representation of the overall structure of the complex of Pin1 and D-PEPTIDE. The D-PEPTIDE bound to the PPIase domain and a PEG400 molecule bound to the WW domain are shown as half-colored bonds with yellow specifying carbon, red oxygen, blue nitrogen, and magenta phosphorus. The R14A Pin1 mutation that improves crystal growth, size, and stability is also shown.
To determine whether the L-PEPTIDE serves as a substrate for Pin1’s PPIase domain, 2D 1H NMR exchange spectra of D- and L-PEPTIDE were measured (Supplementary Table 3 and Supplementary Figure 2). The lack of any detectable cis isomer of the D.Thr2-Pip3 peptide bond prevented further evaluation of the substrate potential of the D-PEPTIDE. However, the inability of oligopeptides with a D-amino acid preceding the peptide bond to undergo PPIase-mediated isomerization has been shown previously (36). In fact, for all PPIases investigated to date, kcat/KM values decreased 102- to 105-fold for substrates bearing a D-amino acid at the position preceding proline (37). This loss in catalytic efficiency for other structural classes of PPIases is consistent with the results presented here for Pin1 and the D- and L-PEPTIDES. The high affinity and selectivity place the D-PEPTIDE among the most promising small molecule inhibitors discovered to date for in vivo studies of Pin1-mediated signaling pathways (Supplementary Table 1).
Crystallization and Structure Determination
To better understand the structural features accompanying high-affinity recognition of the Pin1 PPIase domain by the L-PEPTIDE and D-PEPTIDE, as well as the inhibitory mechanism of the D-PEPTIDE, we solved high-resolution crystal structures of Pin1 complexed to each non-natural peptide (Table 1). An R14A mutant of Pin1 was used in the structural studies because it exhibits higher stability, more predictable crystallization of very large crystals, and no interference with Pin1 phosphopeptide binding (WW domain) or catalytic activity (PPIase domain) (21). While both Pin1 domains are capable of binding to phosphorylated targets, the L-PEPTIDE and D-PEPTIDE specifically bind to Pin1’s PPIase domain, leaving the WW domain untouched (where Arg14 is located) with a poly(ethylene glycol) (PEG) molecule derived from the crystallization conditions bound between the WW and PPIase domain interfaces (Figure 1, panel b). The R14A mutation favors crystal formation and crystal growth in a different space group, P3121, than wild-type Pin1, P43212. Notably, in addition to the other practical properties mentioned above, the new crystal form is much more resistant to damage and cracking introduced during inhibitor soaking than wild-type Pin1.
TABLE 1.
Data collection and refinement statistics
| L-PEPTIDE complex | D-PEPTIDE complex | |
|---|---|---|
| Data collection | ||
| Space group | P3121 | P3121 |
| Cell dimensions | ||
| a, b, c (Å) | 68.84, 68.84, 79.51 | 68.73, 68.73, 79.96 |
| α, β, γ (deg) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 33.0–1.50 (1.36–1.50)a | 35.0 –1.45 (1.49 –1.45)a |
| Rsym or Rmerge (%) | 2.6 (17.0)a | 4.5 (46.9)a |
| I/σI | 44.6 (6.9)a | 41.2 (2.0)a |
| Completeness (%) | 98.4 (96.6)a | 98.6 (97.1)a |
| Redundancy | 3.6 (3.3)a | 4.6 (3.0)a |
| Refinement | ||
| Resolution (Å) | 33.0 –1.50 | 35.0 –1.45 |
| No. reflections | 32653 | 35494 |
| Rwork/Rfree (%)b | 23.6/25.1 | 22.2/23.5 |
| No. atoms | ||
| Protein | 1155 | 1155 |
| Ligand | 67 | 73 |
| Water | 165 | 172 |
| B-factors (Å2) | ||
| Protein | 17.7 | 21.4 |
| Ligand/ion | 23.2 | 30.2 |
| Water | 28.6 | 33.6 |
| Rms deviations | ||
| Bond lengths (Å) | 0.011 | 0.010 |
| Bond angles (deg) | 1.35 | 1.36 |
Highest-resolution shell is shown in parenthesis.
Rfree is calculated with 5% of the data randomly omitted from refinement.
3D Structure of the Pin1–D-PEPTIDE Complex
The structure of the D-PEPTIDE complex was determined and refined to 1.45 Å resolution (Figure 2, panel a; Table 1). The bound peptide forms an extensive interface with the Pin1 PPIase domain that includes van der Waals interactions as well as hydrogen bonds and electrostatic interactions with backbone and side chains of Pin1 (Figure 2, panel c). The extensive and intimate nature of the D-PEPTIDE binding site coupled with the observed nearly trans pipecolinyl peptide bond conformation (ω bond angle = 183°) of the D-P.Thr2-Pip3 dipeptide segment supports the measured inhibitory activity of the peptide for Pin1 (Figure 2, panel c and Supplementary Table 1).
Figure 2.
Recognition of D-PEPTIDE by human Pin1. a) The electron density associated with the D-PEPTIDE shown as a SigmaA-weighted 2Fo − Fc electron density map contoured at 1.5σ with D-PEPTIDE carbon atoms color-coded yellow. The side chain of Phe1 of D-PEPTIDE was not visible at any stage of structure elucidation and refinement. b) Surface representation of the binding pocket surrounding Pip3. One wall of the hydrophobic binding pocket is composed of Leu122, Met130, and Phe134 colored orange. The lower active site wall consisting of His59 and His157 is colored slate. The flexible side chain of Arg68 is colored blue. c) Stereoview of D-PEPTIDE bound to Pin1’s PPIase domain. Hydrogen bonds are shown with intermolecular hydrogen bonds rendered as green dashed cylinders and an intramolecular hydrogen bond in D-PEPTIDE rendered as red dashed cylinders.
The positively charged patch formed by a triad of basic residues, Lys63, Arg68, and Arg69, defines the entrance to the PPIase domain’s active site (Figure 2, panel c), and two of these residues, Lys63 and Arg69, modulate specificity for the anionic side chain, D-P.Thr2, amino-terminal to the proline mimic Pip. Somewhat surprisingly, the D-P.Thr2 residue is cradled in a basic cavity that is more spacious than anticipated, with Lys63 and Arg69 making direct contact to the phosphate moiety and the Arg68 side chain highly disordered with no direct interaction with inhibitor peptide. Indeed, this relatively spacious phosphate binding cavity would allow for “rolling” of the phosphate as the isomerization reaction proceeds while maintaining electrostatic contact to the basic side chains. In addition, the side chain hydroxyl group of Ser154 forms a hydrogen bond to the carbonyl oxygen that would undergo peptide bond isomerization resulting in a complete sequestration of the peptide bond carbonyl at the entrance to the Pin1 active site cavity.
Pip, used as a more spacious mimic of proline, is cradled in a greasy pocket formed by the side chains of Leu122, Met130, and Phe134 (Figure 2, panel b). The six-membered ring of Pip compared to the five-membered ring of proline provides a larger accessible surface, possibly accounting for at least part of the high affinity of the inhibitor for the Pin1 PPIase domain active site. As mentioned previously, Pip is found in natural products like FK506 and rapamycin, and this amino acid is used in other synthetic pharmacophores as a surrogate for proline.
Moreover, the non-natural and aromatic amino acid Nal stacks edge-to-face with the Pip ring and resides in a pocket formed by Leu122, Ala124, Phe125, and Met130. Notably, the thioether side chain of Met130 is sandwiched by Pip on the bottom and Nal on top (Figure 2, panel b). The extensive nature of this Nal-mediated interaction with Pin1 may also explain Pin1’s preference for bulky hydrophobic/aromatic residues in the X + 1 position relative to proline in Pin1 substrates (23). The N-terminal residue abutting P.Thr is consistently disordered in these structures, and other Pin1–substrate complexes (M. Verdecia, Y. Zhang, and J. Noel, Salk Institute, unpublished data), suggesting that the polypeptide chain preceding the phospho-amino acid residue is highly flexible.
Rotation Direction of Isomerized Peptide Bonds in Pin1-Mediated Catalysis
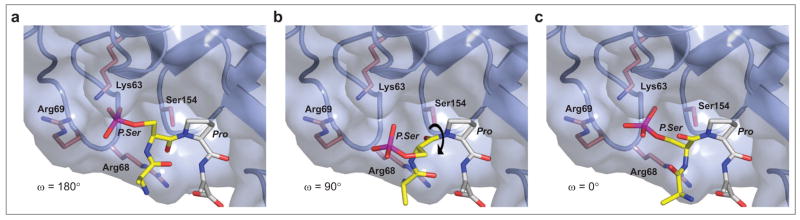
It is worth considering which end of a protein defined centrally by the P.Ser/P.Thr-Pro dipeptide segment complexed in the Pin1 PPIase domain active site will remain fixed and which end will undergo rotation as the peptide bond flips between a cis and trans configuration. The complex of Pin1 with the D-PEPTIDE provides important clues with which to answer this question. Rotation of the C-terminal end of a bound peptide/protein would result in a movement of Pro/Pip out of the proline-binding pocket as well as a loss of a hydrogen bond between the proline carbonyl oxygen and the backbone amide of Gln131. Conversely, rotation of the N-terminal end of a bound peptide/protein would not significantly alter Pin1-substrate recognition due to the spacious phosphate binding pocket that can accommodate a rolling P.Ser/P.Thr side chain (Figure 3) and due to the paucity of interactions between Pin1 and residues amino-terminal to the D-P.Thr residue of D-PEPTIDE as described earlier. Notably, the first amino acid of D-PEPTIDE, Phe1, exhibits no traceable electron density. Furthermore, the N-terminal segment of the bound peptide enters the Pin1 active site near Arg68, the latter of which shows a high degree of positional flexibility as evidenced in previously determined Pin1 structures (12) providing a mobile structural element that can accommodate multiple conformations of N-terminal flanking residues of Pin1 substrates. The C-terminal end of the bound peptide resides in a much more restrictive environment, effectively limiting the range of conformations accessible to a bound substrate.
Figure 3.
3D models depicting a possible isomerization trajectory of the P.Ser-Pro peptide bond. Human Pin1 is shown as a ribbon diagram colored blue with the Pin1-accessible surface depicted as a semitransparent skin. A rendered model of an Ala-P.Ser-Pro-Ala tetrapeptide substrate is shown as half-colored bonds. The carbon atoms of the section of peptide most likely to undergo rotation are colored yellow, while the carbon atoms of the portion of the peptide most likely to remain relatively fixed are colored light gray. Pin1 residues providing possible stabilizing interactions are also shown as half-colored bonds with carbon atoms depicted as dark pink. a) The P.Ser-Pro peptide bond depicted with an ω angle of 180° (trans). b) The P.Ser-Pro peptide bond depicted with an ω angle of 90°. c) The P.Ser-Pro peptide bond depicted with an ω angle of 0° (cis).
Structure of Pin1 with the L-PEPTIDE and Comparison with the D-PEPTIDE Complex
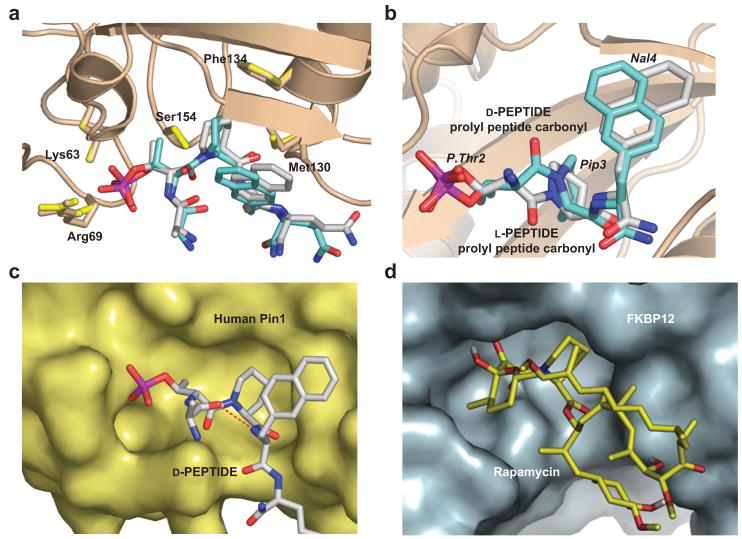
The complex of Pin1 with L-PEPTIDE refined to 1.5 Å resolution (Table 1) is almost identical in structure to that of D-PEPTIDE bound to Pin1 except for a few notable and complementary differences (Figure 4, panel a). The position of the peptide bond carbonyl between L-P.Thr and Pip faces out of the active site cavity and away from the side chains of Arg68, Gln131, and Ser154, resulting in an ω bond angle for the L-P.Thr-Pip peptide bond close to the cis conformation (ω = −19°). In both peptide complexes with Pin1, the nitrogen atom in the Pip heterocyclic ring assumes some degree of distortion from the idealized sp2 arrangement common in peptide bonds and is in fact close to a tetrahedral sp3 hybridization state. This conformation is indicative of a transition of the intervening peptide bond from a partial double bond character to characteristics of a single C–N bond.
Figure 4.
Comparison of the active sites of human Pin1 and FKBP complexes. a) Comparative binding modes of D- and L-PEPTIDE to human Pin1 shown as superimposed images. The D- and L-PEPTIDE-bound structures of Pin1 are identical to each other. The Pin1 amino acid side chains involved in peptide recognition are shown as half-colored bonds with carbon rendered brown for the D-PEPTIDE complex and yellow for the L-PEPTIDE complex. D- and L-PEPTIDE are depicted as half-colored bonds with carbon colored light gray for the D-PEPTIDE and cyan for the L-PEPTIDE. b) A close-up view rendered in a slightly different orientation than panel a. Only the P.Thr2-Pip3-Nal4 subsection of the peptides are shown for clarity. c) The accessible surface of Pin1 complexed with the D-PEPTIDE. The intramolecular hydrogen bond (dashed red cylinders) between the carbonyl oxygen of Phe1 and the amide nitrogen of Nal4 defines a bound peptide conformation that closely resembles the conformation and binding mode of rapamycin bound to FKBP depicted in panel d (PDB code 1fkb). d) Rendered 3D structure of the FKBP12 accessible surface shown bound to rapamycin. In Pin1, the Pip residue forms an intimate set of hydrophobic interactions with Phe134 and Leu122 in the Pin1 active site. This interaction is spatially conserved in the FKBP–rapamycin complex with Phe46 and Trp59 in human FKBP12 sequestering the Pip moiety of rapamycin.
D-PEPTIDE possesses a higher inhibitory activity than L-PEPTIDE, even though the two complexes are highly similar (Figure 4, panel a). One possible distinguishing feature is the carbonyl group of the P.Thr-Pip peptide bond in D-PEPTIDE that forms a hydrogen bond with the Ser154 hydroxyl moiety. Mutation of Ser154 to bulkier residues including cysteine (S154C) or aspartic acid (S154D) results in clashes with the carbonyl group of the P.Thr-Pip peptide bond and an attendant increase of Ki by nearly 100-fold (Supplementary Table 1). However, in contrast, an S154A Pin1 mutant demonstrates that the loss of this hydrogen bond does not compromise the high-affinity binding of the D-PEPTIDE (Supplementary Table 1). Moreover, based upon the 3D structure of the D-PEPTIDE complex of the R14A–S154A mutant, D-PEPTIDE binds in a conformation identical to that with which it binds R14A mutant (Supplementary Table 2 and Supplementary Figure 1). On the other hand, the S154C mutant binds L-PEPTIDE, which does not possess the Ser154 hydrogen bond to the peptide bond carbonyl, with higher affinity than the wild-type Pin1. This favorable energetic result may be the consequence of the hydrophobic interaction between the backbone and the cysteine side chain when the prolyl-peptide bond carbonyl group faces outward (Figure 4, panel c).
Implications for Structure-Based Design
In addition to the enhanced hydrophobic interactions provided by Nal and Pip, one additional structural feature possibly accounting for the high affinity of the D-PEPTIDE and L-PEPTIDE Pin1 ligands and also reminiscent of conformational constraints found in other clinically important PPIase inhibitors is the presence of a well-defined intramolecular hydrogen bond between the carbonyl oxygen of Phe1 and the amide group of Nal4 (Figure 2, panel c, and Figure 4, panel c). The formation of this intramolecular hydrogen bond results in a type I β-turn for both the L-PEPTIDE and D-PEPTIDE Pin1 inhibitors (Figure 4, panel a). This noncovalent, intramolecular bridge shows considerable conformational homology with the cyclic structures of natural PPIase inhibitors, namely, FK506 (Kd = 0.4 nM to FKBP12) and rapamycin (Kd = 0.2 nM to FKBP12). These two natural products of mixed biosynthetic origin (nonribosomal peptide and polyketide) share a similar overall structure with a Pip moiety serving as a proline mimic (Figure 4, panel d). Interestingly, while there is no clear sequence similarity between human Pin1 and human FKBP, these two PPIases share a 3D fold (12). Furthermore, both the Pin1 and FKBP active sites share a conserved shape and cavity volume outside of the recognition elements responsible for selectivity of the residue amino terminal to proline (Figure 4, panels c and d).
This similarity in structure extends to the conformations of FK506/rapamycin bound to FKBP and the L-PEPTIDE/D-PEPTIDE bound to Pin1 (Figure 4, panels c and d). The close similarity in the chemical features of the respective PPIase binding pockets in Pin1 and FKBP and the conformations of high-affinity inhibitors in each case suggest new avenues for future improvements in the inhibitory potency and pharmacological properties of peptide-based Pin1 inhibitors. This includes the use of cyclic peptides or peptidomimetics.
A significant issue that remains in D-PEPTIDE from the drug design perspective concerns the difficulty associated with delivering phosphate groups across the hydrophobic barrier of the plasma membrane. To obtain a lead peptide with better cell permeability, one could take advantage of the greatly enhanced inhibitory potential of D-PEPTIDE due to conformational (type I β-turn) and steric factors (Pip and Nal binding) to eliminate the phosphate group entirely, pay the electrostatic energetic penalty, and recover enough binding energy by replacing the phosphate group with neutral hydrogen bonding partners.
METHODS
Inhibitor Synthesis
Ac-Phe1-P.Thr2-Pip3-Nal4-Gln5-NH2 was synthesized on a Syro II multiple peptide synthesizer (Multi-SynTech). Synthesis was accomplished using commercially available Nα-Fmoc-protected amino acid derivatives (Bachem; NovaBiochem) and a Rink Amide MBHA resin. Acylation was performed using PyBOP as activation reagent except for the activation of Fmoc-Thr(PO(OBzl)OH) that was accomplished by HATU. Ac-Phe-D-P.Thr-Pip-Nal-Gln-NH2 was synthesized using the standard method for “global phosphorylation” of resin-bound peptides and Fmoc-D-Thr(Trt)-OH as building block (38 – 41).
Peptides were cleaved from the resin by treatment with 94% trifluoroacetic acid/2.5% H2O/2.5% ethylene dithiol/1% triisopropylsilane (v/v/v/v), precipitated with diethylether, purified by preparative reversed-phase high-performance liquid chromatography, lyophilized, and stored at −80 °C. Peptide identity was verified by MALDI TOFMS.
Pin1 Activity Assay and Inhibition Constants
PPIase activities were measured at 10 °C in 35 mM HEPES buffer, 1 mM DTT at pH 7.8 with a Hewlett-Packard 8453A UV–vis spectrophotometer using the protease-free assay according to Janowski et al. (42– 44). Suc-Ala-Glu-Pro-Phe-4-nitroanilide in 0.5 M LiCl/trifluoroethanol (anhydrous) was used as the substrate. The concentration of wild-type Pin1 or Pin1 variants ranged between 5 and 200 nM depending on the activity of the respective Pin1 variant. Prior to measurements, all components except substrate were incubated for 300 s at 10 °C, and the measurement was initiated upon addition of the substrate. Kinetic traces of cis/trans isomerization of the substrate were followed at 330 nm for several half-lives of the total reaction. Ki for D- and L-PEPTIDE was obtained by measuring the remaining Pin1 activity at different inhibitor concentrations of the respective peptide in the activity assay mentioned above. Since the affinity of Pin1 for Suc-Ala-Glu-Pro-Phe-pNA is low when compared to L- and D-PEPTIDE, the plot of remaining activity versus concentration can be used to calculate Ki values. In several cases, isotermal titration calorimetry (ITC) of the Pin1–inhibitor pairs served as an independent measure of affinity. Results are given as mean averages ± 10% (n = 3–5).
Protein Purification, Crystallization, and Data Collection
The Pin1 R14A mutant was prepared using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and expressed and purified in a manner similar to wild-type human Pin1 as described previously (12). Crystals grew from 1.9 –2.2 M ammonium sulfate, 50 mM HEPES, pH 7.5, and 1 mM DTT within 2 d using vapor diffusion in 2 μL of sitting drops containing 18 mg ml−1 R14A Pin1. Peptide inhibitors were dissolved in 50 mM NaCl, 100 mM HEPES, pH 7.5, at a concentration of 50 mM. One-tenth volume of inhibitor stocks was added to the mother liquor made up of 2.0 M ammonium sulfate, 50 mM HEPES, pH 7.5, and 1 mM DTT. R14A Pin1 crystals were transferred into 2 μL of mother liquor containing 0.5 mM of either the L-PEPTIDE or the D-PEPTIDE. The crystals were harvested into 2 μL of a cryoprotection solution consisting of 40% (v/v) PEG400 after 48 h of soaking and then transferred to a crystal freezing loop for flash-freezing in liquid nitrogen. Data were collected at wavelength 0.9537 Å at 100 K on beam-line 8.2.2 of the Advanced Light Source (ALS). Diffraction data were processed with HKL2000 (45), and the statistics are summarized (Table 1).
Structure Solution and Refinement
The crystal structures of human Pin1 R14A in complex with D-PEPTIDE and L-PEPTIDE were determined by molecular replacement using wild-type Pin1 (PDB code 1PIN) as the search model and the program AmoRe (46) from the CCP4 package (47). Refinement was first carried out using CNS (48), with a 5% test set (reflections) excluded for Rfree cross-validation (49). Final refinements employed REFMAC in CCP4 (50). SigmaA-weighted 2Fo − Fc and Fo − Fc electron density maps were calculated after each cycle of refinement and inspected to guide model rebuilding using O (51). The locations of the peptides were clear in Fo − Fc maps even after the first round of refinement. The inhibitor model was built into the electron density maps using O (51).
During initial stages of refinement, the prolyl peptide amide nitrogen, while constrained to be sp2, consistently deviated from the sp2 hybridization state in electron density maps. After several rounds of refinement, the geometric restraint was lessened to allow the refinement of the amide nitrogen position to account for the “extra” electron density indicative of a partially pyrimidalized nitrogen. The final models were evaluated using PROCHECK (52). For the Pin1–D-PEPTIDE complex, 94.4% of the residues reside in the most favored regions of the Ramachandran plot and the remaining 5.6% are found in additionally allowed areas. For the Pin1–L-PEPTIDE complex, 94.4% of the residues are found in the most favored regions of the Ramachandran plot, with 4.8% in additionally allowed regions and 0.8% in generally allowed regions with no residues found in the disallowed regions. Refinement statistics are summarized (Table 1). Figures in this manuscript were prepared with PyMOL (53).
NMR
For the 1H assignments, the NMR samples contained 1.17 mM L-PEPTIDE or 1.67 mM D-PEPTIDE adjusted to pH 7.8 in a H2O/D2O mixture (95:5 v/v). Standard homonuclear 1D and 2D (total correlation spectroscopy and rotational frame nuclear Overhauser effect spectroscopy (ROESY)) pulse sequences were collected at 25 °C using a DRX500 spectrometer (Bruker). 1H NMR data were acquired at a 500.13 MHz resonance frequency with a 5 mm inverse triple-resonance probe that possessed XYZ-gradient capability. All spectra were processed with the XWINNMR 3.5 software (Bruker) and referenced to external 2,2-dimethyl-2-silapentane-5-sulfonate. The proton resonances (Supplementary Table 3) were assigned according to classical assignment strategies (54). The cis and trans conformers were distinguishable in the ROESY spectra based on Thr2 Hα–Pip3 Hα and Thr2 Hα–Pip3 Hα connectivities, respectively. The L-PEPTIDE exhibited both a cis and a trans conformation of Pip3 at a 61:39 ratio (Supplementary Figure 2), while the D-PEPTIDE existed solely in the trans form.
Exchange measurements were performed in 100% D2O (pD 7.8) at 25 °C using a 2D Nuclear Overhauser Effect Spectroscopy pulse sequence with the mixing time set to 450 ms. On the basis of signal intensities (Supplementary Figure 3), the rate constant of the isomerization reaction was calculated according to the formula:
| (1) |
where Icc is the intensity of the diagonal peak representing the cis form, Itc is the intensity of the trans-to-cis exchange cross-peak, and xcis is the mole fraction of the cis component (55). In the absence of Pin1, the intensity of the exchange cross-peak was 2.08% of the diagonal peak, resulting in a turnover kuncat of 0.083 ± 0.011 s−1. After addition of Pin1 (4 μM final concentration), the intensity of the exchange cross-peak increased to 2.38% of the diagonal peak, resulting in a turnover kcat of 0.097 ± 0.024 s−1. With the addition of D-PEPTIDE (10 μM final concentration), the intensity of the exchange cross-peak returned to 2.07% of the diagonal peak, resulting in a turnover k of 0.088 ± 0.010 s−1, indicating inhibition of the previously Pin1-catalyzed interconversion of L-PEPTIDE cis/trans isomers. No significant alterations in the turnover numbers were observed upon further addition of D-PEPTIDE (up to 25 μM final concentration).
Supplementary Material
Acknowledgments
We thank the staff of ALS Beamline 8.2.2 for technical support in X-ray data collection. This work was supported by grants from the National Institutes of Health to J. P. Noel and T. Hunter (CA54418) and K-P. Lu (GM58556). Additional funds were provided by the Fonds der Chemischen Industrie to G. Fischer. J. P. Noel is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing Interest Statement: The authors declare that they have no competing financial interests.
Accession Codes: Coordinates and structure factors have been deposited in the Protein Data Bank. PDB accession codes: Pin1–D-PEPTIDE complex, 2ITK; Pin1–L-PEPTIDE complex, 2ITI.
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Schmid FX. Protein folding. Prolyl isomerases join the fold. Curr Biol. 1995;5:993–994. doi: 10.1016/s0960-9822(95)00197-7. [DOI] [PubMed] [Google Scholar]
- 2.Edlich F, Fischer G. Pharmacological targeting of catalyzed protein folding: the example of peptide bond cis/trans isomerases. Handb Exp Pharmacol. 2006;172:359–404. doi: 10.1007/3-540-29717-0_15. [DOI] [PubMed] [Google Scholar]
- 3.Lu KP, Suizu F, Zhou XZ, Finn G, Lam P, Wulf G. Targeting carcinogenesis: a role for the prolyl isomerase Pin1? Mol Carcinog. 2006;45:397–402. doi: 10.1002/mc.20216. [DOI] [PubMed] [Google Scholar]
- 4.Etzkorn FA. Pin1 flips Alzheimer’s switch. ACS Chem Biol. 2006;1:214–216. doi: 10.1021/cb600171g. [DOI] [PubMed] [Google Scholar]
- 5.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 6.Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 7.Rahfeld JU, Schierhorn A, Mann K, Fischer G. A novel peptidyl-prolyl cis/trans isomerase from Escherichia coli. FEBS Lett. 1994;343:65–69. doi: 10.1016/0014-5793(94)80608-x. [DOI] [PubMed] [Google Scholar]
- 8.Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid FX. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi N, Hayano T, Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 10.Siekierka JJ, Hung SH, Poe M, Lin CS, Sigal NH. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature. 1989;341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 11.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 12.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 13.Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 2002;12:164–172. doi: 10.1016/s0962-8924(02)02253-5. [DOI] [PubMed] [Google Scholar]
- 14.Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7:435–441. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- 15.Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, Uchida T, Hunter T, Lu KP. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci USA. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez-Sola D, Dalla-Favera R. PINning down the c-Myc oncoprotein. Nat Cell Biol. 2004;6:288–289. doi: 10.1038/ncb0404-288. [DOI] [PubMed] [Google Scholar]
- 18.Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- 19.Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, Gu L, Tang X, Lu KP, Xiao ZX. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature. 2002;419:849–853. doi: 10.1038/nature01116. [DOI] [PubMed] [Google Scholar]
- 20.Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature. 2002;419:853–857. doi: 10.1038/nature01120. [DOI] [PubMed] [Google Scholar]
- 21.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 22.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 23.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 24.Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- 25.Stukenberg PT, Kirschner MW. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol Cell. 2001;7:1071–1083. doi: 10.1016/s1097-2765(01)00245-3. [DOI] [PubMed] [Google Scholar]
- 26.Sears RC. The life cycle of c-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 27.Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–1737. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CJ, Cho YG, Park YG, Nam SW, Kim SY, Lee SH, Yoo NJ, Lee JY, Park WS. Pin1 overexpression in colorectal cancer and its correlation with aberrant beta-catenin expression. World J Gastroenterol. 2005;11:5006–5009. doi: 10.3748/wjg.v11.i32.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, Wheeler TM, Lu KP, Bao L. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003;63:6244–6251. [PubMed] [Google Scholar]
- 30.Nakashima M, Meirmanov S, Naruke Y, Kondo H, Saenko V, Rogounovitch T, Shimizu-Yoshida Y, Takamura N, Namba H, Ito M, Abrosimov A, Lushnikov E, Roumiantsev P, Tsyb A, Yamashita S, Sekine I. Cyclin D1 overexpression in thyroid tumours from a radio-contaminated area and its correlation with Pin1 and aberrant beta-catenin expression. J Pathol. 2004;202:446–455. doi: 10.1002/path.1534. [DOI] [PubMed] [Google Scholar]
- 31.Rippmann JF, Hobbie S, Daiber C, Guilliard B, Bauer M, Birk J, Nar H, Garin-Chesa P, Rettig WJ, Schnapp A. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ. 2000;11:409–416. [PubMed] [Google Scholar]
- 32.Ryo A, Uemura H, Ishiguro H, Saitoh T, Yamaguchi A, Perrem K, Kubota Y, Lu KP, Aoki I. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin Cancer Res. 2005;11:7523–7531. doi: 10.1158/1078-0432.CCR-05-0457. [DOI] [PubMed] [Google Scholar]
- 33.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 2004;23:3397–3407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suizu F, Ryo A, Wulf G, Lim J, Lu KP. Pin1 regulates centrosome duplication, and its overexpression induces centrosome amplification, chromosome instability, and oncogenesis. Mol Cell Biol. 2006;26:1463–1479. doi: 10.1128/MCB.26.4.1463-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wildemann D, Erdmann F, Alvarez BH, Stoller G, Zhou XZ, Fanghanel J, Schutkowski M, Lu KP, Fischer G. Nanomolar inhibitors of the peptidyl prolyl cis/trans isomerase Pin1 from combinatorial peptide libraries. J Med Chem. 2006;49:2147–2150. doi: 10.1021/jm060036n. [DOI] [PubMed] [Google Scholar]
- 36.Schiene C, Reimer U, Schutkowski M, Fischer G. Mapping the stereospecificity of peptidyl prolyl cis/trans isomerases. FEBS Lett. 1998;432:202–206. doi: 10.1016/s0014-5793(98)00871-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Fussel S, Reimer U, Schutkowski M, Fischer G. Substrate-based design of reversible Pin1 inhibitors. Biochemistry. 2002;41:11868–11877. doi: 10.1021/bi0262395. [DOI] [PubMed] [Google Scholar]
- 38.Andrews DM, Kitchin J, Seale PW. Solid-phase synthesis of a range of O-phosphorylated peptides by post-assembly phosphitylation and oxidation. Int J Pept Protein Res. 1991;38:469–475. doi: 10.1111/j.1399-3011.1991.tb01528.x. [DOI] [PubMed] [Google Scholar]
- 39.Perich JW, Reynolds EC. Fmoc/solid-phase synthesis of Tyr(P)-containing peptides through t-butyl phosphate protection. Int J Pept Protein Res. 1991;37:572–575. doi: 10.1111/j.1399-3011.1991.tb00777.x. [DOI] [PubMed] [Google Scholar]
- 40.Perich JW, Ruzzene M, Pinna LA, Reynolds EC. Efficient Fmoc/solid-phase peptide synthesis of O-phosphotyrosyl-containing peptides and their use as phosphatase substrates. Int J Pept Protein Res. 1994;43:39–46. doi: 10.1111/j.1399-3011.1994.tb00374.x. [DOI] [PubMed] [Google Scholar]
- 41.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 42.Janowski B, Wollner S, Schutkowski M, Fischer G. A protease-free assay for peptidyl prolyl cis/trans isomerases using standard peptide substrates. Anal Biochem. 1997;252:299–307. doi: 10.1006/abio.1997.2330. [DOI] [PubMed] [Google Scholar]
- 43.Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Echeverria C, Kofron JL, Kuzmic P, Rich DH. A continuous spectrophotometric direct assay for peptidyl prolyl cis-trans isomerases. Biochem Biophys Res Commun. 1993;191:70–75. doi: 10.1006/bbrc.1993.1185. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. HKL: processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 47.CCP4. Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 49.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 50.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 51.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 53.DeLano WL. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- 54.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 55.Baine P. Comparison of rate constants determined by two-dimensional NMR spectroscopy with rate constants determined by other NMR techniques. Magn Reson Chem. 1986;24:304–307. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.