Abstract
Olfactomedin 1 (Olfm1) is a secreted glycoprotein belonging to a family of olfactomedin domain-containing proteins. It is involved in the regulation of neural crest production in chicken and promotes neuronal differentiation in Xenopus. Here, we investigate the functions of Olfm1 in zebrafish eye development. Overexpression of full-length Olfm1, and especially its BMY form lacking the olfactomedin domain, increased the thickness of the optic nerve and produced a more extended projection field in the optic tectum compared with control embryos. In contrast, injection of olfm1–morpholino oligonucleotide (Olfm1–MO) reduced the eye size, inhibited optic nerve extension, and increased the number of apoptotic cells in the retinal ganglion cell and inner nuclear layers. Overexpression of full-length Olfm1 increased the lateral separation of the expression domains of eye-field markers, rx3 and six3. The Olfm1–MO had the opposite effect. These data suggest that zebrafish Olfm1 may play roles in the early eye determination, differentiation, optic nerve extension, and branching of the retinal ganglion cell axon terminals, with the N-terminal region of Olfm1 being critical for these effects. Injection of RNA encoding WIF-1, a secreted inhibitor of Wnt signaling, caused changes in the expression pattern of rx3 similar to those observed after Olfm1–MO injection. Simultaneous overexpression of WIF-1 and Olfm1 abolished the WIF-1 effect. Physical interaction of WIF-1 and Olfm1 was demonstrated by coimmunoprecipitation experiments. We concluded that Olfm1 serves as a modulator of Wnt signaling.
Keywords: eye development, axonal extension, neuronal differentiation, Wnt signaling, WIF-1, morpholino oligonucleotides
Introduction
Olfactomedin 1 (Olfm1), also known as noelin in chicken and Xenopus, and as pancortin in mouse and rat, is a secreted glycoprotein that was originally identified in the bullfrog olfactory neuroepithelium (Snyder et al., 1991; Nagano et al., 1998; Barembaum et al., 2000). A conserved domain in the C-terminal part of Olfm1 was named the olfactomedin domain and is found in a family of proteins consisting of at least 13 members in mammals (Zeng et al., 2005). Some family members, such as latrophilins and gliomedin, are membrane-bound proteins containing the olfactomedin domain in the extracellular N-terminal regions, whereas the intracellular C-terminal domains of these proteins are essential for the transduction of extracellular signals to intracellular signaling pathways (Volynski et al., 2004; Eshed et al., 2005). Other family members, similar to Olfm1, are secreted glycoproteins whose functions are mostly unknown. Mutations in one of the secreted olfactomedin-domain containing proteins, myocilin, contribute to open-angle glaucoma (Stone et al., 1997), an eye disease characterized by the death of retinal ganglion cells (RGCs), degeneration of axons in the optic nerve, and a specific deformation of the optic nerve head. Most glaucoma-causing mutations in myocilin are located in the olfactomedin domain and lead to the accumulation of the mutated protein inside the cells with subsequent apoptotic cell death (Joe et al., 2003; Liu and Vollrath, 2004).
In Xenopus, olfm1 mRNA was detected mainly in postmitotic neurogenic tissues in the developing central and peripheral nervous systems, first appearing after neural tube closure (Moreno and Bronner-Fraser, 2001, 2002, 2005). In chicken, Olfm1 is involved in generation of the neural crest cells (Barembaum et al., 2000). Four structurally different mRNAs, named AMY, BMY, AMZ, and BMZ, are produced from the Olfm1 gene (Danielson et al., 1994). They share a common central region (M) and have two different 5′-regions (A or B) transcribed from distinct promoters, and two different 3′-regions (Y and Z) produced by alternative splicing (Danielson et al., 1994). The AMY and BMY forms encode a shorter form of Olfm1 lacking the olfactomedin domain that is encoded by two exons found in the AMZ and BMZ forms.
The zebrafish genome contains two olfm1 genes, olfm1a and olfm1b, located on chromosomes 5 and 21, respectively. At least four transcripts are produced from each olfm1 gene (Nakaya and Tomarev, 2007). The distributions of different transcripts of olfm1 genes during embryonic development [16–55 h postfertilization (hpf)] have been reported previously in detail (Nakaya and Tomarev, 2007).
In this study, we investigate possible roles of the olfm1 genes in zebrafish eye development using gene silencing by morpholino oligonucleotides (MOs) and overexpression of different Olfm1 isoforms. We demonstrate that suppression of Olfm1 expression leads to multiple developmental defects, including reduction of eye size and eye pigmentation, retinal degeneration, and inhibition of the optic nerve growth. In contrast, the overexpression of Olfm1, and especially Olfm1 variants lacking the olfactomedin domain, produces a thicker optic nerve with an extended terminal area in the optic tectum. We show that Olfm1 physically interacts with WIF-1, a secreted inhibitor of Wnt signaling. Olfm1 and WIF-1 have antagonistic effects, and their interaction may fine tune Wnt activity.
Materials and Methods
Husbandry of fishes.
Wild-type zebrafish were maintained as described by Westerfield (2000). Embryos were produced by natural matings. For some experiments, pigmentation of embryos was reduced by adding 0.2 mm 1-phenyl-2-thiourea to water. All experiments using animals were approved by the National Eye Institute Animal Use and Care Committee.
Frozen sections and immunofluorescence.
Zebrafish embryos were dechorionated and fixed with 4% paraformaldehyde in 0.1 m PBS, pH 7.4, at 4°C overnight. For frozen sectioning, fixed embryos were equilibrated in 30% sucrose and embedded into OCT Compound (Electron Microscopy Sciences). Frozen sections (5 μm) were produced using a cryostat (CM3050; Leica). Frozen sections were blocked in 5% normal goat serum and then incubated with anti-Olfm1 antibody (1:500 dilution), anti-Hu-C/D (1:400 dilution; Abcam), Zn-5 (1:500; Oregon Monoclonal Bank), anti-PKCβ1 (1:300 dilution; Santa Cruz Biotechnology), or Zpr-1 (1:200 dilution; Zebrafish International Resource Center). Slides were washed in PBS and then incubated with Alexa488- or Alexa595-conjugated goat anti-mouse IgG (1:400 dilution; Invitrogen) and with 4′,6′-diamidino-2-phenylindole (DAPI) for nuclear staining. Images were collected using a confocal laser microscope (TCS SP2; Leica) or an Axoplan2 microscope (Carl Zeiss).
In situ RNA hybridization.
Zebrafish olfm1 TR2 and TR4-specific probes were produced as described previously (Nakaya and Tomarev, 2007). TrkC1, six3, and rx3 cDNAs were purchased from Open Biosystems. Digoxigenin (DIG)-labeled cRNA probes were synthesized by in vitro transcription of the corresponding linearized plasmids using SP6, T7, or T3 polymerases and DIG–RNA labeling mixture (Roche). Whole-mount in situ hybridization was performed as described previously (Toyama et al., 1995). After visualization of in situ hybridization signals, some embryos were embedded in OCT, and 10 μm frozen sections were prepared. The mounted sections were observed under an Axoplan2 microscope.
Olfm1 antibody production.
Mouse anti-Olfm1 antibody was produced against the KFKQVEESHKQHLARQ peptide using the Custom Antibody Production Service of the University of Virginia. Specificity of the antibody was validated by Western blot experiments with two types of cell lysates. First, lysates of COS-7 cells and COS-7 cells transfected with mouse Olfm1, mouse optimedin, and mouse myocilin were used. Second, eye extracts were prepared from wild-type mice and null mice that lacked exons 2 and 3 encoding the M part of Olfm1 (Cheng et al., 2007).
Western blot.
Whole embryos were homogenized in a lysis buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 1% Triton X-100, 5 mm NaF, 0.5 mm sodium orthovanadate, 10% glycerol, 1 mm dithiothreitol, 1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin) for 20 min on ice. After centrifugation, the soluble fraction was collected and protein concentration was determined using the BCA assay kit (Pierce). The extracted proteins (15 μg) were separated on an 12% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane (Invitrogen). A membrane was incubated with anti-Olfm1 antibody (1:10 dilution) followed by anti-mouse IgG antibody conjugated to horseradish peroxidase (HRP) (1:5000 dilution; GE Healthcare). The HRP signals were detected using a chemiluminescence detection kit (SuperSignal Femto Dura Extended Duration Substrate; Pierce) and a BioMax MS film (Eastman Kodak).
Whole-mount immunofluorescence.
Anti-acetylated tubulin antibody was purchased from Sigma-Aldrich. Alexa 488-conjugated secondary antibody was purchased from Invitrogen. Zebrafish embryos were dechorionated and fixed with 4% paraformaldehydes in PBS at 4°C overnight. Embryos were incubated in blocking buffer (1% bovine serum albumin, 0.1% Triton X-100, 0.05% dimethylsulfoxide, and 2% goat serum in PBS) at 4°C for 1 h and then at 4°C overnight in blocking buffer containing the primary antibody. Embryos were washed three times with PBS-T (0.1% Triton X-100 in PBS) for 1 h and then incubated with the fluorescence-labeled secondary antibody at 4°C overnight. Embryos were washed three times with PBS-T for 1 h and mounted on a glass slide in 0.5% glycerol. Images were collected using a Carl Zeiss Axoplan2 fluorescent microscope equipped with a deconvolution software (Axovision; Carl Zeiss).
Terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling staining.
Zebrafish frozen sections (6 μm) were incubated with rabbit anti-ssDNA antibody (IBL USA) at 4°C overnight. After washing in PBS, sections were incubated with Alexa 488 anti-rabbit IgG/DAPI. Immunofluorescence images were collected with a Leica TCS SP2 confocal laser microscope.
Morpholino and RNA injections.
The MO antisense oligonucleotide to knockdown Olfm1 expression in embryos was designed and produced by Gene Tools. The sequences of oligonucleotides used in these experiments were as follows: control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; Olfm1–MO, 5′-AGCAAAGGCACCGACATCTCTGCTC-3′; missense MO, 5′-AGGAAACGCACCCACATGTCTCCTC-3′.
The MOs were dissolved in distilled water, mixed with 2× injection buffer (0.05% phenol red, 240 mm KCl, and 40 mm HEPES, pH 7.4), and injected into egg yolk of cell stage 1–4 embryos by a Pneumatic PicoPump (PV820; World Precision Instruments). For rescue experiments, the insert of cDNA clone CO250150 encoding the full-length AMZ form of zebrafish Olfm1b obtained from Open Biosystems was subcloned into the pCS2+ vector. Mouse Olfm1 deletion constructs, N1–293 and N1–152, were prepared by PCR amplifications from the full-length mouse Olfm1 cDNA clone BC026547 using the following primers: ataagcttgccgccaccatgtcggtgccgctgctgaa [forward (FW)], attctagaatgtcggtgccgctgctgaa [N1–293 reverse (RV)], and attctagaagcctacttgaactgcctggctagat (N1–152-RV). Zebrafish wif-1 was amplified by PCR from cDNA clone BC091959 (Open Biosystems) and cloned into the pCS2+ vector. Identity of all selected clones was confirmed by sequencing. RNA was synthesized using a mMESSAGE mMACHINE kit (Ambion) and different pCS2 constructs as templates. Synthesized RNAs (0.02–0.1 ng) were injected into one-cell stage embryos with/without the indicated MOs.
Quantitative reverse transcription-PCR.
Ten embryos from each developmental stage were dechorionated and homogenized in 500 μl of Trizol (Invitrogen) using a Polytron (PT-MR2100; Kinematica) at the maximum power for 1 min. Total RNA was isolated from the lysates following the protocol of the manufacturer. RNA was dissolved in distilled water at concentrations of 0.1–0.3 μg/μl. Quantitative (Q) PCR was performed using 0.5 μg of total RNA as described previously (Hemish et al., 2003). Briefly, RNA was converted to cDNA using Taqman Reverse Transcription Reagents (Applied Biosystems) and then dispensed into a 96-well PCR plate. Forward and reverse primers for a target gene and SYBR Green PCR Master Mix (Applied Biosystems) were added into each wells, and PCR reaction was performed and monitored using a 7900HT Real Time Thermocycler (Applied Biosystems). Elongation factor 1α (EF1α) was used for normalization after a confirmation that its expression was not dramatically changed during developmental stages under study. Primers used for the Q-PCR were as follows: ef1α (FW), 5′-CAGTGCTGGATTGCCACACT-3′; ef1α (RV), 5′-CCAGAACGACGGTCGATCTT-3′; TR2 (FW), 5′-CGATGATCACCAACTGGATGTC-3′; TR2 (RV), 5′-GCCACGCTTAATTTGGTGCTA-3′; TR4 (FW), 5′-GGTGCTCAGCACCATGGC-3′; TR4 (RV), 5′-GCGGTCAGTTTGGTGGTGTT-3′; Olfm1a Y form (FW), 5′-GCTGCAGAGACTGAAGAACAAGTT-3′; Olfm1a Y form (RV), 5′-AACTGAATGATGCGATCAGTTGAC-3′; Olfm1a Z form (FW), 5′-CCACAACAGGGTGTCTAATCTTGA-3′; Olfm1a Z form (RV), 5′-AGTCAGCTTCCCACAGGCTAAT-3′; Olfm1b Y form (FW), 5′-GAGAGCCAGAGGCCAGAAGAA-3′; Olfm1b Y form (RV), 5′-CGTGACATAAACAGCGTGACTGA-3′; Olfm1b Z form (FW), 5′-TGCAGAATCTGACTGCGAGTCT-3′; Olfm1b Z form (RV), 5′-GGAGTGCAGGTCATCATAGTCATAA-3′.
Luciferase assay.
Super8XTOPFlash vector (37.5 pg) and renilla luciferase plasmid (phRL-SV40, 2.5 pg; Promega) were injected to one-cell stage embryos together with or without enhanced green fluorescent protein (EGFP), wif-1, and/or olfm1 RNAs (25 pg). The luciferase activities in 24 hpf embryos were determined using a Luciferase Assay kit (Promega) as in the protocol of the manufacturer. Briefly, 10 embryos were homogenized in the lysis buffer (100 μl), and the supernatant was collected after spinning at 15,000 × g for 15 min. The firefly and renilla luciferase activities in the supernatant were sequentially determined by adding the Luciferase Assay Substrate and the STOP&GROW solutions and using a multiplate luminometer (VICTOR-light; PerkinElmer Life and Analytical Sciences).
Optic nerve analysis.
DiI injection was performed using the previously described method (Yoda et al., 2004). Briefly, embryos were fixed with 4% paraformaldehyde and placed in 1.2% low melting agarose. DiI (0.5 nl, 75 mg/ml) was injected into two positions diagonal to each other (nasodorsal and temporovental) in the left eye. The embryos were incubated at 28°C overnight, and the projection of the optic nerve stained with DiI was observed and photographed using a dissection fluorescence microscope (Stemi SV11; Carl Zeiss). The diameter of the optic nerves was measured and calculated using the NIH Image software.
Coimmunoprecipitation.
HEK-293 cells were grown in 60 mm plates and transfected with Flag-tagged Olfm1 (Torrado et al., 2002) and WIF-1 tagged with the human heavy chain IgG (Hsieh et al., 1999) cDNA constructs using Lipofectamine 2000 (Invitrogen). (WIF-1 constructs were kindly provided by Dr. Jen-Chih Hsieh, State University of New York at Stony Brook, Stony Brook, NY). At 2 d after transfection, conditioned medium was collected and centrifuged at 15,000 × g for 15 min at 4°C. To reduce background, conditioned medium was incubated with proteinG agarose (Roche) beads for 1 h at 4°C in a rotary shaker. The beads were removed by centrifugation, and the monoclonal Flag antibody beads were added to precleared conditioned medium for 2 h at 4°C in a rotary shaker. The beads were precipitated and washed three times with lysis buffer. Bound proteins were eluted by a 3XFLAG peptide (Sigma) and analyzed by Western blotting as described above. The peroxidase-conjugated antibodies against human IgG (1:1000 dilution) were used for detection of WIF-1 protein.
Statistical analysis.
Data were analyzed using the unpaired Student's t test preceded by F test for variances.
Results
Overexpression of Olfm1 increases thickness of the retina and the optic nerve
Olfm1 is a conserved protein showing a 85% identity of amino acid sequence between zebrafish and mouse (Nakaya and Tomarev, 2007). To test effects of overexpression of different forms of Olfm1, mRNAs encoding the full-length AMZ form of zebrafish Olfm1 (zOlfm1b) and truncated forms of mouse Olfm1 (Fig. 1A) were injected into zebrafish embryos. N1–293 mRNA encodes the BMZ form of mouse Olfm1 without olfactomedin domain, whereas N1–152 mRNA encoded the BMY form of mouse Olfm1. The nomenclature of transcripts of zebrafish olfm1 genes is shown in Figure 1B. Injection of RNAs encoding zOlfm1 or N1–293 into embryos induced synthesis of a protein migrating as a single band with apparent molecular weight of 70 or 50 kDa, respectively (Fig. 1C, left). These apparent molecular weights were higher than the predicted molecular weights of 52.5 and 33.4 kDa, respectively, indicating carbohydrate modification of proteins encoded by these constructs. The levels of endogenous Olfm1 are low at this stage of development, and Olfm1 bands were masked by the background bands. RNA encoding the N1–152 form produced two protein bands on the gel, with apparent molecular weights of 24.6 and 15.9 kDa, most probably corresponding to modified and unmodified proteins, respectively. Because N1–152 is contained within the N1–293 protein, this indicates that the N1–152 protein is not modified as efficiently as longer proteins containing full or partial Z region. When expression constructs were transfected into HEK-293 cells, proteins with apparent molecular weights that were higher than calculated molecular weights were also produced. Treatment of cell lysates with a mixture of glycosidases [PNGaseF, β-N-acetylglucosaminidase, β-1–4-galactosidase, o-glycosidase and α(2–3,6,8,9)neuraminidase] reduced the apparent molecular weights of different Olfm1 forms to predicted molecular weights, confirming that modification of Olfm1 is mainly attributable to glycosylation (data not shown).
Figure 1.
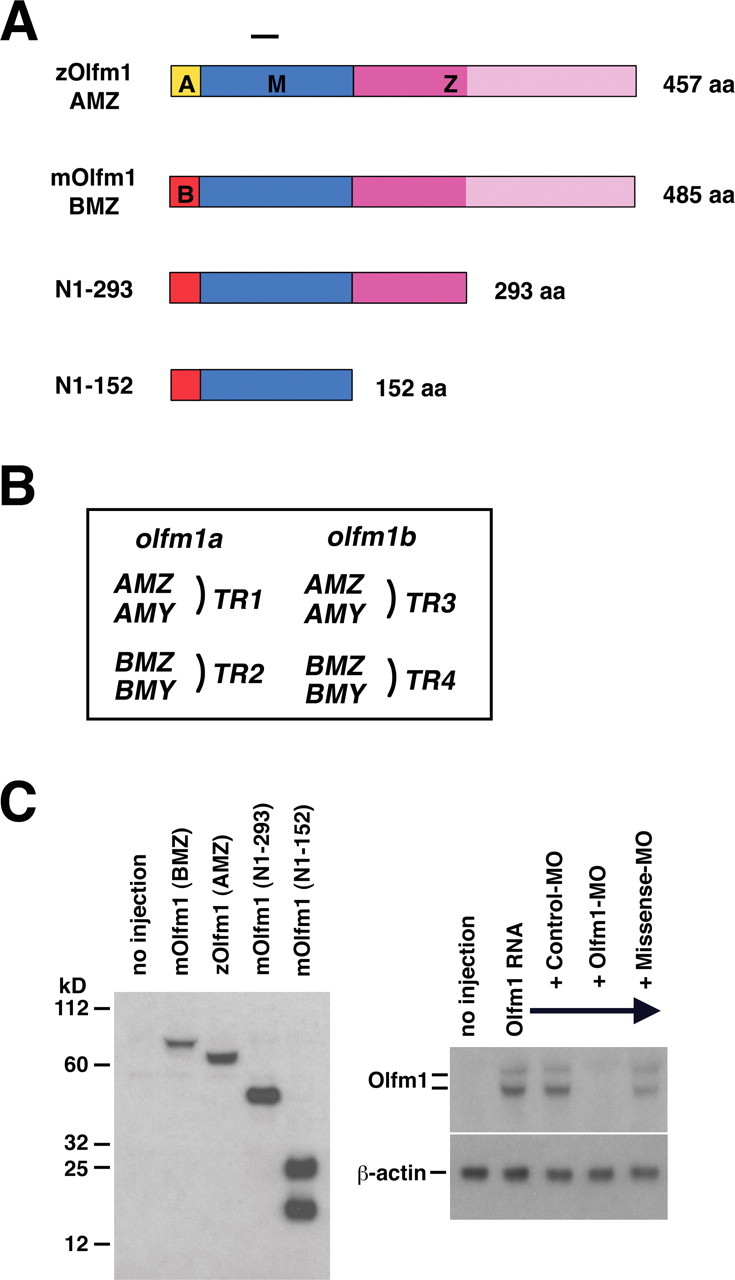
A, Schematic diagram of the Olfm1 constructs used in this study. A, M, and Z denote different protein regions. The A or B regions (shown in yellow or red, respectively) contain a signal peptide, the M region (shown in blue) is common between different Olfm1 forms, and the Z region (shown in dark and light pink) contains the olfactomedin domain (shown in light pink). B, The nomenclature of transcripts of zebrafish olfm1a and olfm1b genes. C, Expression of different Olfm1 forms 24 h after injection of 0.1 ng of corresponding RNAs at cell stage 1. Total soluble proteins were analyzed by Western blotting using the monoclonal antibody against the peptide in the M domain (left). The right demonstrates inhibition of Olfm1 by Olfm1–MO coinjection. Mouse Olfm1 RNA preceded by the zebrafish sequence sensitive to Olfm1–MO was synthesized and injected into cell soma of cell stage 1 embryos with or without an injection of the Olfm1–MO, control MO, and missense MO into the egg yolk. The Olfm1–MO completely inhibited the Olfm1 protein expression (appeared as upper glycosylated and lower unglycosylated bands) from the injected RNA in the bud stage embryos.
Injection of 20 pg of RNAs encoding different forms of zebrafish or mouse Olfm1 did not induce obvious morphological changes. The general structure of the eye or retina did not change significantly after zOlfm1 RNA injection in 72 hpf embryos (data not shown). However, the full-length zOlfm1, but not truncated mouse forms, produced a statistically significant increase in the thickness of the inner nuclear layer and the total retina compared with EGFP injected embryos (69.12 ± 2.37 vs 60.07 ± 0.56 μm, p < 0.05 and 150.04 ± 3.73 vs 132.27 ± 3.30 μm, p < 0.01, respectively).
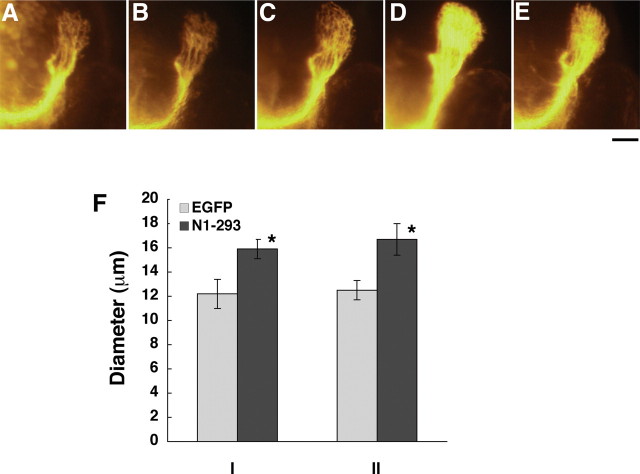
To analyze changes in the optic nerve after overexpression of Olfm1, DiI was injected into the eyes of 72 hpf embryos, and the thickness and the arborization of the optic nerve was analyzed 16 h later. Injection of zOlfm1 and especially truncated N1–152 and N1–293 constructs increased the thickness of the optic nerve and produced a broader projection field in the optic tectum compared with control embryos (Fig. 2A–E). Measurements of the optic nerve at two positions (exit from the optic globe and optic chiasm level) demonstrated its increased thickness after injection of the N1–293 truncated construct (Fig. 2F). The extended projection field of the optic nerve terminals in the optic tectum could be attributed to the acceleration of optic nerve growth, an increase in the branch number of the optic nerve axons, or facilitated arborization. The fact that truncated forms of Olfm1 are more effective in the optic nerve implies that the N-terminal part of Olfm1 has a role to regulate the optic nerve extension different from the eye size regulations.
Figure 2.
A–E, Facilitation of the optic nerve extension by injections of RNAs encoding different forms of Olfm1. A total of 0.1 ng of RNAs encoding the full-length zOlfm1 (C), N1–293 (D), N1–152 olfm1 (E), EGFP RNAs (B), or buffer control (A) were injected at one-cell stage. Embryos were fixed with 4% paraformaldehyde at 72 hpf. DiI was injected into one side of the eye, and embryos were incubated overnight at 28°C. The fluorescent images of the frontal view of head were collected to visualize the terminals of the optic nerve at the optic tectum. Scale bar, 0.05 mm. F, The comparison of optic nerve diameters after injection of RNAs encoding EGFP or N1–293 at the level of exit from globe (I) and optic chiasm (II) (n = 5). *p < 0.05.
Knockdown of zebrafish Olfm1 synthesis by morpholino oligonucleotide injections
To examine effects of inhibition of Olfm1 synthesis during development, we used antisense MO injection. In a previous study, we demonstrated that BMZ and BMY transcripts of the olfm1a and olfm1b genes (TR2 and TR4 forms, respectively) were detected by in situ hybridization starting from 16 hpf, whereas AMZ and AMY transcripts of both genes (TR1 and TR3 forms) were detected later (Nakaya and Tomarev, 2007). We used a Q-PCR analysis to identify olfm1 variants that were the most abundant at earlier developmental stages. Expression of the TR2 forms (the olfm1a gene) was detected at the tail-bud stage of development (12 hpf), whereas the appearance of TR4 (the olfm1b gene) was delayed by ∼4 h until the 5-somites stage (16 hpf) (Fig. 3A). Relative amounts of the long (AMZ and BMZ) and short (AMY and BMY) forms of olfm1a and olfm1b mRNAs were also estimated by the Q-PCR using forward primers and reverse primers located in exon 4′ or 4 (Fig. 3B,C). These experiments demonstrated that the long forms were preferentially transcribed from the olfm1a gene in early development (16 hpf). Conversely, both longer and shorter forms were transcribed from the olfm1b gene from 16 hpf and onward. In summary, the BMZ transcript of olfm1a predominate at early developmental stages. The earliest expression of olfm1a was detected in the ventral region of the future head at the tail-bud stage (Fig. 3D,E).
Figure 3.
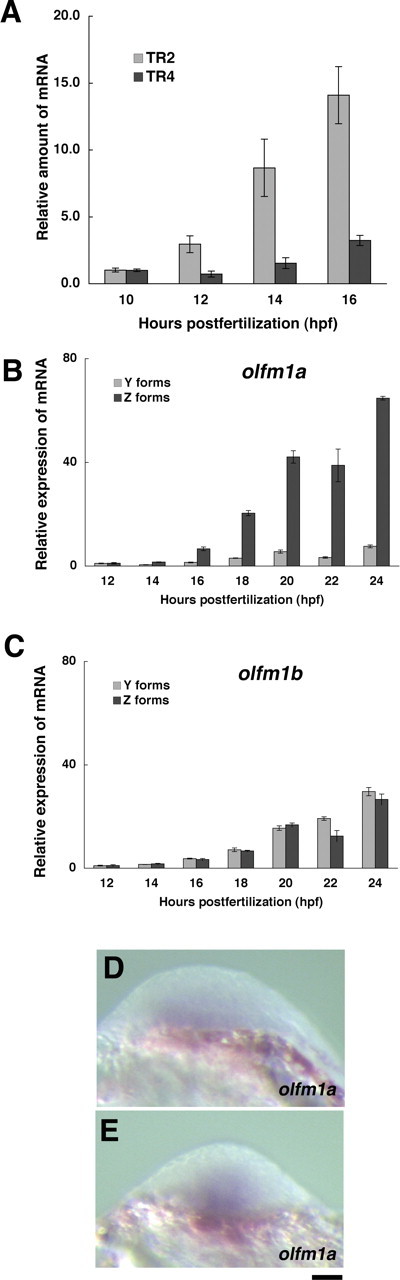
Expression of olfm1 mRNAs in the developing zebrafish embryos. A, Expression kinetics of olfm1a (the TR2 form corresponding to BMZ and BMY transcripts) and olfm1b mRNAs (the TR4 form corresponding to BMZ and BMY transcripts) in the course of early development. Total RNA was isolated from 10 embryos for each time point. Primers specific for different groups of transcripts were used for Q-PCR analysis. The amplified DNA fragments produced single bands as judged by agarose gel electrophoresis. Values on the y-axis represent the amount of mRNA for each time point shown relative to that of 10 hpf embryos. The average amounts of amplification cycles were 28.6 and 32.3 for TR2 and TR4, respectively, for 16 hpf embryos. Normalization was performed relative to the EF1a gene expression. B, C, Expression kinetics of shorter (Y) and longer (Z) forms of olfm1 mRNAs in the developing zebrafish embryos. Values on the y-axis represent the amount of mRNA for each time point shown relative to that of 12 hpf embryos. The average amounts of amplification cycles were 30.8, 28.6, 32.0, 32.3, 29.3, 27.5, 29.8, and 30.6 for TR1–TR4, olfm1a Y and Z forms, and olfm1b Y and Z forms, respectively, for 16 hpf embryos. D, E, Expression pattern of olfm1a mRNA in the head of a tail-bud stage embryo. Antisense probe for whole-mount in situ hybridization recognized olfm1a TR2 transcripts. Scale bar, 0.05 mm.
To see the effect of simultaneous suppression of multiple Olfm1 isoforms, we injected Olfm1–MO, which was complementary to a stretch of identical nucleotides in the region encoding the initiator methionine in TR2 and TR4 transcripts. Efficiency of inhibition was tested in control experiments by injection of 5 ng of this MO together with mouse Olfm1 RNA containing the zebrafish sequence complimentary to Olfm1–MO. The Olfm1–MO completely inhibited detectable Olfm1 protein expression in the tail-bud stage embryos. A control MO and a mismatch MO differing from Olfm1–MO by five nucleotide replacements did not inhibit Olfm1 (Fig. 1C, right). Analysis of morphological changes of the developing embryos demonstrated that 1.25–5 ng of control MO did not produce pronounced morphological defects, whereas increasing concentrations of Olfm1–MO produced dose-dependent, damaging effects. When 5 ng of Olfm1–MO was injected, the first detectable morphological changes were detected at 20 hpf. These included flattening of the head and the delay of eye development (data not shown). At 24 hpf, morphological defects included some cloudiness of the brain, indicating its degeneration, defective eye development, distorted body axis, the heart expansion, the rough surface of the skin, and cell dissociations from the dorsal surface of the body (data not shown). At later developmental stages (later than 35 hpf), Olfm1–MO-injected embryos demonstrated reduced pigmentation of the eye and the body, smaller eyes, and shorter body length compared with embryos injected with control MO (Figs. 4A–D, 5A,B). Analysis of sections through the cranial part of 72 hpf MO-injected embryos revealed a hypoplastic tectum with strongly damaged marginal zone and the loss of mandibular and hyosymplectic cartilages (Fig. 4C,D).
Figure 4.
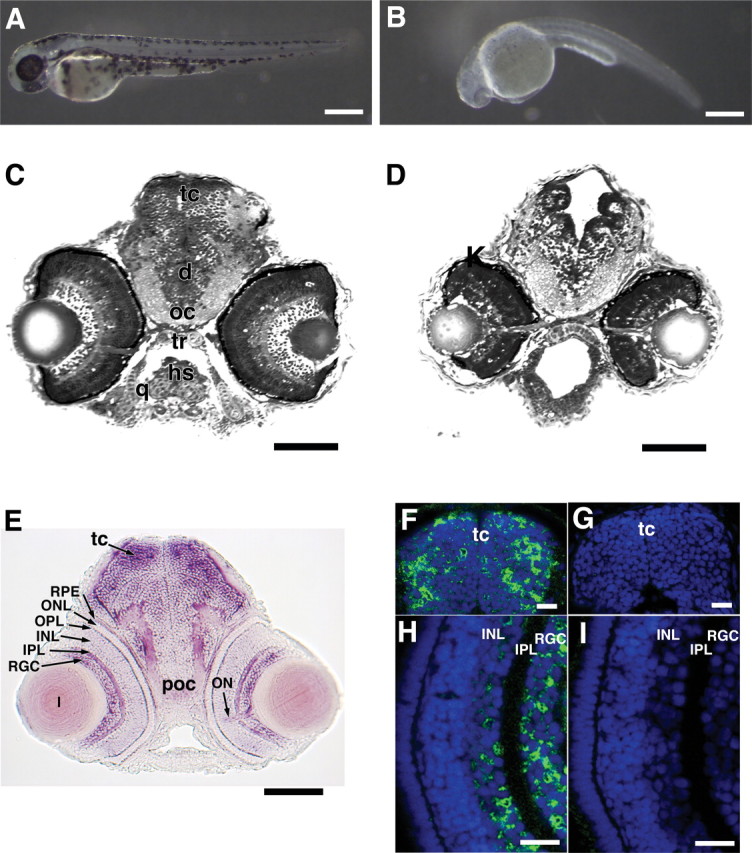
Effect of Olfm1 downregulation on the development of zebrafish embryos. A total of 5 ng of Olfm1–MO, which inhibits translations of TR2 and TR4, or control MO were injected into the egg yolk of one- to four-cell stage embryos. Control (A) and Olfm1–MO-treated (B) 3 days postfertilization embryos. Note dramatically reduced pigmentation as well as arched body axis after Olfm1–MO treatment. Scale bar, 0.25 mm. C and D show 0.5 μm plastic sections of control (C) and Olfm1–MO treated (D) 72 hpf embryos at the eye level. There is severe hypoplasia of the optic tectum, damage to the retina, and loss of cranial cartilages. Scale bar, 0.1 mm. E, A transverse section of 72 hpf embryos at the eye level after whole-mount in situ hybridization with antisense TR4-specific RNA. Signals were seen in brain, especially in the optic tectum, RGC layer, inner and outer plexiform layers, optic nerve, and retinal pigmented epithelium. Scale bar, 0.1 mm. F–I, Distribution of Olfm1 protein in the head of 72 hpf embryos. Head frozen sections were stained with anti-Olfm1 monoclonal antibody (F, H) or with no primary antibodies (G, I). Scale bar, 0.025 mm. d, Diencephalon; hs, hyosymplectic; oc, optic chiasm; q, quadrate; tc, optic tectum; tr, trabeculae; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer; Poc, post optic commissure; RPE, retinal pigmented epithelia; RGC, RGC layer.
Figure 5.
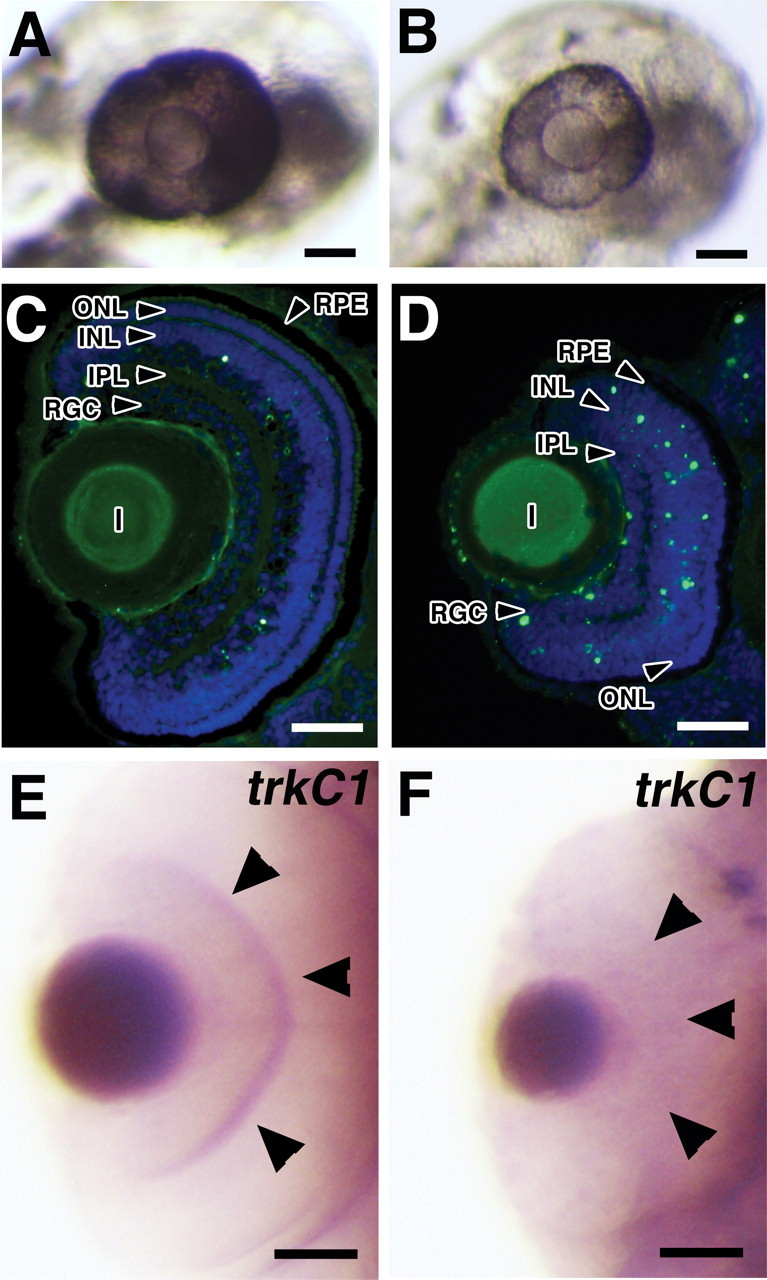
Effects of Olfm1–MO injections on the eyes of 72 hpf embryos. A, B, Embryos were injected with 5 ng of control (A) or Olfm1–MO (B). Olfm1–MO-injected embryos showed a reduction of eye size and pigmentation compared with embryos injected with control MO. C, D, TUNEL staining of eye sections from embryos injected with 5 ng of control MO (C) or Olfm1–MO (D). E, F, In 3 days postfertilization embryos, Olfm1–MO (F) inhibited trkc1 expression in both RGC and inner plexiform layers compared with control MO (E). Scale bar, 0.05 mm. IPL, Inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; RPE, retinal pigmented epithelium; RGC, retinal ganglion cells.
Sectioning of the embryos demonstrated that the size of the retina was dramatically reduced, and all retinal layers were affected. The RGC layer was disorganized and contained fewer cells than in control retina. The inner nuclear layer contained degenerating cells, which were not observed in the control (Fig. 5C,D). Inner plexiform and outer nuclear layers appeared to be thinner in Olfm1–MO-injected retinas. Pigmentation of the retinal pigment epithelium was reduced in Olfm1–MO-injected embryos. Terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) staining of the eye sections showed that there were more apoptotic nuclei in the RGC and inner nuclear cell layers of Olfm1–MO-treated 72 hpf embryos than control embryos (Fig. 5C,D), indicating that the apoptosis might play an essential role in the reduction of the retina size in Olfm1–MO-injected embryos. Among genes tested, Olfm1–MO injection dramatically reduced the expression of a gene encoding tyrosine receptor kinase C1 (trkC1) in the RGCs and the inner plexiform layer (Fig. 5E,F), without affecting its expression in other parts of the embryo (data not shown). Similarly, immunostaining of the eye sections with antibodies specific for different retinal cell subtypes demonstrated (Fig. 6) substantial reduction of staining in the RGC (Hu-C/D and Zn-5 positive) and inner nuclear (Hu-C/D and PKCβ1-positive) layers, whereas staining in the outer nuclear layer were less affected (Zpr-1 positive). Staining of neuronal fibers in the optic nerve fiber layer (Zn-5) and bipolar cells (PKCβ1) was reduced more dramatically than staining of the cell bodies with DAPI.
Figure 6.
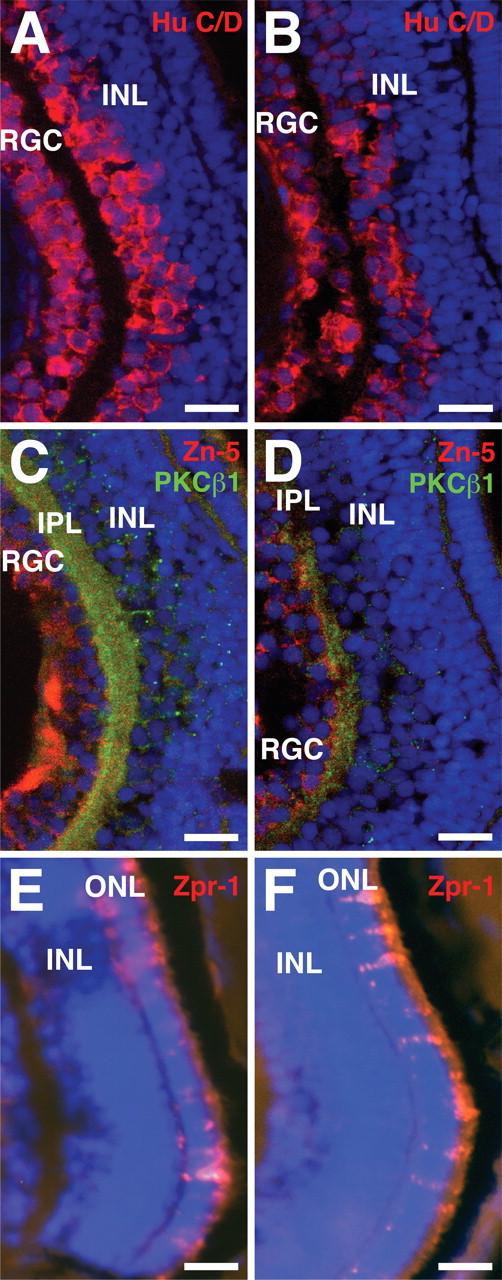
Effects of Olfm1–MO injection on the expression of retinal marker proteins. Control MO (A, C, E) or Olfm1–MO (B, D, F)-injected embryos were fixed at 72 hpf, and the frozen sections at the eye level were prepared. The sections were stained for Hu-C/D (A, B, red), PKCβ1 (C, D, green), Zn-5 (C, D, red), and Zpr-1 (E, F, red) together with DAPI (blue). Scale bar, 0.05 mm. IPL, Inner plexiform layer; INL, inner nuclear layer; ONL, outer nuclear layer; RGC, retinal ganglion cells.
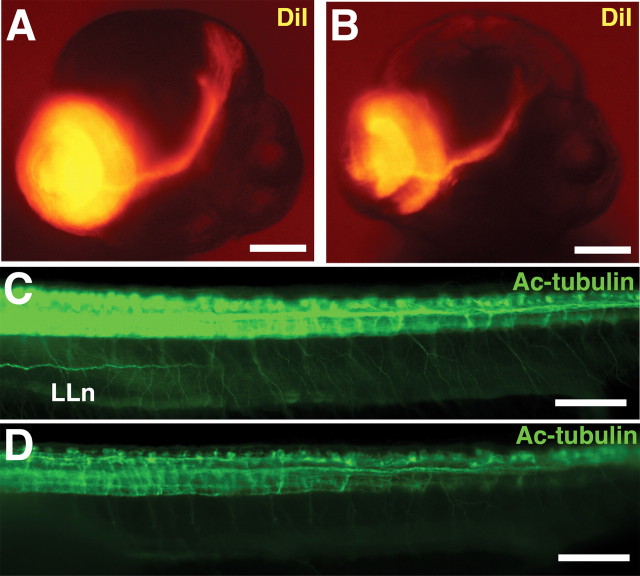
As a consequence of disorganization and reduction of cell number in the RGC layer, the optic nerve contained fewer nerve fibers, and these were not so densely packed (fasciculated) as in control eyes. These damaged fibers appeared less efficient in the transport of DiI to the optic tectum, the main target of the RGC axons, as was demonstrated by DiI injection (Fig. 7A,B). A reduction of the optic tectum size in Olfm1–MO-injected embryos also may contribute to the observed defects in arborization of the optic nerve axon to the tectum. Olfm1–MO injection also reduced the axonal network in the spinal cord and the lateral line nerve extension as demonstrated by the staining of neuronal axons with antibodies against acetylated tubulin (Fig. 7C,D). Sites of the most pronounced damage, the eye and brain, approximately corresponded to sites of Olfm1 expression at later developmental stages (72 hpf). olfm1a and olfm1b showed similar expression patterns, with the olfm1b producing the strongest signals (Fig. 4E) (data not shown) and expressed in the RGC layer, inner plexiform layer, and optic nerve head. Expression was also detected in midbrain, especially in the optic tectum. Areas enriched in neuronal fibers such as the post-optic commissure and optic nerve stalk were also positive, suggesting the olfm1 mRNAs are transported from cell bodies into these axons. Confocal immunofluorescence revealed that the Olfm1 protein was present in the same place where the mRNA localized. The Olfm1 antibody-specific immunofluorescent signals were detected in the entire forebrain, with especially strong signals in the upper and peripheral optic tectum (Fig. 4F). In the eye, the protein was localized mainly in the RGCs and inner nuclear layer cells (Fig. 4H). Olfm1 showed cytoplasmic and probably extracellular localization and was excluded from nuclei. Staining without the primary antibody did not show any positive signal (Fig. 4G,I).
Figure 7.
Olfm1 is required for axonal extensions. A, B, DiI-labeled optic nerve of 80 hpf control (A) and Olfm1–MO-injected embryos (B). A total of 1.25 ng of Olfm1–MO produced inhibition of axon growth to the optic tectum. C, D, Control (C) and Olfm1–MO-treated (D) 35 hpf embryos were stained with antibodies against acetylated tubulin. Note strong inhibition of neurons and their networks in Olfm1–MO-injected embryos. LLn, Lateral line nerve. Scale bar, 0.1 mm.
Effects of Olfm1–MO appeared to be specific because the inhibitory effect of MO on the eye size as well as defects to the body and brain were partially rescued by coinjection of RNA encoding the N1–152 Olfm1 construct (Table 1). We tried several factors to rescue Olfm1–MO effects on the eye size. Among injected RNA tested, runxb RNA but not runxa RNA produced a significant rescue effect (data not shown). Zebrafish runx genes encode runt-related transcription factors that are, similar to the olfm1 genes, strongly expressed in sensory neuronal tissues such as trigeminal ganglia and Rohon-Beard neurons. However, runx genes have not been reported to be expressed in the eye (Kataoka et al., 2000; Burns et al., 2002). Injection of runxb RNA changed the levels of olfm1 mRNAs: expression of the olfm1a gene at 24 hpf increased (154 ± 12% for TR2 and 312 ± 26% for TR1), whereas expression of the olfm1b gene did not change or decreased (118 ± 24% for TR3 and 74 ± 21% for TR4). This suggests that runxb acts upstream of olfm1 and may regulate the olfm1 gene expression. In conclusion, these data support the idea that Olfm1 has an important developmental role in the differentiation of neural retina and may affect the axon growth.
Table 1.
The eye size regulation by olfm1 and wif-1
| Dose (ng) | Size of eyes (mm) |
||
|---|---|---|---|
| 2.5 dpf | 4.5 dpf | ||
| 0.274 ± 0.002 | |||
| Olfm1–MO | 5 | 0.185 ± 0.003** | |
| EGFP + Olfm1–MO | 0.1 + 5 | 0.191 ± 0.003 | |
| Olfm1 + Olfm1–MO | 0.1 + 5 | 0.202 ± 0.010 | |
| N1–293 + Olfm1–MO | 0.1 + 5 | 0.199 ± 0.005 | |
| N1–152 + Olfm1–MO | 0.1 + 5 | 0.206 ± 0.005*,‡ | |
| 0.309 ± 0.002 | 0.361 ± 0.002 | ||
| WIF-1 | 0.01 | 0.316 ± 0.002* | 0.366 ± 0.002 |
| WIF-1 | 0.03 | 0.319 ± 0.003** | 0.363 ± 0.002 |
| WIF-1 | 0.05 | 0.323 ± 0.003** | 0.375 ± 0.002** |
| WIF-1 | 0.10 | 0.327 ± 0.003** | 0.373 ± 0.003** |
| 0.316 ± 0.002 | |||
| EGFP | 0.05 | 0.305 ± 0.003 | |
| WIF-1 | 0.05 | 0.327 ± 0.004* | |
| WIF-1 + EGFP | 0.05 + 0.05 | 0.328 ± 0.002 | |
| WIF-1 + Olfm1 | 0.05 + 0.05 | 0.312 ± 0.004*,† | |
| WIF-1 + N1–293 | 0.05 + 0.05 | 0.330 ± 0.003 | |
| WIF-1 + N1–152 | 0.05 + 0.05 | 0.331 ± 0.005 | |
*p < 0.05, **p < 0.01.
†Against WIF-1-injected embryos.
‡Against Olfm1–MO-injected embryos. dpf, Days postfertilization.
The effect of Olfm1 on the expression of selected genes involved in eye development
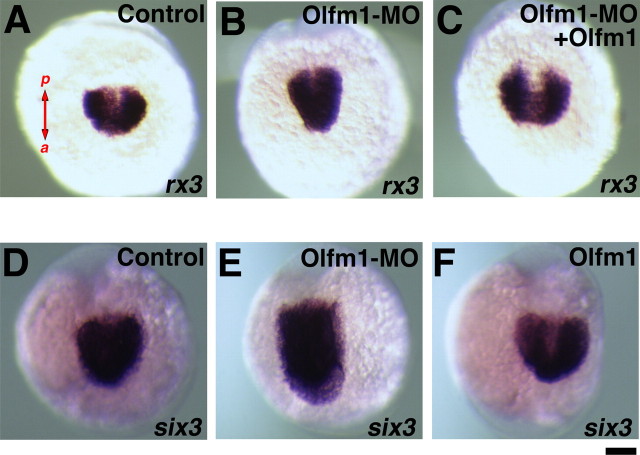
Because experimental changes in Olfm1 expression alter eye size, we checked whether the expression domains of early eye field markers in the anterior neural plate are also affected. The gene expression patterns of rx3, which is the earliest eye field marker, and six3, which acts downstream of rx3, were analyzed by in situ hybridization. Similar to published results (Chuang et al., 1999), the rx3 expression was detected in the lateral optic primordia and ventral medial diencephalon from 12 to 15 hpf embryos (Fig. 8A). Injection of Olfm1–MO reduced the lateral separation of the optic primordia expressing rx3 and increased the anteroposterior length of rx3 expression (Fig. 8B). Coinjection of zOlfm1 RNA together with Olfm1–MO restored the rx3 expression pattern and made it similar to that observed in control embryos (Fig. 8C). This indicates that the effect of Olfm1–MO injection on rx3 expression is specific and attributable to reduction of Olfm1 level. Expression of six3 was dramatically extended in the anteroposterior direction after Olfm1–MO injection (Fig. 8E). The olfm1 RNA injection did not affect anteroposterior extent of six3 expression but increased the lateral separation of optic primordia (Fig. 8F).
Figure 8.
Changes in the expression patterns of early eye markers after olfm1 RNA or Olfm1–MO injection. Embryos were injected with 0.1 ng of zOlfm1 RNA (F), 5 ng of Olfm1–MO, and 0.1 ng of EGFP RNA (B, E) or a mixture of zOlfm1 RNA and Olfm1–MO (C) and were analyzed for rx3 (A–C), and six3 (D–F) mRNA expression at 13 hpf. Control embryos (A, D) were injected with 0.1 ng of EGFP RNA. Shown are dorsal views of the future heads. a, Anterior; p, posterior. Scale bar, 0.1 mm.
Formation of the anteroposterior axis of the early forebrain is regulated by a number of genes, including wnt (Houart et al., 2002; Pézeron et al., 2006; Stigloher et al., 2006; Tendeng and Houart, 2006). It has been shown that injection of RNA encoding the Wnt inhibitor sFRP1 expands the area of six3 expression (Kim et al., 2007). Similarly, injection of RNA encoding another inhibitor of Wnt signaling WIF-1 leads to the anteroposterior expansion of rx3 expression comparable with the effect of Olfm1–MO, although it did not show any obvious effect on the distance between both future eye fields (Fig. 9A,B). The effect of the wif-1 RNA injection on the rx3 expression was reversed by coinjection with olfm1 RNA. The anteroposterior expression of rx3 returned to normal size in wif-1- and olfm1-injected embryos (Fig. 9C). wif-1 RNA injection not only expanded the expression of early eye markers but also enhanced the size of eyes at two developmental stages tested (Table 1). The increase in eye size by wif-1 was inhibited by a coinjection with olfm1 RNA but not with RNA encoding truncated forms of Olfm1. Together, these data indicate that WIF-1 and Olfm1 may affect the same signaling pathways and that these proteins may interact with identical targets and/or possibly with each other.
Figure 9.
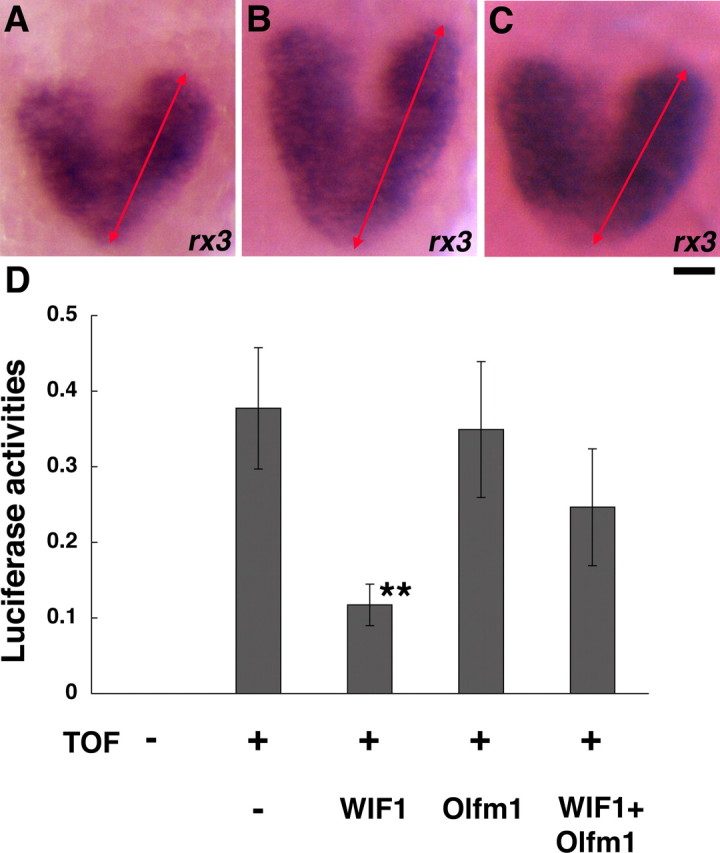
Effects of WIF-1 and Olfm1 on the rx3 expression and the TOPFlash reporter activity. A–C, Embryos were injected with 0.1 ng of EGFP RNA (A), wif-1 plus EGFP RNA (B), or wif-1 plus olfm1 RNAs (C), and rx3 expression was analyzed by in situ hybridization. The lengths of rx3 anteroposterior expression (represented by red arrows) were 197.8 ± 4.9 μm (n = 20), 263.7 ± 2.8 μm (**; n = 20), and 228.8 ± 4.53 μm (**,++; n = 12) for EGFP, wif-1 plus EGFP, and wif-1 plus Olfm1 RNA injections, respectively. **p < 0.0001 against EGFP; ++p < 0.00001 against wif-1 plus EGFP. Scale bar, 0.05 mm. D, Embryos were injected with EGFP (25 pg) plus wif-1 RNA (25 pg), EGFP RNA (25 pg) plus olfm1 RNA (25 pg), or wif-1 RNA (25 pg) plus olfm1 RNA (25 pg) together with Super8XTOPFlash (37.5 pg) and phRL-SV40 (2.5 pg). Embryos were homogenized in the lysis buffer 24 h after the injections, and the luciferase activities were measured. Data were normalized for renilla luciferase activity. **p < 0.01.
Interaction of Olfm1 and WIF-1
We used a TopFlash reporter system to check effects of WIF-1 and Olfm1 on canonical Wnt signaling in zebrafish. A reporter plasmid, which contains luciferase preceded by the multimerized consensus sequence for the lymphocyte enhancer factor/T-cell factor, was injected into embryos, and the luciferase activity was measured in the lysates 24 h after injections (Fig. 9D). The basal level of luciferase activity attributable to endogenous wnts was markedly inhibited by the wif-1 RNA injection, as expected. olfm1 RNA alone did not have an effect on the luciferase activity. However, coinjection of olfm1 RNA together with wif-1 RNA reduced the inhibitory effect of wif-1, suggesting that there may be an antagonistic interaction between WIF-1 and Olfm1.
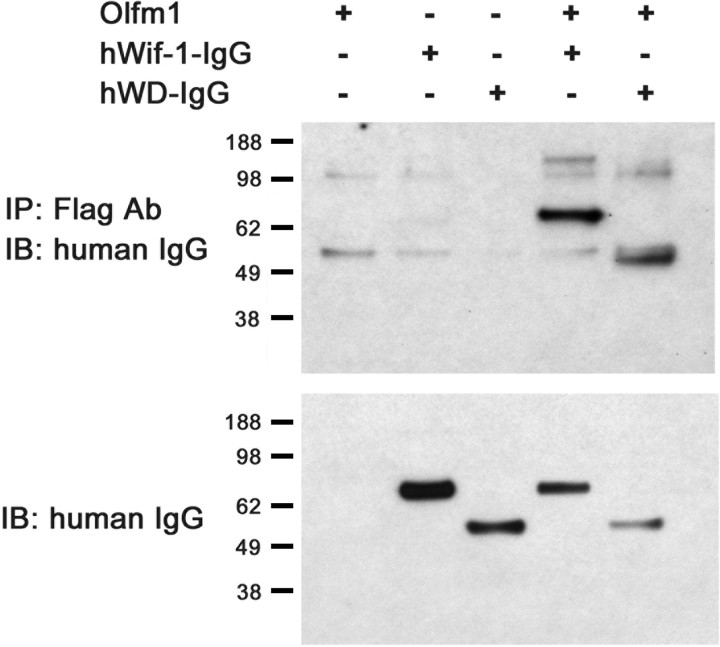
Possible physical interaction of Olfm1 and WIF-1 was tested in cell culture experiments. NIH3T3 cells were cotransfected with cDNAs encoding Olfm1 and WIF-1 or its WD domain fused to the IgG heavy chain. The WIF-1 WD domain is essential for interactions with Wnt proteins (Patthy, 2000). Because both Olfm1 and WIF-1 are secreted proteins, coimmunoprecipitation was performed from conditioned medium of transfected cells. The results demonstrated (Fig. 10) that Olfm1 interacts with WIF-1 and that the WD domain is sufficient for this interaction. We tested the truncated forms of Olfm1 for the interaction. Interestingly, the Flag-tagged truncated Olfm1 are much less secreted in cell culture, and, because of the amount of protein in the culture medium, they did not show a detectable interaction with the WIF-1 compared with the full-length form. Although the immunoprecipitation was also performed using in vivo tissue, it did not give detectable results because of the inability of anti-Olfm1 and anti-WIF-1 antibodies to detect the endogenous intact protein. It must be confirmed by producing other antibodies in future.
Figure 10.
Physical interaction of Olfm1 and WIF-1. HEK-293 cells were cotransfected with indicated constructs. Conditioned medium was collected 48 h after transfection, and protein complexes were precipitated with Flag antibody beads. Proteins eluted from the beads were separated by SDS-PAGE and probed with peroxidase-conjugated antibodies against human IgG (top). Bottom shows Western blot of conditioned medium before immunoprecipitation probed with peroxidase-conjugated antibodies against human IgG.
Discussion
Zebrafish is a convenient system to analyze functions of genes and proteins in development. Olfactomedin domain-containing proteins play important roles in normal development and disease. There are two olfm1 genes in zebrafish, and, in this work, we studied the effects of their inhibition by MOs and overexpression by injection of Olfm RNAs. Overexpression of the AMZ form but not the BMY form of Olfm1 led to mild enlargement of the retina. It was reported previously that injection of RNA encoding the AMY form of olfm1 into Xenopus embryos induced expansion of neural territories, especially in the retina (Moreno and Bronner-Fraser, 2005). Conversely, a decrease in Olfm1 levels by MO led to a reduction in eye size, inhibition of optic nerve extension, and reduced pigmentation of the eye. Reduction in eye size has also been reported after inhibition of Olfm2, another olfactomedin domain-containing protein, by a MO injection (Lee et al., 2008). Other changes observed after the Olfm1–MO injection into zebrafish included defects in the brain and the mandibular and the hyosymplectic cartilages, reduced body pigmentation, and distorted body axis. It is interesting to note that the injection of Olfm2–MO also produced defects in cartilaginous structures, but, in this case, frontal arches (mandibular and hyoid) were less affected than caudal branchial arches (Lee et al., 2008). Most of Olfm1–MO effects were partially rescued by the coinjection of the N1–152 Olfm1 construct, whereas other constructs were less effective. We could not achieve a complete rescue even by the injection of higher amount of the olfm1 RNA. Similarly, the damaging effects of Olfm2–MO injections could not be restored by injection of olfm2 RNA (Lee et al., 2008). Some defects caused by the Olfm1–MO injection could be predicted on the basis of previous observations in Xenopus and chicken (Barembaum et al., 2000; Sakuragi et al., 2006). For example, the reduced pigmentation and the loss of cranial cartilages caused by the Olfm1–MO injection indicate its possible effects on neural crest cell differentiation into melanocytes and cranial cartilages. Brain and eye defects may result from the involvement of Olfm1 in neurogenesis. At the same time, it should be mentioned that the elimination of the central M part from all isoforms of Olfm1 in mice did not produce visible phenotype (Cheng et al., 2007). One possible explanation for this observation is that the elimination of the M part from all Olfm1 forms does not prevent the formation of alternatively spliced forms of Olfm1 that were lacking the M part but contained the olfactomedin domain. These products may perform some functions of full-length Olfm1. Our preliminary experiments also indicate that, in rodents, Olfm2 shows an expression pattern very similar to that for Olfm1 and may produce a compensatory effect (V. V. Senatorov and S. I. Tomarev, unpublished observations). This is also true for zebrafish olfm1 and olfm2 genes that show overlapping but not identical expression patterns (Nakaya and Tomarev, 2007; Lee et al., 2008).
A possible explanation for the observed eye defects may be the change in the pattern of expression of rx3 and six3. These genes are essential for early eye development. Domains of their expression in the optic primordia were more laterally separated by the olfm1 RNA injection and narrowed by the Olfm1–MO injection. A change in the lateral separation of the eye primordia was also observed for the rx2 gene expression (data not shown). This effect can be caused by changes in sonic hedgehog (SHH) expression, which is produced by the notochord and floor plate in the midline and is involved in the separation of the eye fields (Ekker et al., 1995; Martí et al., 1995; Shimamura et al., 1995; Li et al., 1997; Rorick et al., 2007), although Olfm2–MO did not change the pattern of SHH expression (Lee et al., 2008).
Most probably, Olfm1, similar to some other secreted glycoproteins, interacts with cellular receptor(s) and stimulates or inhibits intracellular signaling pathways. It has been reported that gliomedin interacts specifically with neurofascin and neuron–glia-related cell adhesion molecule (Eshed et al., 2005). hGC-1, also known as GW112, OLFM4, and hOlfD, may interact with cell surface lectins and cadherins (Liu et al., 2006). Finally, amassin, an extracellular glycosylated protein found in the coelomic fluid of sea urchin, may bind to an unidentified cell surface protein (Hillier and Vacquier, 2003). Recent data indicate that different forms of Olfm1 may interact with β-dystrobrevin (Veroni et al., 2007). β-Dystrobrevin is a component of the dystrophin-associated protein complex that may link the actin cytoskeleton to the extracellular matrix and may serve as a scaffold for signaling proteins.
We found that Olfm1 may interact with WIF-1, a secreted inhibitor of Wnt signaling. WIF-1 is highly expressed in mammalian retina (Hunter et al., 2004). In the embryonic mouse retina (embryonic day 16), it is expressed in the RGCs and the inner nuclear layer, whereas in the adult retina, it is more highly expressed in the inner and outer nuclear layers (Hunter et al., 2004). Therefore, WIF-1 and Olfm1 show overlapping expression patterns in the developing and adult eyes. Wnt signaling is one of the major regulatory cascades important for normal embryogenesis of multicellular organisms. It has been demonstrated that the interaction of Wnt and its antagonists plays a critical role at early stages of eye development (Houart et al., 2002; Zilinski et al., 2004; Pézeron et al., 2006; Stigloher et al., 2006; Tendeng and Houart, 2006). Wnts produced at the hindbrain work as a posteriorizing signal toward the forebrain, whereas its inhibitors counteract to Wnt action and stimulate the anteriorization to establish the forebrain and future eye area. Our study indicates that Olfm1 may modulate Wnt signaling by binding the Wnt inhibitors and Wnt receptors. Identification of Wnt receptors interacting with Olfm1 is currently in progress.
One of the key observations of this paper concerns possible functions of Olfm1. Injection of RNA encoding the olfactomedin domain-lacking isoforms or long (BMZ) form of Olfm1, which was a predominant transcript in the course of early zebrafish development (Fig. 3), increased the thickness of the optic nerve and produced more extended projection field in the tectum in 72 hpf embryos (Fig. 2). The olfactomedin domain-lacking isoforms produced more pronounced effects than full-length Olfm1. It has been reported previously that short and long forms of Olfm1 may have different functional roles (Moreno and Bronner-Fraser, 2005). Although the Olfm1–MO injection inhibited the neurite extension from the motor and Rohon-Beard neurons, the injection of olfm1 RNA did not lead to detectable changes in the axon network produced by these neurons, as judged by staining with antibodies against acetylated tubulin (data not shown). At present, we do not fully understand why the truncated forms of olfactomedin are more effective in increasing thickness of the optic nerve than their full-length counterparts. Available data suggest that truncated and full-length forms of olfactomedin may interact with different proteins. For example, the BMY but not BMZ form of olfactomedin interacts with WAVE1 (Cheng et al., 2007), an actin regulating protein that is present along the leading edge of the neuronal growth cones. It is possible that such interaction promote the optic nerve growth. Another possibility is that short and long forms of olfactomedin are secreted with different efficiencies and therefore may exercise different effects on the optic nerve growth via interactions with presumptive receptors and extracellular matrix. Injections of RNAs encoding short forms of Olfm1 may also shift the balance between short and long forms compared with injection of the long forms. It is feasible to suggest that Olfm1, by modulating Wnt signaling pathways, may change the activity of several factors, which were shown to be involved in the axon branching or it may affect interactions of the growing axons with the supporting glia cells. Our preliminary results indicate that Olfm1 may induce neurite outgrowth in different experimental systems, including nerve growth factor-stimulated PC12 cells (H.-S.L., N.N., and S.I.T, unpublished observations) and embryonic rat hippocampal neurons (N.N. and S.I.T, unpublished observations). Altogether, these data indicate that Olfm1 has an essential role in axon extension. Available data suggest that Olfm1 may be essential for neural development and neural crest generation (Barembaum et al., 2000; Moreno and Bronner-Fraser, 2001). One of the forms of Olfm1, BMY (also known as pancortin-2), may serve as a mediator of ischemia-induced apoptosis of neurons in the adult cerebral cortex (Cheng et al., 2007). Recent data suggest that olfactomedin-2 is an important regulator of development of the anterior nervous system (Lee et al., 2008). Other olfactomedin domain-containing proteins, for example, Xenopus tiarin, may participate in the specification of the dorsal neural tube (Tsuda et al., 2002). Gliomedin appears to mediate Schwann cell–axon interaction and molecular assembly of the nodes of Ranvier (Eshed et al., 2005). Sea urchin amassin mediates the massive intercellular adhesion of coelomocytes, primitive immune cells found in echinoderms (Hillier and Vacquier, 2003).
In summary, our data demonstrate that the olfm1 genes are essential for development of several organs, including eye and brain. In the eye, Olfm1 may be essential for the early eye determination, RGC differentiation, and the optic nerve axons growth and targeting of the optic tectum. Although mutations in the olfm1 gene have not been found yet, mutations in two other family members, myocilin and olfm2, appear to be contributing factors to human glaucoma (Stone et al., 1997; Funayama et al., 2006). Expression of olfm1 in RGCs and in the optic nerve makes olfm1 a good candidate gene for mutations that might also contribute to human glaucoma.
Footnotes
This work was supported by the National Eye Institute intramural research program. We thank David Rawnsley for his participation in some experiments during his summer student training program at the National Eye Institute. We thank Drs. Heiner Westphal, Igor Dawid, and Reiko Toyama for their suggestions and Drs. Olof Sundin and Janine Davis for suggestions and critical reading of this manuscript. We thank Dr. Hiroko Kataoka for providing runx constructs, Drs. Aiwu Cheng and Mark Mattson for providing eyes from Olfm1 null mice, Drs. J. Nathans and J. C. Hsieh for providing WIF-1–IgG and WD–IgG fusion constructs, Dr. R. Fariss for help with the confocal microscope, and Dr. R. Moon for providing the Super8XTOPFlash reporter construct.
References
- Barembaum M, Moreno TA, LaBonne C, Sechrist J, Bronner-Fraser M. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat Cell Biol. 2000;2:219–225. doi: 10.1038/35008643. [DOI] [PubMed] [Google Scholar]
- Burns CE, DeBlasio T, Zhou Y, Zhang J, Zon L, Nimer SD. Isolation and characterization of runxa and runxb, zebrafish members of the runt family of transcriptional regulators. Exp Hematol. 2002;30:1381–1389. doi: 10.1016/s0301-472x(02)00955-4. [DOI] [PubMed] [Google Scholar]
- Cheng A, Arumugam TV, Liu D, Khatri RG, Mustafa K, Kwak S, Ling HP, Gonzales C, Xin O, Jo DG, Guo Z, Mark RJ, Mattson MP. Pancortin-2 interacts with WAVE1 and Bcl-xL in a mitochondria-associated protein complex that mediates ischemic neuronal death. J Neurosci. 2007;27:1519–1528. doi: 10.1523/JNEUROSCI.5154-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84:195–198. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- Danielson PE, Forss-Petter S, Battenberg EL, deLecea L, Bloom FE, Sutcliffe JG. Four structurally distinct neuron-specific olfactomedin-related glycoproteins produced by differential promoter utilization and alternative mRNA splicing from a single gene. J Neurosci Res. 1994;38:468–478. doi: 10.1002/jnr.490380413. [DOI] [PubMed] [Google Scholar]
- Ekker SC, McGrew LL, Lai CJ, Lee JJ, von Kessler DP, Moon RT, Beachy PA. Distinct expression and shared activities of members of the hedgehog gene family of Xenopus laevis. Development. 1995;121:2337–2347. doi: 10.1242/dev.121.8.2337. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Feinberg K, Poliak S, Sabanay H, Sarig-Nadir O, Spiegel I, Bermingham JR, Jr, Peles E. Gliomedin mediates Schwann cell-axon interaction and the molecular assembly of the nodes of Ranvier. Neuron. 2005;47:215–229. doi: 10.1016/j.neuron.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Funayama T, Mashima Y, Ohtake Y, Ishikawa K, Fuse N, Yasuda N, Fukuchi T, Murakami A, Hotta Y, Shimada N. SNPs and interaction analyses of noelin 2, myocilin, and optineurin genes in Japanese patients with open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47:5368–5375. doi: 10.1167/iovs.06-0196. [DOI] [PubMed] [Google Scholar]
- Hemish J, Nakaya N, Mittal V, Enikolopov G. Nitric oxide activates diverse signaling pathways to regulate gene expression. J Biol Chem. 2003;278:42321–42329. doi: 10.1074/jbc.M308192200. [DOI] [PubMed] [Google Scholar]
- Hillier BJ, Vacquier VD. Amassin, an olfactomedin protein, mediates the massive intercellular adhesion of sea urchin coelomocytes. J Cell Biol. 2003;160:597–604. doi: 10.1083/jcb.200210053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houart C, Caneparo L, Heisenberg C, Barth K, Take-Uchi M, Wilson S. Establishment of the telencephalon during gastrulation by local antagonism of Wnt signaling. Neuron. 2002;35:255–265. doi: 10.1016/s0896-6273(02)00751-1. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, Nusse R, Dawid IB, Nathans J. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- Hunter DD, Zhang M, Ferguson JW, Koch M, Brunken WJ. The extracellular matrix component WIF-1 is expressed during, and can modulate, retinal development. Mol Cell Neurosci. 2004;27:477–488. doi: 10.1016/j.mcn.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe MK, Sohn S, Hur W, Moon Y, Choi YR, Kee C. Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem Biophys Res Commun. 2003;312:592–600. doi: 10.1016/j.bbrc.2003.10.162. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Ochi M, Enomoto K, Yamaguchi A. Cloning and embryonic expression patterns of the zebrafish Runt domain genes, runxa and runxb. Mech Dev. 2000;98:139–143. doi: 10.1016/s0925-4773(00)00445-7. [DOI] [PubMed] [Google Scholar]
- Kim HS, Shin J, Kim SH, Chun HS, Kim JD, Kim YS, Kim MJ, Rhee M, Yeo SY, Huh TL. Eye field requires the function of Sfrp1 as a Wnt antagonist. Neurosci Lett. 2007;414:26–29. doi: 10.1016/j.neulet.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Lee JA, Anholt RR, Cole GJ. Olfactomedin-2 mediates development of the anterior central nervous system and head structures in zebrafish. Mech Dev. 2008;125:167–181. doi: 10.1016/j.mod.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tierney C, Wen L, Wu JY, Rao Y. A single morphogenetic field gives rise to two retina primordia under the influence of the prechordal plate. Development. 1997;124:603–615. doi: 10.1242/dev.124.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Chen L, Zhu J, Rodgers GP. The glycoprotein hGC-1 binds to cadherin and lectins. Exp Cell Res. 2006;312:1785–1797. doi: 10.1016/j.yexcr.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Liu Y, Vollrath D. Reversal of mutant myocilin non-secretion and cell killing: implications for glaucoma. Hum Mol Genet. 2004;13:1193–1204. doi: 10.1093/hmg/ddh128. [DOI] [PubMed] [Google Scholar]
- Martí E, Takada R, Bumcrot DA, Sasaki H, McMahon AP. Distribution of Sonic hedgehog peptides in the developing chick and mouse embryo. Development. 1995;121:2537–2547. doi: 10.1242/dev.121.8.2537. [DOI] [PubMed] [Google Scholar]
- Moreno TA, Bronner-Fraser M. The secreted glycoprotein Noelin-1 promotes neurogenesis in Xenopus. Dev Biol. 2001;240:340–360. doi: 10.1006/dbio.2001.0472. [DOI] [PubMed] [Google Scholar]
- Moreno TA, Bronner-Fraser M. Neural expression of mouse Noelin-1/2 and comparison with other vertebrates. Mech Dev. 2002;119:121–125. doi: 10.1016/s0925-4773(02)00308-8. [DOI] [PubMed] [Google Scholar]
- Moreno TA, Bronner-Fraser M. Noelins modulate the timing of neuronal differentiation during development. Dev Biol. 2005;288:434–447. doi: 10.1016/j.ydbio.2005.09.050. [DOI] [PubMed] [Google Scholar]
- Nagano T, Nakamura A, Mori Y, Maeda M, Takami T, Shiosaka S, Takagi H, Sato M. Differentially expressed olfactomedin-related glycoproteins (Pancortins) in the brain. Brain Res Mol Brain Res. 1998;53:13–23. doi: 10.1016/s0169-328x(97)00271-4. [DOI] [PubMed] [Google Scholar]
- Nakaya N, Tomarev S. Expression patterns of alternative transcripts of the zebrafish olfactomedin 1 genes. Gene Expr Patterns. 2007;7:723–729. doi: 10.1016/j.modgep.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthy L. The WIF module. Trends Biochem Sci. 2000;25:12–13. doi: 10.1016/s0968-0004(99)01504-2. [DOI] [PubMed] [Google Scholar]
- Pézeron G, Anselme I, Laplante M, Ellingsen S, Becker TS, Rosa FM, Charnay P, Schneider-Maunoury S, Mourrain P, Ghislain J. Duplicate sfrp1 genes in zebrafish: sfrp1a is dynamically expressed in the developing central nervous system, gut and lateral line. Gene Expr Patterns. 2006;6:835–842. doi: 10.1016/j.modgep.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Rorick AM, Mei W, Liette NL, Phiel C, El-Hodiri HM, Yang J. PP2A:B56epsilon is required for eye induction and eye field separation. Dev Biol. 2007;302:477–493. doi: 10.1016/j.ydbio.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Sakuragi M, Sasai N, Ikeya M, Kawada M, Onai T, Katahira T, Nakamura H, Sasai Y. Functional analysis of chick ONT1 reveals distinguishable activities among olfactomedin-related signaling factors. Mech Dev. 2006;123:114–123. doi: 10.1016/j.mod.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Snyder DA, Rivers AM, Yokoe H, Menco BP, Anholt RR. Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry. 1991;30:9143–9153. doi: 10.1021/bi00102a004. [DOI] [PubMed] [Google Scholar]
- Stigloher C, Ninkovic J, Laplante M, Geling A, Tannhäuser B, Topp S, Kikuta H, Becker TS, Houart C, Bally-Cuif L. Segregation of telencephalic and eye-field identities inside the zebrafish forebrain territory is controlled by Rx3. Development. 2006;133:2925–2935. doi: 10.1242/dev.02450. [DOI] [PubMed] [Google Scholar]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- Tendeng C, Houart C. Cloning and embryonic expression of five distinct sfrp genes in the zebrafish Danio rerio. Gene Expr Patterns. 2006;6:761–771. doi: 10.1016/j.modgep.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Torrado M, Trivedi R, Zinovieva R, Karavanova I, Tomarev SI. Optimedin: a novel olfactomedin-related protein that interacts with myocilin. Hum Mol Genet. 2002;11:1291–1301. doi: 10.1093/hmg/11.11.1291. [DOI] [PubMed] [Google Scholar]
- Toyama R, O'Connell ML, Wright CV, Kuehn MR, Dawid IB. Nodal induces ectopic goosecoid and lim1 expression and axis duplication in zebrafish. Development. 1995;121:383–391. doi: 10.1242/dev.121.2.383. [DOI] [PubMed] [Google Scholar]
- Tsuda H, Sasai N, Matsuo-Takasaki M, Sakuragi M, Murakami Y, Sasai Y. Dorsalization of the neural tube by Xenopus tiarin, a novel patterning factor secreted by the flanking nonneural head ectoderm. Neuron. 2002;33:515–528. doi: 10.1016/s0896-6273(02)00590-1. [DOI] [PubMed] [Google Scholar]
- Veroni C, Grasso M, Macchia G, Ramoni C, Ceccarini M, Petrucci TC, Macioce P. beta-dystrobrevin, a kinesin-binding receptor, interacts with the extracellular matrix components pancortins. J Neurosci Res. 2007;85:2631–2639. doi: 10.1002/jnr.21186. [DOI] [PubMed] [Google Scholar]
- Volynski KE, Silva JP, Lelianova VG, Atiqur Rahman M, Hopkins C, Ushkaryov YA. Latrophilin fragments behave as independent proteins that associate and signal on binding of LTX(N4C) EMBO J. 2004;23:4423–4433. doi: 10.1038/sj.emboj.7600443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. A guide for the laboratory use of zebrafish (Danio rerio) Eugene, OR: University of Oregon Press; 2000. The zebrafish book. [Google Scholar]
- Yoda H, Hirose Y, Yasuoka A, Sasado T, Morinaga C, Deguchi T, Henrich T, Iwanami N, Watanabe T, Osakada M, Kunimatsu S, Wittbrodt J, Suwa H, Niwa K, Okamoto Y, Yamanaka T, Kondoh H, Furutani-Seiki M. Mutations affecting retinotectal axonal pathfinding in Medaka, Oryzias latipes. Mech Dev. 2004;121:715–728. doi: 10.1016/j.mod.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Zeng LC, Han ZG, Ma WJ. Elucidation of subfamily segregation and intramolecular coevolution of the olfactomedin-like proteins by comprehensive phylogenetic analysis and gene expression pattern assessment. FEBS Lett. 2005;579:5443–5453. doi: 10.1016/j.febslet.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Zilinski C, Brownell I, Hashimoto R, Medina-Martinez O, Swindell EC, Jamrich M. Expression of FoxE4 and Rx visualizes the timing and dynamics of critical processes taking place during initial stages of vertebrate eye development. Dev Neurosci. 2004;26:294–307. doi: 10.1159/000082271. [DOI] [PubMed] [Google Scholar]