S-Adenosyl-L-methionine (SAM) is a ubiquitous molecule that participates in various biochemical reactions, second only to ATP as the most frequently used enzyme substrate.[1] The recently described SAM-dependent halogenases involved in the biosynthesis of secondary metabolites in actinomycetes represent a new family of SAM-binding proteins[2, 3] that catalyze the nucleophilic displacement of L-methionine (L-met) from SAM with halides to form halogenated 5′-deoxyadenosine (5′-XDA).[4, 5] These enzymes belong to a family of over 100 archaeal and bacterial proteins with no assigned function (pfam 01887, DUF62). Here we report that a DUF62 member from the recently sequenced marine bacterium Salinispora arenicola CNS-205 (SaDUF62, Sare_1364, genome accession number NC_ 009953) has no significant halogenase, but instead SAM hydrolase (adenosine-forming) activity in vitro.
SAM is biosynthesized from ATP and L-met by SAM synthetase MetK. Besides its well-known and essential role as a methyl donor to nucleic acids and proteins,[6] the electrophilic character of the carbons surrounding the sulfonium group of SAM makes it also a source of methylene, amino, ribosyl, and aminoalkyl groups, as well as 5′-deoxyadenosyl radicals.[1] Furthermore, SAM acts as a molecular effector in the riboswitch-mediated feedback regulation of metK and methionine biosynthesis genes in bacteria[7, 8] and plants.[9] Yet, the regulatory role of SAM is not limited to the met operon. SAM levels have been shown to influence morphological differentiation and secondary-metabolite biosynthesis in the soil bacteria Streptomyces—high levels of SAM cause antibiotic overproduction and inhibit sporulation—at least in part by promoting transcription of regulatory genes.[10–12]
We recently reported that biosynthesis of the marine bacterial natural product salinosporamide A from Salinispora tropica involves a SAM-dependent chlorinase[5] similar to fluorinase from the fluoroacetate-producer Streptomyces cattleya.[4] These two enzymes represent yet another example of the versatility of SAM-binding proteins,[2,3] and are the only members characterized from a family of over 100 bacterial and archaeal homologues available in GenBank and assigned as “protein of unknown function, DUF62, pfam 01887”. Given their wide distribution in the bacteria and archaea domains of life (in the eukarya, only two proteins are found from Entamoeba species; for E. histolytica lateral gene transfer of bacterial genes into its genome has been suggested;[13] accession numbers XP_ 651697, E. histolytica, and XP_001734670, E. dispar) and their relatedness to the newly described SAM-dependent halogenases, we were intrigued about their function (Figure 1A). Recently, Rao et al.[14] reported the crystal structure of a DUF62 protein at 2.5 Å resolution, MJ1651, from the archaeon Methanococcus jannaschii (PDB ID: 2F4N) and showed that it had no chlorinating activity. Two other crystal structures from the Riken Structural Genomics/Proteomics Initiative have been deposited in the Protein Data Bank, as PH0463 from Pyrococcus horikoshii (1WU8, PhDUF62) and TTHA0338 from Thermus thermophilus (2CW5). However, to the best of our knowledge, no catalytic activity has been demonstrated to date.
Figure 1.
Relatedness of SAM-dependent halogenases to DUF62 proteins from different bacteria and archaea. A) Phylogenetic tree. Archaeal proteins were used as outgroup to root the neighbor joining tree. The scale bar indicates 0.1 changes per amino acid. Values on nodes are estimates of the reliability of a particular grouping, that is, bootstrap scores in % out of 2000 replicates. Chlorinase SalL from S. tropica, fluorinase FlA from S. cattleya, DUF62 Sare_1364 from S. arenicola (SaDUF62) and PH0463 from P. horikoshii (PhDUF62) are highlighted. Note that the characterized halogenases form a distinct clade in the actinobacteria group. B) Partial alignment. Conserved active site residues unique for DUF62 proteins are highlighted. Values on top of the alignment correspond to SaDUF62 residue numbering. Asp11 and Asn188, involved in contacts with the ribose and the adenine ring, respectively, are conserved in all proteins (data not shown). Accession codes for DUF62 proteins are Cm, Caldivirga maquilingensis IC-167 (ZP_01710958.1), Ec, E. coli 101-1 (ZP_00924576), Fs, Frankia sp. CcI3 (YP_481014), Hp, Helicobacter pylori 26695 (NP_207503), Ph, P. horikoshii OT3 (NP_142440, PDB ID 1WU8), Pi, Pyrobaculum islandicum DSM 4184 (YP_929529), Sa, S. arenicola CNS-205 (ZP_01648926), Tf, Thermobifida fusca YX (YP_288283), Tk, Thermococcus kodakarensis KOD1 (YP_182475), and for chlorinase (ABP73643 and YP_001157878, PDB ID 2Q6I) and fluorinase (CAE46446, PDB ID 1RQP).
Our previous structural studies on chlorinase SalL[5] together with those by Dong et al. on fluorinase FlA[4] showed that catalytic-site residues of SAM-dependent halogenases include 1) conserved Asp11 (chlorinase numbering) and Asn188 involved in contacts with the ribose and adenine moieties, respectively, and 2) Gly131 (Ser158 in fluorinase), Tyr70 (Thr77 in fluorinase), and Trp129 (Phe156 in fluorinase) in the halide binding pocket. Moreover, a unique 23-residue loop in the N-terminal domain of fluorinase also appears to affect the active-site architecture and halide specificity in the two enzymes.
Site-directed mutagenesis on chlorinase[5] demonstrated that replacement of Tyr70 by a less-bulky threonine residue allowed water to enter the active site, thereby reducing turnover to 0.07 % in comparison to the wild-type enzyme. Similarly, mutation of Ser158 to glycine in fluorinase also leads to hydration of the halide binding pocket and reduction in activity.[15] Sequence alignments (Figure 1B) show that Gly131 is conserved in DUF62 proteins, while Tyr70 is replaced by less-bulky residues including Val, Gly and Ile in 127, 13 and 11 cases, respectively. Therefore, we anticipate that DUF62 proteins will most likely not act as efficient nucleophilic halogenases due to the predicted size of the halide binding pocket and expected presence of water in the active site. Furthermore, their wide distribution in bacteria and archaea suggests a role in primary rather than secondary metabolism. In fact, one deposited structure of an archaeal family member from P. horikoshii (PhDUF62, PDB ID: 1WU8) with 28 % identity to chlorinase revealed adenosine bound in the active site. That alone does not necessarily disclose its enzymatic activity, since chlorinase also copurifies with adenosine, but it does indeed suggest that DUF62 proteins have a nucleoside derivative such as SAM as substrate. The PhDUF62 crystal structure at 2.6 Å resolution also reveals a water molecule that hydrogen bonds with Gly128, analogous to the chloride-coordinating Gly131 described for chlorinase.[5] Together with the phylogenetic analysis of members of the superfamily (Figure 1), which shows that the characterized halogenases form a distinct clade in the actinobacteria group, these data suggest divergent evolution and possibly different enzyme activity.
We decided to test the enzymatic activity of a DUF62 family member from the marine actinomycete S. arenicola closely related to chlorinase (SaDUF62, 35 % sequence identity to SalL on the amino acid level). Upon incubation with SAM and fluoride, chloride, bromide or iodide ions, no halogenase activity could be detected, except for the very slow formation of 5′-iodo-5′-deoxyadenosine (5′-IDA) with a turnover (kcat = 0.5 h−1) about two orders of magnitude lower than that of SalL (Scheme 1). The Km for iodide at 20 mM is 4 × 104-fold higher than the iodide concentration in seawater (0.5 μM); this indicates that the iodinase activity most likely has no biological relevance in S. arenicola.
Scheme 1.

Reactions catalyzed by SaDUF62 from S. arenicola. The respective apparent kinetic constants are an average of at least two independent measurements. Reactions catalyzed by chlorinase from S. tropica (X = Cl, Br, I)[5] and fluorinase from S. cattleya (X = F, Cl)[4, 16] are shown for comparison.
However, incubation of SaDUF62 and SAM showed the pH-dependent formation (optimum at pH 8) of adenosine, as monitored by reversed-phase HPLC and LC/MS. Its turnover (kcat = 0.5 min −1) was comparable to the 5′-chloro-5′-deoxyadenosine (5′-ClDA) synthase activity of SalL (kcat = 0.9 min−1) and had a similar Km for SAM (1.6 μM). Interestingly, chloride inhibits this reaction with a Ki of 1 mM. S-adenosyl-L-homocysteine and 5′-deoxy-5′-methylthioadenosine were also tested as substrates, but no activity was detected.
The reaction catalyzed by chlorinase and fluorinase has been shown to be reversible (Scheme 1).[5,16] In contrast, the reversibility of SaDUF62 to form SAM from adenosine and L-met could not be demonstrated; this reflects the fact that hydroxide is a poor leaving group. Moreover, ATP and AMP were evaluated, but to no avail. When testing against 5′-IDA or 5′-ClDA with L-met, however, SaDUF62 similarly catalyzed the synthesis of SAM.
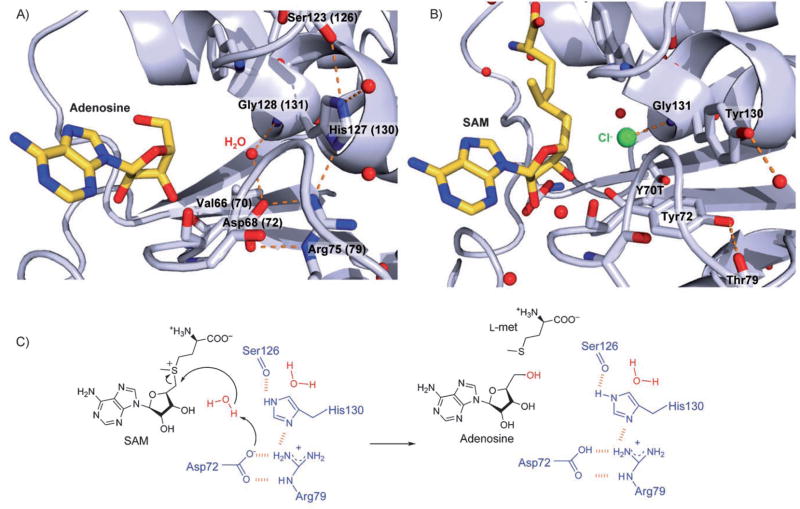
Neither chlorinase wild-type nor its Y70T mutant, which allowed hydration of the halide binding pocket, showed any detectable SAM hydrolase activity. This indicates that during evolution (assuming that SAM-dependent halogenases evolved from SAM hydrolases) not only the halide binding pocket was restricted to avoid hydration and favor nucleophilic substitution, but also residues responsible for acid–base catalysis and hydrolase activity were possibly mutated. As highlighted in Figure 1B, DUF62 proteins show several highly conserved and unique residues compared to chlorinase and fluorinase. Inspection of the available crystal structure of PhDUF62 reveals that these residues are appropriately positioned near C5′ of adenosine, allowing not only hydration of the active site (smaller residues), but also offering a possibility for acid/base catalysis (polarizable residues). Notably, Asp68 (Tyr in halogenases) is within hydrogen bonding distance (3.1 Å) to a water molecule that coordinates Gly128 (Figure 2A and B). Asp68 in turn forms hydrogen bonds to Arg75 (Thr in halogenases), which coordinates to His127 (Tyr). The other two available DUF62 crystal structures (2F4N and 2CW5) are unbound and show no electron density for the respective His127, while Asp and Arg have the same architecture as in PhDUF62. Moreover, homology modeling[17] of SaDUF62 (35 % amino acid identity to PhDUF62) shows similar organization of the triad. Thus, the highly conservative and perfect geometry of this Asp-Arg-His triad (Figures 1B and 2A) suggests its role in the formation of a reactive hydroxide ion for SAM hydrolysis. The exact mechanism for water activation remains to be demonstrated. A simplified proposal in which Asp acts as a base to abstract a proton from water is depicted in Figure 2C.
Figure 2.
Comparison of DUF62 protein and chlorinase active sites and proposed mechanism for hydrolytic activity. A) PhDUF62 from P. horikoshii with bound adenosine (PH0463, PDB ID: 1WU8) showing the putative Asp-Arg-His catalytic triad. Residue numbering for SaDUF62 is shown in parenthesis. Hydrogen bonds are depicted as orange dashes, and water molecules as red dots. B) Chlorinase SalL Y70T from S. tropica with bound SAM and chloride (PDB ID: 2Q6O).[5] Replacement of Tyr70 by threonine allows water to enter the active site (analogous to VaL66 (70) of DUF62 proteins). The putative Asp-Arg-His catalytic triad is replaced by Tyr-Thr-Tyr. C) Proposed reaction mechanism of SaDUF62. pKa values predicted with PROPKA[30] are Asp, 2.1, Arg, 11.6 and His, 1.3. We propose that Asp72 acts as a base to abstract a proton from water and that Arg79 and His130 facilitate the basicity of Asp72. The available crystal structure of PhDUF62 depicts adenosine and a remaining water molecule H-bonded to Asp72; this suggests that more than one water molecule might be present in the active site upon SAM binding. For simplicity, we show only one reactive water in the above mechanism.
Our in vitro studies on SaDUF62 together with in silico analysis of other members of this newly discovered superfamily of SAM-binding proteins suggest that most DUF62 enzymes will act as SAM adenosylhydrolases, while a small subclass, represented by chlorinase and fluorinase (EC 2.5.1.63), have a more specialized function in secondary metabolism. It is noteworthy that the salinosporamide A producer S. tropica[18] also contains a DUF62 member besides SalL (StDUF62, Strop_1405, 86 % identity to SaDUF62 and 35 % identity to SalL on the amino acid level), and that inactivation of salL completely abolishes salinosporamide A production,[5] thus demonstrating that StDUF62 is unable to replace SalL’s function in vivo. Again, this observation suggests different enzymatic activity for the two relatives and supports our hypothesis of divergent evolution. Therefore, as with other halogen-incorporating enzymes that evolved from hydroxylation logic,[19] SAM-dependent halogenases appear to have originated from a common SAM interactive hydrolase. To the best of our knowledge, the only other SAM hydrolase reported to date is the 17 kDa SAMase (5′-methylthioadenosine-forming) from coliphage T3 (E.C. 3.3.1.2), which is involved in overcoming host restriction.[20–22]
Although of no physiological relevance, SaDUF62 does have very slow iodinase activity in vitro. Of all halides, iodide is the weakest nucleophile. However, its nucleophilicity should be the least impaired by hydration. In fact, while the kcat of chlorinase SalL wild-type for iodide is threefold lower than that for chloride, this observation is reversed (sixfold) in SalL Y70T, which presents a hydrated halide binding pocket. Furthermore, the Km of SaDUF62 for iodide is tenfold lower than that of chlorinase, thus indicating higher affinity, likely due to the presence of polar residues, for example, Asp-Arg-His triad. The observation that chloride inhibits hydrolase activity (Ki is 45-fold lower than the Km of SalL) also indicates relatively high affinity of SaDUF62 for halides. Therefore, it appears that exclusion of water from the halide binding pocket and elimination of hydrolase activity in halogenases, through bulkier and less polarisable residues, improved halogenating turnover considerably at the expense of increasing Km only tenfold, with favourable overall catalytic efficiency (kcat/Km).
Analysis of gene neighborhood in various members of archaea and bacteria with the Integrated Microbial Genome tool from the Joint Genome Institute (http://imgweb.jgi-psf.org/cgi-bin/w/main.cgi) identifies no universal gene cluster. However, a common feature is the presence of proteins involved in both nucleotide and amino acid biosynthesis, or DNA, RNA or protein methylation. ABC transporters and regulatory genes are also frequent, thus indicating that a possible role for this new SAM hydrolase activity could be in regulating SAM levels by returning its chemical components to simple building blocks that might be reassembled back to SAM when conditions are warranted.
Experimental Section
Chemicals
SAM-p-toluenesulfonate, 5′-chloro-5′-deoxyadenosine (5′-ClDA), L-methionine, S-(5′-adenosyl)-L-homocysteine (SAH), 5′-deoxy-5′-(methylthio)adenosine (MTA), adenosine-5′-triphosphate (ATP) and adenosine-5′-monophosphate (AMP) were purchased from Sigma, 5′-IDA and adenosine were from Acros Organics, and 5′-FDA and 5′-BrDA were a gift from D. O’Hagan, University of St. Andrews. All other chemicals were of analytical grade.
SaDUF62 purification
The SaDUF62 gene from S. arenicola CNS-205 (protein accession number YP_001536 256, genome accession number NC_009 953) was amplified by PCR from total genomic DNA using the forward primer 5′-CGT GGT TCC CAT GGC ATG GCG TCG ACG CCC TGG ATC-3′ (NcoI site underlined) and the reverse primer 5′-GCT CGA ATT CAA GCT TTC AGC CGG CGG TGA CGC GGA-3′ (HindIII site underlined), designed for ligation into pHIS8,[23] yielding plasmid pJH1. Expression in Escherichia coli BL21(DE3), purification and removal of the amino-terminal His8-tag were carried out as previously described for recombinant chlorinase SalL.[5] The yield of soluble protein was approximately 1 mg L−1, and it was stored in phosphate buffer (50 mM, pH 7.9) and 50 % (v/v) glycerol at −20 °C.
SaDUF62 activity assay
SaDUF62 and SalL reactions were investigated by incubating the enzyme (10 to 2000 nM) with 1) SAM (0.5–500 μM), 2) SAM and KX (X = F, Cl, Br, I, 10–400 mM), or 3) L-met (0.5–100 mM) and 5′-XDA (X=Cl, I, 0.3–100 μM) in phosphate buffer (50 mM, pH 7.9) at 37°C. After the reaction mixture had been boiled for 2 min and centrifuged for 30 min to eliminate precipitated protein, the clear supernatant (100 μL) was analyzed by HPLC, as described.[24] The identity of the products was confirmed by comparing the retention times with authentic standards and by LC/(+)ESI MS. The detection limit of the HPLC assay was >1 pmol. The following compounds were also tested as substrates by using SaDUF62 (2 μM), but no activity was detected: 1) L-met (50–100 mM) and either ATP, AMP or adenosine (0.2–0.5 mM), 2) SAH (0.4 mM), 3) MTA (0.4 mM).
Protein in silico analysis
Homology searches were carried out with BLAST,[25] homology modeling was generated with ESy-Pred3D,[17] protein structures were visualized with PyMol,[26] and phylogenetic trees were created from sequence alignments by using ClustalX[27] and visualized with TreeView.[28]
Acknowledgments
We are grateful to Dr. F. Pojer for valuable discussions that contributed to this manuscript and for help with gel filtration analysis. We thank Prof. D. O’Hagan (University of St. Andrews) for kindly providing 5′-FDA and 5′-BrDA standards and Prof. W. Fenical and Dr. P. Jensen (UCSD) for providing S. arenicola. A.S.E. is a Tularik postdoctoral fellow of the Life Sciences Research Foundation. J.P.N. is an investigator of the Howard Hughes Medical Institute. This work was further supported by the NIH (CA127622 to B.S.M.) and the National Science Foundation (MCB-023602 to J.P.N.).
Footnotes
Note Added in Proof
During revision of this manuscript, a similar study by Deng et al.[29] describing the in vitro characterization of PhDUF62 was published. The authors show that while PhDUF62 is unable to catalyze fluorination or chlorination reactions, it does convert SAM to adenosine in vitro. Assay of the enzyme in H218O results in [5′-18O]adenosine without labeling of the protein; this indicates that a mechanism of the type depicted in Figure 2 is operating rather than a second possibility through an enzyme-bound intermediate. The enzyme is alternatively named S-adenosyl-L-methionine:hydroxide adenosyl-transferase.
References
- 1.Fontecave M, Atta M, Mulliez E. Trends Biochem Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Kozbial PZ, Mushegian AR. BMC Struct Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loenen WAM. Biochem Soc Trans. 2006;34:330–333. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- 4.Dong C, Huang F, Deng H, Schaffrath C, Spencer JB, O’Hagan D, Naismith JH. Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 5.Eustáquio AS, Pojer F, Noel JP, Moore BS. Nat Chem Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schubert HL, Blumenthal RM, Cheng X. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs RT, Grundy FJ, Henkin TM. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 8.Epshtein V, Mironov AS, Nudler E. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba Y, Sakurai R, Yoshino M, Ominato K, Ishikawa M, Onouchi H, Naito S. Proc Natl Acad Sci USA. 2003;100:10225–10230. doi: 10.1073/pnas.1831512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DJ, Huh JH, Yang YY, Kang CM, Lee IH, Hyun CG, Hong SK, Suh JW. J Bacteriol. 2003;185:592–600. doi: 10.1128/JB.185.2.592-600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto S, Lezhava A, Hosaka T, Okamoto-Hosoya Y, Ochi K. J Bacteriol. 2003;185:601–609. doi: 10.1128/JB.185.2.601-609.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin SK, Xu D, Kwon HJ, Suh JW. FEMS Microbiol Lett. 2006;259:53–59. doi: 10.1111/j.1574-6968.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- 13.Tsukamoto S, Yokosawa H. Curr Med Chem. 2006;13:745–754. doi: 10.2174/092986706776055571. [DOI] [PubMed] [Google Scholar]
- 14.Rao KN, Burley SK, Swaminathan S. Proteins: Struct Funct Bioinf. 2008;70:572–577. doi: 10.1002/prot.21646. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Robinson DA, McEwan AR, O’Hagan D, Naismith JH. J Am Chem Soc. 2007;129:14597–14604. doi: 10.1021/ja0731569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng H, Cobb SL, McEwan AR, McGlinchey RP, Naismith JH, O’Hagan D, Robinson DA, Spencer JB. Angew Chem. 2006;118:773–776. doi: 10.1002/anie.200503582. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:759–762. doi: 10.1002/anie.200503582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert C, Leonard N, De B, Depiereux XE. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 18.Udwary DW, Zeigler L, Asolkar R, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 20.Studier FW, Movva NR. J Virol. 1976;19:136–145. doi: 10.1128/jvi.19.1.136-145.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spoerel N, Herrlich P. Eur J Biochem. 1979;95:227–233. doi: 10.1111/j.1432-1033.1979.tb12957.x. [DOI] [PubMed] [Google Scholar]
- 22.Spoerel N, Herrlich P, Bickle TA. Nature. 1979;278:30–34. doi: 10.1038/278030a0. [DOI] [PubMed] [Google Scholar]
- 23.Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 24.Schaffrath C, Deng H, O’Hagan D. FEBS Lett. 2003;547:111–114. doi: 10.1016/s0014-5793(03)00688-4. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMol Molecular Graphics System. DeLano Scientific; Palo Alto, CA: 2002. http://www.pymol.org. [Google Scholar]
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Page RD. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 29.Deng H, Botting CH, Hamilton JT, Russell RJ, O’Hagan D. Angew Chem. 2008;120:5437–5441. doi: 10.1002/anie.200800794. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:5357–5361. doi: 10.1002/anie.200800794. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Robertson AD, Jensen JH. Proteins: Struct, Funct, Bioinf. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]