Abstract
Physical differences in small molecule binding between enzymes and non-enzymes were found through mining the protein-ligand database, Binding MOAD (Mother of All Databases). The data suggest that divergent approaches may be more productive for improving the affinity of ligands for the two classes of proteins. High-affinity ligands of enzymes are much larger than those with low affinity, indicating that the addition of complementary functional groups is likely to improve the affinity of an enzyme inhibitor. However, this process may not be as fruitful for ligands of non-enzymes. High- and low-affinity ligands of non-enzymes are nearly the same size, so modest modifications and isosteric replacement might be most productive. The inherent differences between enzymes and non-enzymes have significant ramifications for scoring functions and structure-based drug design. In particular, non-enzymes were found to have greater ligand efficiencies than enzymes. Ligand efficiencies are often used to indicate druggability of a target, and this finding supports the feasibility of non-enzymes as drug targets. The differences in ligand efficiencies do not appear to come from the ligands; instead, the pockets yield different amino acid compositions, despite very similar distributions of amino acids in the overall protein sequences.
Introduction
Both enzymatic and non-enzymatic proteins can bind small molecules, but enzymes catalyze reactions and have a fundamentally different role from non-enzymes, which may have an impact on their recognition of ligands. Do these two types of binding events have the same physical characteristics? Furthermore, are there any differences between high-affinity complexes and weaker binding events that can be linked to their physical contacts? To answer these questions, physicochemical patterns were mined from our protein-ligand database Binding MOAD (Mother of All Databases), where MOAD is pronounced “mode” as a pun on a ligand’s mode of binding.1,2
Binding MOAD is the largest curated database of high-resolution protein-ligand complexes from the Protein Data Bank (PDB).3 Though it only reflects proteins that can be crystallized, these are the exact systems where structure-based insights will be used. The PDB is the source of all structures used for docking and scoring development by academics. However, the data used here are significantly larger than most sets used to develop existing scoring functions, which are typically sets of <300 complexes of <50 unique proteins. We use 2214 structures: 1790 enzymes and 424 non-enzymes (512 unique enzymes and 176 unique nonenzymes). This study provides an important benchmark of the current landscape available from structural biology (incomplete and/or biased as it may be).
For this study, we have compared distributions of various properties between four classes of protein complexes. Distribution analysis is used widely in many fields, and it is important to stress that it does not define “absolute rules”, nor are the data presented as such. These are general guidelines, and of course, there will be exceptions to those trends. Distribution analysis can show that “men are taller than women” and “women live longer than men.” Those trends are true even though some women are 6’ tall and some men live to 100.
Empirically derived rules can be very useful in discovering and applying new principles in chemistry. One of the most well known examples is Lipinski’s Rule of Five, which describes the physical properties of orally-available drugs.4,5 These rules provide general guidelines for size, lipophilicity, and hydrogen-bonding characteristics that correlate with the likelihood that a molecule can be orally absorbed into the body. The findings are based on distribution data of the chemical characteristics of orally absorbed molecules going into Phase-II testing. The dataset is biased by issues outside of pharmacokinetics such as the need for good synthesis (not just accessible chemistry, but few steps in high yield) and market considerations (completely economic, no basis in the thermodynamics of protein-ligand binding). The rules do not hold for natural products, actively transported molecules, molecules that require metabolism for activation, or most antibiotics, antifungals, vitamins, and cardiac glycosides. There are plenty of molecules in Lipinski space that are not drugs, and many molecules outside that space that are. Despite these limitations and biases, the Rule of Five is used widely in the pharmaceutical industry.
We hope that the present work will also aid drug discovery. In this study, we provide new patterns which describe high-affinity, protein-ligand binding and outline differences between enzymes and non-enzymes. Of course, there will be examples that fall outside the typical pattern, but these relationships provide a good description of the general landscape that structural biology can provide at this time. We expect that our understanding will grow as more structures become available through the various protein structure initiatives.6 These guiding principles may be useful in designing targeted libraries for drug discovery and improving scoring functions. They are also important to advancing our fundamental understanding of chemical biology, protein-ligand binding, and the biophysics that dictate molecular recognition.
Non-covalent, small molecule binding is a tradeoff between the enthalpy gained by making specific contacts between functional groups of the ligand and the protein and entropy lost by forcing the ligand and protein into a specific conformation.7,8 Since this study uses crystal structures it is difficult to fully account for the effect caused by entropy. However, it is possible to determine the physical characteristics of the small molecule and the protein which leads to the binding affinity.
Other studies9,10 have noted an inherent limitation in mining protein structures for physical characteristics of binding. When a pocket is discovered on a protein surface, it is difficult to identify whether it is a true binding site or if it is capable of high-affinity binding appropriate to represent drug-like binding. This study does not suffer from these limitations; all sites have been curated to assure that they are true binding pockets, and the high-affinity complexes are separated from those with low affinity.
Only complexes with binding data (Kd, Ki, or IC50) were used for this study. No complexes in MOAD are annotated with Km data, so almost all ligands are inhibitors, agonists, or antagonists (a small number are cofactors, 5%, included only for systems were affinity data is appropriate). We specifically focused on the contacts between the ligand and the protein, excluding any structure with poorly defined contacts such as missing atoms from under-resolved density or ligands and side chains resolved in multiple orientations. Distributions of ligand size, buried surface area (BSA), exposed surface area (ESA), and other physical characteristics were examined for statistically significant differences between four subsets of the complexes: high-affinity binding to enzymes, high-affinity non-enzymes, low-affinity enzymes, and low-affinity non-enzymes. A common metric to evaluate lead compounds is ligand efficiency.11–14 In this study, ligand efficiencies for the different classes of proteins are reported as affinity per size (−ΔGbind divided by the number of non-hydrogen atoms) and per the degree of contact between the ligand and the pocket (−ΔGbind/BSA).
Here, we focus on the most significant differences between molecular recognition of tight and weak binding to enzymes and non-enzymes.
Methods
Data for this study come from the largest comprehensive database of protein-ligand crystal structures with binding data, Binding MOAD. The latest version of Binding MOAD was created from structures released on 12/31/2006 or earlier; it contains 9836 complexes, comprised of 3151 unique protein families binding 4659 unique ligands. The great care taken in curating this dataset has been outlined elsewhere,1 but it should be noted for these purposes that ~9,000 crystallography papers have been examined to determine the appropriateness of every ligand (crystallographic additives, post-translational modifications, and covalently bound ligands are excluded from consideration). From these efforts, binding affinity data is available for 30% of the entries, with a preference for Kd data over Ki data over IC50 values. The affinities were converted to free energies of binding by ΔGbind = RT×ln(Kd) or simply approximated by ΔGbind = RT×ln(Ki or IC50) with a temperature of 298 K.
High-affinity binding was defined Kd, Ki, or IC50 ≤ 250 nM (ΔGbind ≤ −9 kcal/mol), which is approximately the average of all the complexes with binding data in Binding MOAD. Enzyme complexes were defined from the Enzyme Classification number in the PDB file. The nonenzymes were annotated by hand using keywords reported in the remarks section of the PDB entry. Binding MOAD’s high-affinity non-enzymes and enzymes are listed in the Supporting Information. All complexes and binding data are available at the Binding MOAD website, www.BindingMOAD.org.
To calculate surface areas, BSA and ESA were calculated with GoCAV using radii based on united-atom OPLS parameters.2 This code reports buried molecular surface area (MSA) of the pocket and also defines ESA of the binding site, bounded by the 3D coordinates of the ligand.
The SlogP for the ligands was calculated using MOE,15 based on the method developed by Wildman and Crippen.16 For the 2D and 3D descriptors calculated with MOE, the idealized SDF files from the PDB were used if available; otherwise, the coordinates of the ligand from the protein’s structure were taken. Hydrogens were added with MOE. In an effort to identify any differences, all 2D and 3D ligand characteristics available within MOE were compared for the four groups of complexes: high-affinity enzyme, low-affinity enzyme, high-affinity non-enzyme and low-affinity non-enzyme.
Statistical Analysis
Statistical significance was assessed with the programs SAS17 and JMP18. Initial assessments used JMP to calculate all pair-wise correlations for the over 200 descriptors calculated. For the descriptors showing interesting trends, the significance of the differences between the distributions of physical properties were determined by the Wilcoxon rank-sum test, which is most appropriate given the non-Gaussian distributions of the data. We also performed one-way ANOVA, two-way ANOVA, and Tukey-Kramer HSD tests between the four classifications. Since these second series of tests require near-normal distributions, the square-root transform was applied to reduce the skew and bring the distributions closer to normal. For the important descriptors, distribution analyses from JMP are included in the Supporting Information (Supporting Information, Figures S1-S7), and each includes the mean, median, quantiles, distribution histogram, and outlier box plot. The results of the Tukey-Kramer HSD test are presented in Supporting Information (Supporting Information, Tables S1-S5).
Histograms of the distributions of ligand size were binned in increments of 5 heavy atoms. Distributions of BSA and ESA were binned by 50 Å2. Those plotting ligand efficiency were binned by 0.1 kcal/mol-atom for affinity per size or 10 cal/mol-Å2 for affinity per degree of contact. Distributions of SlogP were binned by 2 log units. These bin sizes were in proportion to the size of the datasets and were consistent with those automatically generated by JMP.
Results and Discussion
Considerable effort was made to determine direct mathematical relationships between affinity and surface area, ligand size, or other characteristics of protein-ligand interactions, but there was no global correlation across all complexes. Recent work by Coleman and Sharp19 based on the PDBbind dataset20 also found no correlation between affinity and surface area or depth of the binding pocket. Inspired by analyses of distributions of ligand efficiencies from screening data,11 we changed our approach and focused on distributions of the properties between subsets of protein-ligand complexes.
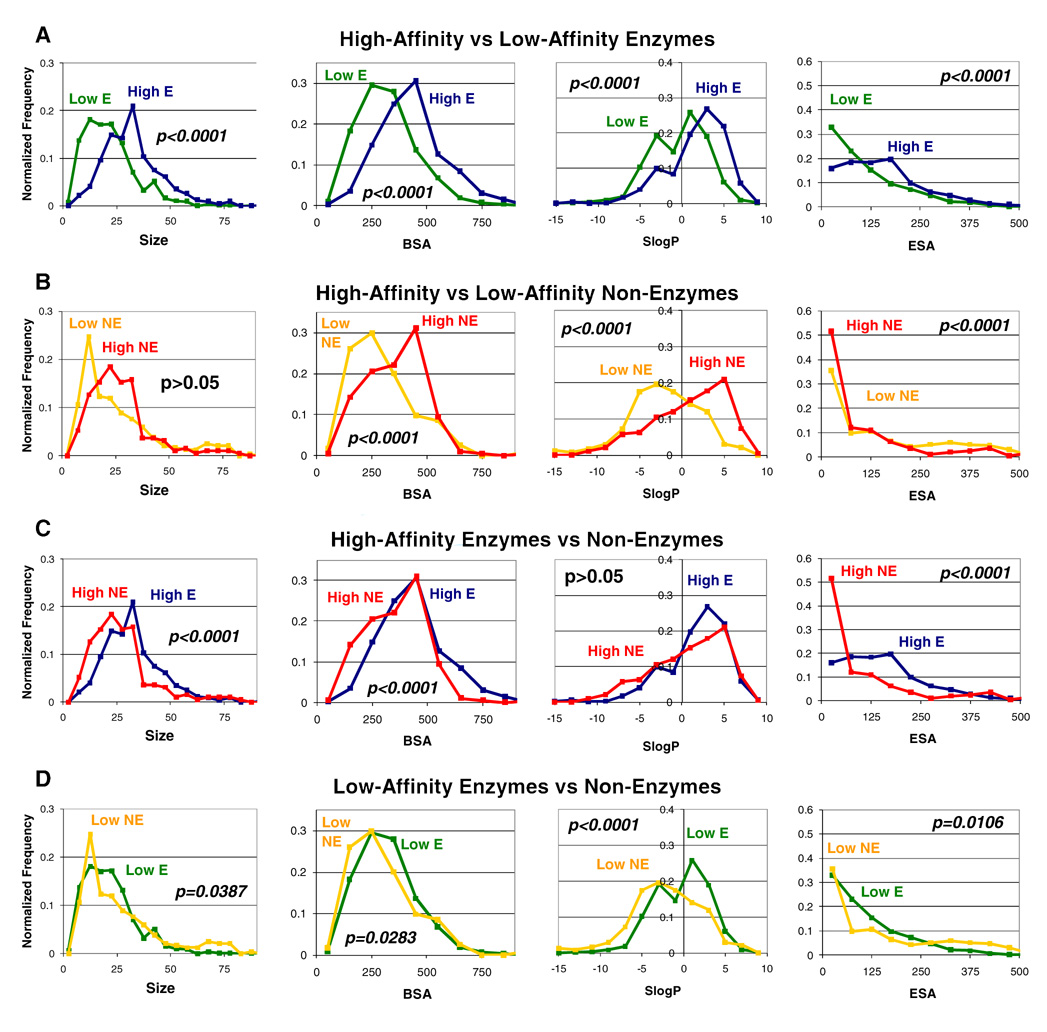
Table 1 outlines the characteristics that differ between high-affinity and low-affinity binding for enzymes and non-enzymes; all emphasized differences in the datasets have a statistical significance >99.99% (p<0.0001) based on a two-tailed, Wilcoxon rank-sum test. Figure 1 shows a comparison between each of the subsets of complexes, examining the distribution of ligand sizes, BSA, SlogP, and ESA. Many of the low-affinity complexes have ~300 Å2 of BSA, but the high-affinity complexes display more contact. It has been estimated that drug-like binding sites have ~300 Å2 of solvent-accessible surface area (SASA).9 Our measurement for BSA is based on MSA, and so, the slightly higher values of the high-affinity complexes are appropriately comparable.9
Table 1.
Characteristics of Protein-Ligand Binding for Enzymes and Non-Enzymes in the Full Dataset.a
| Median | Low Affinity | High Affinity | Comparisonb |
|---|---|---|---|
| Physical | >250 nM | ≤250 nM | |
| Properties | ΔGbind > −9 kcal/mol | ΔGbind ≤−9 kcal/mol | |
| Enzymes | 1048 complexes | 742 complexes | High-affinity ligands are 52% larger and more hydrophobic |
| ΔGbind | −6.6 kcal/mol | −10.9 kcal/mol | |
| Sizec | 21 atoms | 32 atoms | |
| BSA | 305 Å2 | 419 Å2 | |
| ESA (%ESA)d | 87 Å2 (22%) | 144 Å2 (24%) | |
| SlogP | 0.3 | 2.4 | |
| −ΔGbind/atom | 0.31 kcal/mol-atom | 0.36 kcal/mol-atom | |
| -ΔGbind/BSA | 21 cal/mol-Å2 | 26 cal/mol-Å2 | |
| Non-Enzymes | 234 complexes | 190 complexes | Low-affinity ligands are three times more exposed and more hydrophilic |
| ΔGbind | −7.2 kcal/mol | −10.4 kcal/mol | |
| Sizec | 22 atoms | 25 atoms | |
| BSA | 265 Å2 | 361 Å2 | |
| ESA (%ESA)d | 118 Å2 (33%) | 45 Å2 (11%) | |
| SlogP | −2.2 | 1.5 | |
| −ΔGbind/atom | 0.28 kcal/mol-atom | 0.41 kcal/mol-atom | |
| −ΔGbind/BSA | 22 cal/mol-Å2 | 31 cal/mol-Å2 | |
| Comparisonb | Non-enzymes have 17% greater ligand efficiencies | ||
Values presented are medians for each population.
All differences noted in the comparisons sections have a statistical significance of >99.99% (p<0.0001).
Ligand size is given in the number of non-hydrogen atoms.
Percent exposure is ESA/(ESA+BSA).
Figure 1.
Comparisons of (A) enzyme complexes, (B) non-enzyme complexes, (C) high-affinity complexes and (D) low-affinity complexes are presented. High-affinity enzymes are shown in dark blue, and low-affinity enzymes are in green. High-affinity non-enzymes are in red, and low-affinity non-enzymes are in gold. Distribution of ligand sizes (number of non-hydrogen atoms), buried surface area of the pocket (Å2), SlogP, and exposed surface area (Å2) are given in normalized percent frequencies. P-values show the significance of the difference in the medians of the distributions, as determined by a two-tailed Wilcoxon rank-sum evaluation (insignificant differences have p>0.05).
Different approaches for improving inhibitors of enzymes versus non-enzymes
For enzymes, there is a significant difference in the size of the ligands in high- and low-affinity complexes (Figure 1a). High-affinity ligands are much larger (11 more non-hydrogen atoms). However, non-enzymes display very little difference in the size of the ligands between high-affinity and low-affinity complexes (Table 1, Figure 1b). These differences do not come from any influence of the inclusion of cofactors in the set. The medians are nearly unchanged if they are removed from the dataset (see Supporting Information, Table S6).
Sizes of the ligands point to a strong difference in the complexes, particularly in how to improve an inhibitor for enzymes versus non-enzymes. To improve the affinity of an enzyme inhibitor, it appears fruitful to add functional groups to increase the complementary contact between the inhibitor and the protein. In contrast, improving ligands for non-enzymes may best involve conservative changes which maintain the ligand’s size. Tight binders for non-enzymes are less exposed than the low-affinity ligands, making them more sequestered from the surrounding solvent (Table 1). Distributions of the calculated octanol/water partition ratios (Figure 1a,b) show that high-affinity ligands are more hydrophobic than those with low affinity, but there is no significant difference between enzymes and non-enzymes in this regard. It appears that “adding grease” equally improves binding to both enzymes and non-enzymes, consistent with a general desolvation effect.7
The above trends for improving inhibitors for enzymes versus non-enzymes come from observing patterns across different proteins (inter-protein relationships), but information to improve inhibitors for a specific target must come from observing trends of one protein binding a variety of ligands (intra-protein binding trends). This is a more difficult comparison to make because few proteins are crystallized with a significant range of bound ligands. For the few that exist, we must divide them into enzymes and non-enzymes, further reducing the sizes of the available datasets. The findings below are qualitative in nature. Overall, our data show that enzymes appear to have better correlations between size and affinity than non-enzymes.
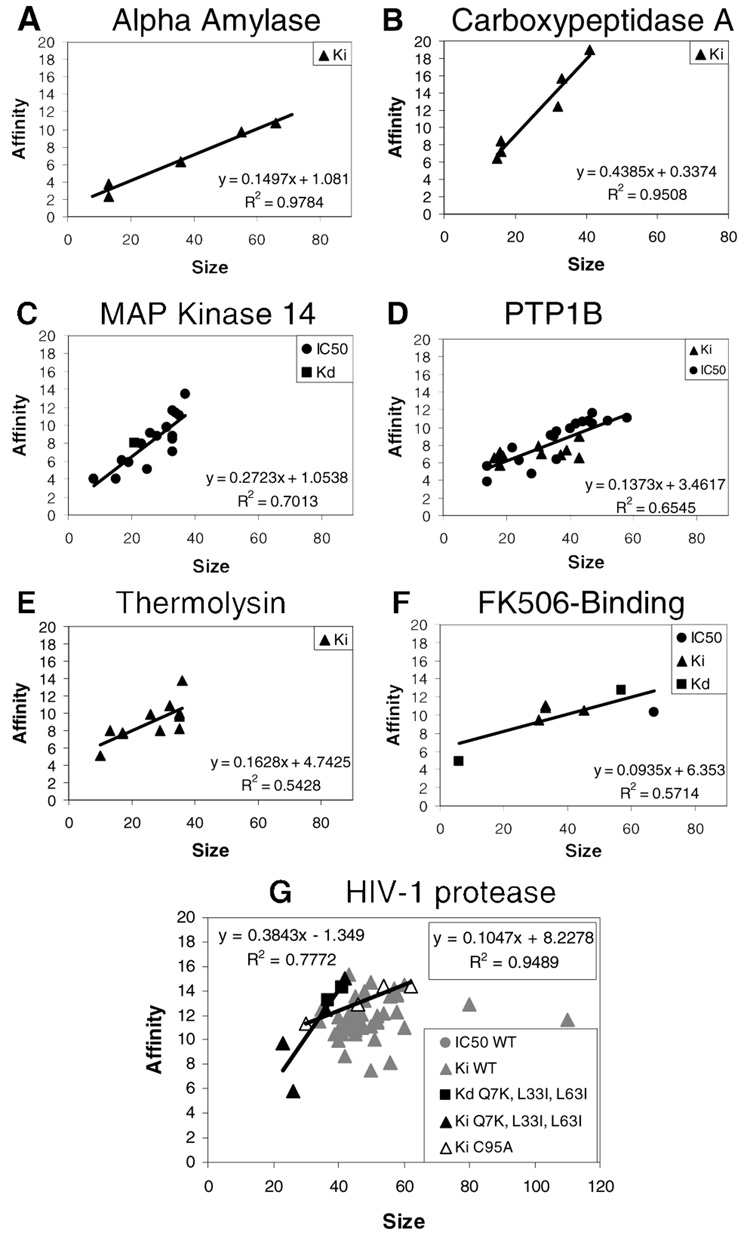
In order to determine a relationship between ligand size and affinity within a protein family (Figure 2 and Figure 3), the complexes were grouped by 100% sequence identity. This organization ensures that changes in affinity are the result of changes in the ligand and not a mutation within the binding site. (For a few proteins, we were able to combine two sets when the mutations were far from the active site and inconsequential.) Groups that contained ≥5 complexes were examined. For non-enzymes, there were only a few proteins available: oligopeptide-binding protein, glutamate receptor 2, estrogen receptor alpha, estrogen receptor beta, arabinose-binding protein, mannose-binding protein, maltose-binding protein, and src SH2-binding domain. For most of the non-enzymes, the ligands are very similar in size and affinity. Six of the eight proteins have a small range of ligand sizes which shows little correlation to affinity (Figure 2 a, b). The small range of observed ligand sizes supports the idea that conservative changes are most appropriate for trying to improve ligands for non-enzymes. However, the lack of a distinct trend between ligand size and affinity does not necessarily prove that a trend could not be observed. It is unclear if the small range of ligands is the result of the specificity of the protein systems or whether more diverse complexes are simply not available from the PDB.
Figure 2.
Limited correlation is seen between size and affinity in non-enzymes (A and B). The proteins with “clusters” of points have smaller binding sites and no ligands over 40 non-hydrogen atoms. The ligands have similar sizes and affinities for oligopeptide-binding protein (OBP), glutamate receptor 2 (GluR2) and mannose-binding protein (MBP), arabinose-binding protein (ABP), and estrogen receptor (ER) alpha and beta. The only non-enzymes with a range of ligand sizes are maltose-binding protein and the non-enzymatic site on the SH2 domain of pp60src tyrosine kinase (C and D, respectively).
Figure 3.
Many examples are available of enzyme complexes that show a strong correlation between size and affinity of the ligands; seven are given here (A-G). HIV-1 protease (G) demonstrates that a large collection of ligands may show no correlation, but subsets of data may reveal strong trends (data for the C95A and Q7K/L33I/L63I mutants). It is interesting that even small binding sites with ligands of 40 non-hydrogen atoms or less (B,C,D) show a linear trend with affinity; this was not seen for non-enzymes with small binding sites.
Only maltose-binding protein (Figure 2c) and the non-enzymatic site on the SH2 domain of pp60src tyrosine kinase (Figure 2d) have a significant range of ligand sizes. The maltose-binding protein complexes contain sugar chains of varying length. Almost all bind with roughly the same affinity, and this may be explained by the fact that the larger ligands show little difference in the BSA contact, despite the very large range of sizes. The non-enzymatic site on the SH2 domain of pp60src tyrosine kinase is the only non-enzyme complex showing some correlation between ligand size and binding affinity. It is interesting that the only exception in non-enzymes is a regulatory site on an enzyme. These linear correlations reflect a trend across several ligands, Δ(ΔGbind/size), which is slightly different than the ligand efficiency of an individual ligand, ΔGbind/size. In the discussions below, we will use the term “trend” or “correlation” when comparing across several ligands bound to the same protein, Δ(ΔGbind/size).
In the case of enzymes in MOAD, thirty-seven proteins were available with five complexes or more. Unlike non-enzymes, over half of the families showed correlations between size and affinity. For brevity, only seven examples of MOAD’s enzymes are given in Figure 3. One of the most interesting features of the data in Figure 3 is that the slopes – the overall trend for each set – significantly vary! Though a linear correlation can be found for a good number of enzymes, the additive contributions of more functional groups appear to be system dependent, with some contributions being rather small. The trends range from 0.44 kcal/mol-atom for carboxypeptidase A (Figure 3b) to 0.09 kcal/mol-atom for FK506-binding protein (Figure 3f). Most scoring functions use additive terms, and these findings underscore the difficulty in developing a universal scoring function, appropriate for all protein systems. Yang et al. have also noted these difficulties in development of their M-Score scoring function21.
However, for 11 enzymes, there was no correlation; the ligands had roughly comparable affinity and sizes, much like the non-enzyme examples. Three enzymes showed a very small range of ligand sizes and a large range in binding affinity (Supporting Information). It is debatable whether these trends are exceptional examples of the correlation expected for enzymes or whether they indicate cases where only conservative changes in sizes are allowed, as would be expected for non-enzymes. It is also possible that they result from an unusual set of ligands from one chemical class.
Though Babaoglu and Shoichet have used fragments of inhibitors of β-lactamase to show that ligand efficiency is not necessarily additive within a binding site,22 fragment-based design often couples these small building blocks in the pursuit of high-affinity ligands.23 From our data above, one might expect greater success for this strategy when targeting enzymes where increasing size generally leads to increasing affinity. A recent study by Hajduk compared fragment-based design for 14 enzymes and four non-enzymes to show that ligand efficiency remained rather constant as the optimal leads were increased in size.24 The contributions were roughly additive for the best functional groups. The average trend across these systems was 0.3 kcal/mol-atom, with individual systems showing trends from approximately 0.23 to 0.51 kcal/mol-atom (reported as binding efficiency indices of 11–28 pKd units per MW in kDa). It is encouraging that the values are comparable to the ligand efficiencies reported in Table 1.
Hajduk’s trends were presented for the most efficient ligands for each protein, emphasizing the most ideal cases of improving a ligand.24 However, his data for Bcl-xL, a non-enzyme with a large binding cleft, showed that many changes will not be optimal. A detailed analysis for >2300 additional molecules showed that many had significantly lower efficiencies. In fact, he suggests that chemical modifications that reduce the ligand efficiency by >10% deviate too much from the ideal and indicate that either the location or chemical nature of the modification is less desirable.
The HIV-1 protease data (Figure 3g) shows that there is a large scatter of inhibitor sizes and affinities, but two subsets of data (from mutants of HIV-1 protease) show strong linearity. This could demonstrate the same issue seen in Hajduk’s detailed analysis of Bcl-xL.24 The full set of data shows wide scatter and little trend, but a carefully chosen subset could reveal idealized trends for a particular protein system or class of ligands from a specific synthetic series. For HIV-1 protease, the compensation between enthalpy and entropy can be hard to control. Lafont et al. have demonstrated that an increase in size from the inhibitor (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-[[(2R)-2-(2-isoquinolin-5-yloxyethanoylamino)-3-methylsulfanyl-propanoyl]amino]-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide (KNI-10033) to the inhibitor (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-[[(2R)-2-(2-isoquinolin-5-yloxyethanoylamino)-3-methylsulfonyl-propanoyl]amino]-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide (KNI-10075) did not increase binding affinity despite a more favorable enthalpy from a strong hydrogen bond.25 The entropic penalty of changing a thio ether (two heavy atoms) in KNI-10033 to a sulfonyl group KNI-10075 (four heavy atoms) is responsible for the lack of change in binding affinity. That study noted that, although others have been able to optimize certain HIV-1 protease inhibitors with respect to enthalpy, the enthalpy-entropy compensation could make optimization of affinity impossible for some chemical series.
An important caveat should be considered in the preceding discussion. It is possible that strong correlations between size and affinity can only be easily determined for large binding sites. Large ligands can be truncated to provide smaller, weaker ligands that bind to subsites. This would give a wide range of ligand sizes and affinities, allowing a definite size-affinity relationship to emerge from the data. It may be more difficult to determine a trend for a small binding site. This would still imply that enzyme inhibitors are more likely to be improved through the addition of functional groups, simply because the binding sites in enzymes are generally larger than those of non-enzymes. However, if this were the case, the trend would be due to the size of the binding site and not necessarily the protein’s basic function.
Though the size argument above is important to note, it is most likely not the cause of the difference between enzymes and non-enzymes. Several examples of smaller binding sites, characterized by ligands of 40 non-hydrogen atoms or less, are presented in Figure 2 and Figure 3. For small non-enzymes, there are no proteins which show a correlation between size and affinity. Conversely, there are several enzymes with small binding sites which do show a good correlation of increased affinity with increased size.
Ligand Efficiencies
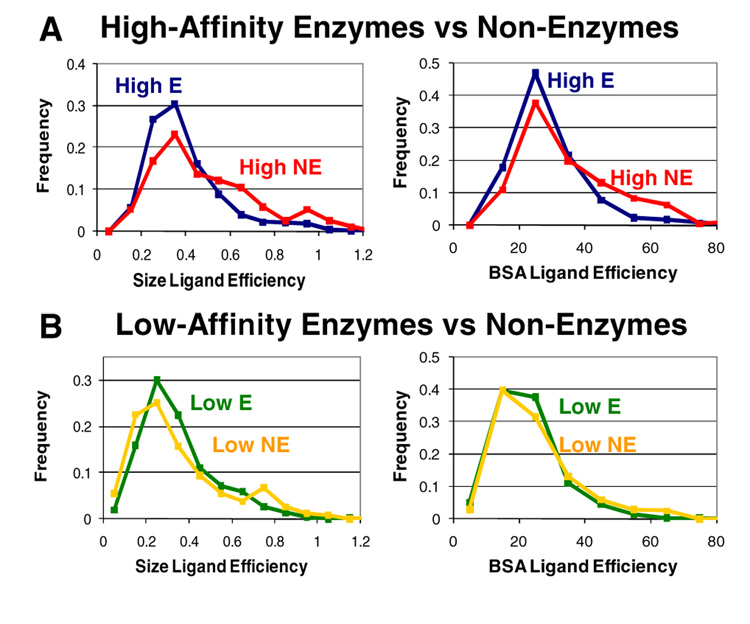
Distributions of ligand efficiencies are given in Figure 4. Ligand efficiency based on contact (−ΔGbind/BSA) can be compared to established values for the desolvation effect. The free energy of transferring a hydrophobic molecule from a hydrophobic solvent into water has been estimated as 24–47 cal/mol-Å2, with the higher value being the most widely accepted.26–28 Honig and coworkers have noted this is lower than the value of 72 cal/molÅ2, derived from the surface tension of a hydrocarbon-water interface.28 Only 0.8% of the complexes in this study have ligand efficiencies that exceed 72 cal/mol-Å2 (i.e., greater than Honig’s value), and many have efficiencies ranging between 20–40 cal/mol-Å2. The low-affinity complexes are roughly bounded by the 47 cal/mol-Å2 value (only 4.1% have greater efficiencies), but the high-affinity complexes have large populations greater than that value. Although, the complexes in Binding MOAD are not exclusively driven by hydrophobic association, these values provide a yardstick for comparisons. However, it should be noted that the range of values from the literature are based on SASA of small molecules in differing environments (ligands), and our values are based on MSA of the contacts within the pockets. While the comparison is not ideal, MSA-based values for ligands are not prevalent in the literature, and SASA of a pocket is not equivalent to SASA of a ligand.
Figure 4.
Distribution of ligand efficiencies per size (–kcal/mol-atom) and per contact (–kcal/mol-Å2), given in normalized percent frequencies. Distributions present comparisons of (A) high-affinity complexes (p<0.0001 in both cases) and (B) low-affinity complexes. High-affinity enzymes are shown in dark blue, and low-affinity enzymes are in green. High-affinity nonenzymes are in red, and low-affinity non-enzymes are in gold.
For low-affinity complexes, the ligand efficiencies are basically the same for enzymes and non-enzymes (Table 1, Figure 4b). However, the differences are significant in high-affinity complexes (p <0.0001 for both efficiencies). The ligand efficiencies for high-affinity, non-enzyme complexes are ~17% greater than those of high-affinity, enzyme complexes (Table 1). Non-enzymes in Figure 4a show a broader distribution of efficiencies and much higher populations above 0.4 kcal/mol-atom (55% of high-affinity non-enzyme complexes vs 37% of high-affinity enzyme complexes) and 30 cal/mol-Å2 (51% of non-enzymes vs 35% of enzymes).
On average over the high-affinity complexes, every atom and square Ångstrom of buried cavity surface is worth more free energy in non-enzymes!
The differences in efficiencies between high-affinity enzymes and non-enzymes are not dependent on the choice of cutoff between high- and low-affinity complexes. Even if the full set of enzymes is compared to the full set of non-enzymes, the ligand efficiencies are better for non-enzyme complexes. For the 1790 enzyme complexes, the median ligand efficiencies are 0.33 kcal/mol-atom and 23 cal/mol-Å2; the median ligand efficiencies for the 424 non-enzymes are 0.36 kcal/mol-atom and 26 cal/mol-Å2.
The same patterns for enzymes and non-enzymes are observed when redundancy is removed (Supporting Information, Table S7, Figures S8 and S9). This is important because it corrects for some biases in the dataset by using only one complex of a protein (some proteins have hundreds of entries and are heavily represented in the PDB). The non-redundant dataset in Binding MOAD is obtained by grouping the proteins into families of 90% sequence identity and representing that family by the single complex with the highest-affinity ligand – in essence, the optimal binding event available for that individual protein. There are 688 unique complexes in this dataset, 512 enzymes and 176 non-enzymes. Again, the high-affinity enzymes (235 complexes) have poorer ligand efficiency than the high-affinity non-enzymes (85 complexes). For the non-redundant datasets, the median ligand efficiencies for high-affinity enzyme complexes are 0.39 kcal/mol-atom and 28 cal/mol-Å2. The median ligand efficiencies for the non-redundant, high-affinity, non-enzyme complexes are still larger at 0.44 kcal/mol-atom and 34 cal/mol-Å2. The smaller number of complexes produces nearly identical distributions, and although the p-value of the comparison is slightly poorer (p = 0.04), it is still significant (96%).
Efficiencies, evolution, and druggability
The significant differences in ligand efficiencies suggest a differentiation in the binding sites of these two classes of proteins, based on their function. This may reflect the different evolutionary pressures upon enzymes and non-enzymes. The higher ligand efficiencies of non-enzymes make them, in essence, more responsive to low concentrations of ligand molecules. This is fitting, given their roles in signaling and regulatory control of cellular function in response to stimuli. Conversely, enzymes are optimized to bind molecules, change them, and release them again.
Ligand efficiencies are one key factor in describing the druggability of a target. Does this imply that non-enzymes may be more druggable? In general, higher ligand efficiencies mean that drug-like affinities can be obtained with smaller molecules. Smaller molecules would tend to provide better oral absorption and fewer functional groups for toxicity concerns.10,29–31 Of course, ligand efficiencies reflect “bindability”, and it is important to recognize that there are additional properties that make a protein a suitable drug target. It must be essential to the disease state. Leads must show selectivity to avoid any negative consequences of off-target binding events. There are a myriad of ADME and pharmacokinetic properties to be considered. However, the differences in ligand efficiencies do indicate a greater likelihood to have better drug-like properties for inhibitors, agonist, and antagonists of non-enzyme targets.
Many non-enzymes are the subject of intense drug discovery efforts in both the private and public sectors; for instance, hormone receptors, signaling proteins, and transcription regulators are targets for anticancer treatment.32,33 Recent discussions on the druggability of protein-protein interfaces note that these difficult targets may be more amenable than originally thought.34,35 Small molecules have been developed that bind to key hot-spot regions with greater efficiencies and deeper burial than the natural partner. Furthermore, many of the non-enzymes not represented in the PDB are membrane-bound receptors. Even though they are not included here, it is likely that the additional information would support the hypothesis that non-enzymes are more druggable, since they are the target of many drugs. G-protein coupled receptors alone constitute 30% of the drugs on the market,30 and genomic analysis has indicated many more receptors are druggable.36
Our results are also in good agreement with a recent study that estimated the druggability of 1096 non-redundant human proteins.10 The predictions used a statistical model trained on NMR-screening data using a small fragment library.37 Four of the top six classes were nonenzymes: vitamin-binding, steroid-binding, lipid-binding, and nucleotide-binding proteins.10 The non-enzymes that were predicted to be the least druggable were large macromolecular complexes and are not reflected in Binding MOAD and this study.
What produces the higher ligand efficiencies in non-enzymes?
Obviously, the root cause of the disparity in ligand efficiencies between enzymes and non-enzymes is of paramount interest. Though the ligands for non-enzymes are smaller, the SlogP characteristics are roughly the same for high-affinity ligands of enzymes and non-enzymes (Figure 1c). If the ligands are chemically similar, then the difference in efficiencies must come from the protein pocket. The most significant difference is the degree of exposure for ligands of non-enzymes versus enzymes. High-affinity ligands have a median exposure of only 11% in non-enzymes, but 25% in enzymes (note that %ESA are used instead of ESA to correct for the difference in sizes of the ligands). Low-affinity ligands for non-enzymes are significantly more exposed (median of 33%), even more than the low-affinity ligands for enzymes (22%). Tight and weak inhibitors have the same degree of exposure in enzymes, but tight ligands for non-enzymes are much more encapsulated than the weak ligands (p<0.0001). Other 2D and 3D ligand descriptors displayed no significant patterns. This comparison was cognizant of correlations between characteristics; for instance, differences in surface area are correlated to size and were not “double counted” as additional differences between high-affinity ligands of enzymes vs non-enzymes.
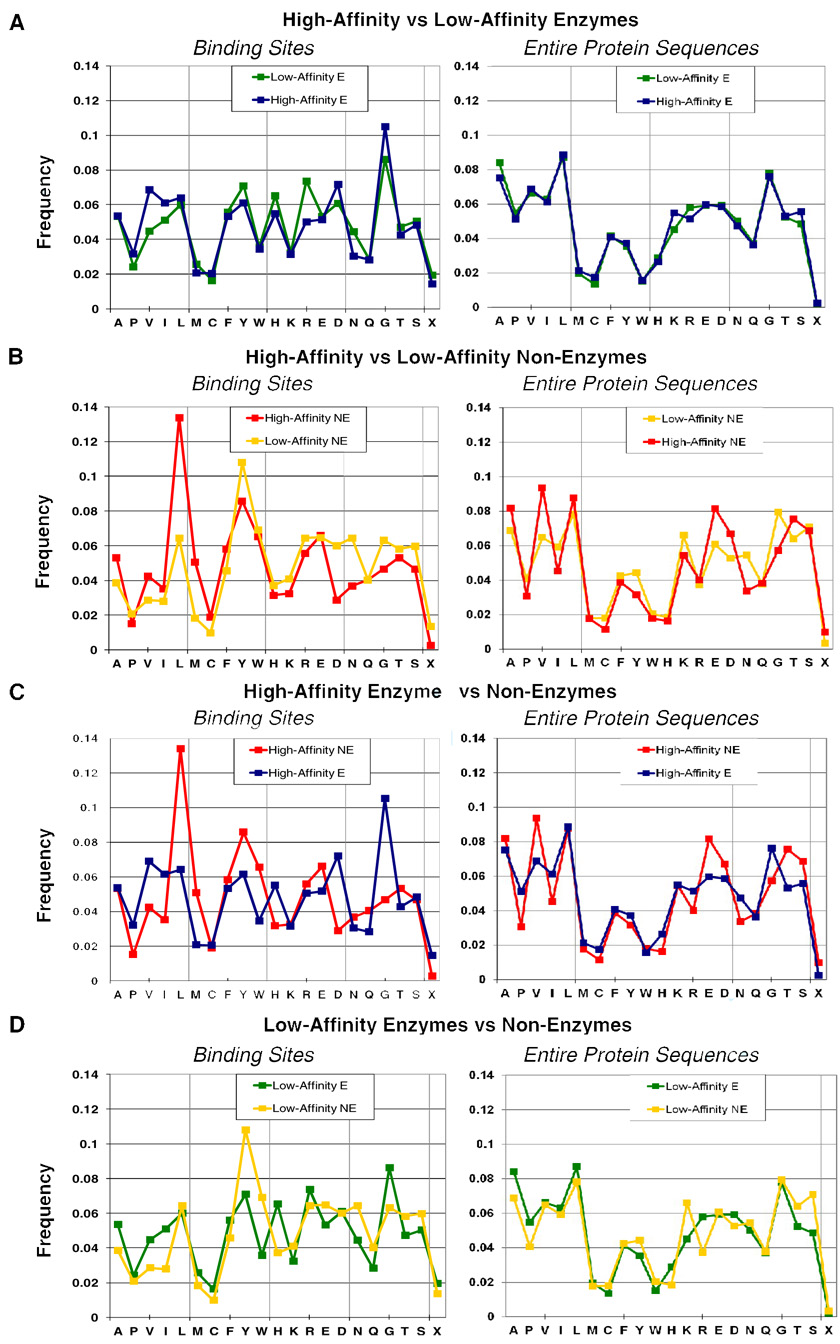
Amino acid composition of the binding sites was examined (Figure 5, left column). There is little difference between the binding sites of high- and low-affinity enzyme complexes. The largest differences are an increase in Val content in high-affinity enzymes and an increase in Arg in the low-affinity complexes. For enzymes, the hydrophobic residues (Ala through Trp) on Figure 5 are 47.0% of the binding sites for high-affinity complexes, but 43.9% for low-affinity ones. This is fitting with the aforementioned finding that the high-affinity ligands are slightly more hydrophobic. The comparison between binding sites of high- and low-affinity non-enzyme complexes shows more pronounced variation, but also holds the general pattern of high-affinity complexes having more hydrophobic content. The Ala-Trp residues are 55.9% of the binding sites for high-affinity complexes, but 43.2% for low-affinity ones. What is most interesting is the comparison between enzymes and non-enzymes, particularly for the high-affinity complexes. The hydrophobic content is higher for non-enzymes (55.9% vs 47.0%), but the reader should recall that there is no significant difference in the SlogP of the ligands (in fact, the median value for non-enzymes is more hydrophilic). Why are more hydrophobic sites recognizing slightly more hydrophilic molecules with better affinity? The answer may lie in the fact that the amino acids making the contacts are significantly different. In high-affinity non-enzymes, Leu and Met provide a large portion of the hydrophobic contacts, at the expense of Val and Ile. The nonenzyme’s preference for Glu over Asp is reversed in high-affinity enzyme complexes, yet the use of Lys and Arg is the same. Leu, Met, and Glu are larger than their counterparts Val, Ile, and Asp. It is possible that those residues are slightly more polarizable. (Confirmation will have to come from in-depth examinations of fully modeled complexes, inclusive of added hydrogens, detailed atom typing, and possibly polarizable force fields. To do this for thousands of complexes is a sizable effort, and outside the scope of the present study.) It should be noted that differences in the binding sites are not correlated with differences in the overall amino acid content; the reader should compare the left and right columns in Figure 5. Leu, Met, Phe, Tyr, and Trp make up nearly the same percentage of residues in the protein sequences, but not the binding sites. This selective placement of differing residues within binding pockets may have direct relevance to analyses of hot-spot regions and potential binding sites on proteins.38–40
Figure 5.
The binding sites (left) and the entire protein sequences (right) are analyzed for amino acid content. Distributions are given in normalized frequencies percent frequencies. Amino acids within 4Å of the ligands are considered to comprise the binding site. Distributions of (A and B) low- and high-affinity complexes of the same class show smaller differences than comparisons between enzymes and non-enzymes (C and D). Amino acids are listed by hydrophobic, aromatic, cationic, anionic, and hydrophilic nature. “X” denotes contacts with cofactors, unnatural amino acids, and covalent modifications on the protein.
Most druggable enzymes
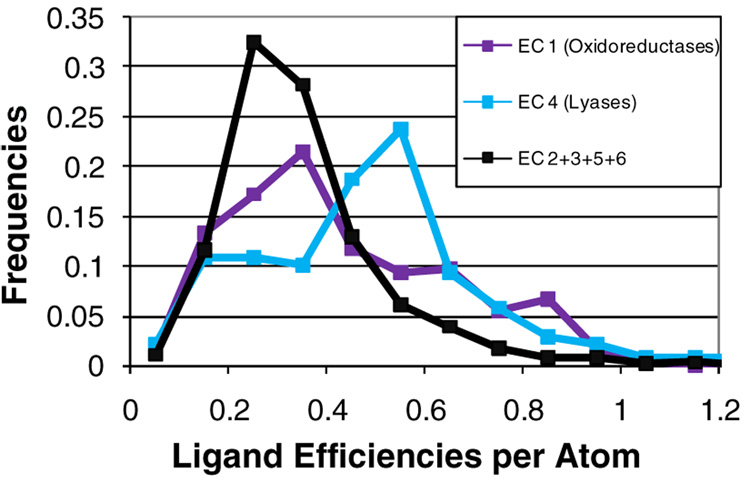
Of course, many pharmaceutically relevant targets are enzymes. By no means is it suggested that they are not appropriate drug targets, especially when they constitute 47% of the drugs on the market30 and a large percentage of new targets identified through genomic analysis.36 The distribution of ligand efficiencies for the enzyme classes suggests that lyases and oxidoreductases are the most druggable enzymes, Figure 6. The distribution of lyases is significantly shifted to higher efficiencies, standing out from the other data. The better efficiencies for oxidoreductases come from an increased population in the tail of the distribution. The median ligand efficiencies for the 139 lyases are 0.50 kcal/mol-atom and 33 cal/mol-Å2; and the median ligand efficiencies for the 256 oxidoreductases are 0.39 kcal/mol-atom and 26 cal/mol-Å2. The 1395 enzymes from the other four classes have median efficiencies of 0.31 kcal/mol-atom and 23 cal/mol-Å2, which are significantly lower (significance of ≥99.99% using the Wilcoxon test). It should be noted that the two enzymes which were predicted to be most druggable in the aforementioned study were also lyases and oxidoreductases, in that order.10
Figure 6.
Distribution of ligand efficiencies (–kcal/mol-atom) for enzymes, given in percent frequencies normalized for the different number of complexes in each enzyme class. The distribution of transferases (EC 2, 468 complexes), hydrolases (EC 3, 843 complexes), isomerases (EC 5, 60 complexes), and ligase (EC 6, 17 complexes) are the same and have been added together for this example (black line). Oxidoreductases (EC 1, purple line, 256 complexes) have larger populations in the higher efficiencies (p<0.0001). The distribution of lyases (EC 4, blue line, 139 complexes) is notably shifted (p<0.0001).
Recently, a new method was introduced to predict druggability of a binding site by estimating the site’s maximum Kd based on the percent hydrophobic SASA and a scaling factor for efficiency that is dependent on the curvature of the site.9 The model was trained on 8 enzymes and applied to 63 structures, comprised of complexes of 26 enzymes and a single structure of the non-enzyme mdm2.41 An important goal of the study was to fit a predictive equation to assess druggability of a site based on protein-ligand structures of orally available compounds. This feature of the study is important to note because the contributions of various physical characteristics within the model should reflect both high-affinity binding and oral bioavailability of the ligand. The model was fit under the assumption that hydrophobic desolvation is the major driving force of binding, so terms based on electrostatics were not included. The model was able to properly rank the training set, noting that outliers were compounds with strong electrostatic components, prodrugs, or ligands that are actively transported. The model was then used to identify new, druggable structures from the PDB. It was interesting that the two newly identified targets were both enzymes. With only two new targets presented, it is not clear whether the model preferentially identifies enzymes over non-enzymes, but a preference towards enzymes may be expected from their model given the training and test sets used. Our data indicate that enzymes and non-enzymes may require different models in such analyses. Furthermore, many of the ligand efficiencies in our set exceed the established values for hydrophobic association, indicating that the most efficient complexes have additional factors which contribute to their affinity. The affinity of these complexes may not be well described by models based solely on hydrophobic SASA.
Conclusion
We have presented a substantial mining study of Binding MOAD, the largest public database of curated protein-ligand structures with binding data. Physical characteristics of bound ligands were compared between enzymes and non-enzymes as well as high-affinity and low-affinity complexes. The comparison between ligand sizes for low-affinity versus high-affinity binding shows that divergent approaches are likely needed to improve the affinity of enzyme inhibitors versus those for non-enzymes. The traditional approach of adding functional groups to fill more of the pocket may work for enzymes, but it may not be as appropriate for non-enzyme systems. However, making ligands more hydrophobic appears to aid binding in both enzymes and non-enzymes.
Non-enzymes have higher ligand efficiencies than enzymes, which may be a reflection of their biological roles. This is also encouraging when considering the druggability of nonenzymes. In the pharmaceutical industry, ligand efficiencies have become a metric for evaluating hits from screening campaigns and even candidate compounds.12 Our results would caution against applying a rigid standard across all protein targets. At the very least, a cutoff based on ligand efficiency should differ between enzymes and non-enzymes. Ideally, cutoffs would differ between protein families and only be considered as one of several guidelines in a selection process.
Binding MOAD provides strong support of several mathematical models cited above,10,24,41 particularly those of Hajduk and coworkers. Our results have implications for the development of scoring functions for docking and predicting druggability of a binding site.42–45 The differences between non-enzymes and enzymes, as well as the differences across enzymatic systems, underscore the challenges of developing universal functions that perform well across all systems. Modest improvement might be achieved by developing separate functions for enzymes and non-enzymes, with even greater improvement expected for functions trained on specific protein families.
Supplementary Material
The following information is available in the supporting information: box plots and distribution analysis from JMP; Tukey-Kramer HSD analysis; patterns obtained from the non-redundant dataset and the full dataset without protein-cofactor complexes; data for the three enzymes with a large range in affinities for a small range of ligand sizes; listing of the high-affinity complexes.
Acknowledgements
This work was funded by a Beckman Young Investigator Award, an NSF CAREER Award (MCB-0546073), and the National Institutes of Health (HG003890). RDS thanks the Molecular Biophysics Training Program for support (GM008270). NAK thanks the Training Program in Bioinformatics for support (GM070449). We thank Chemical Computing Group, Inc. for their generous donation of MOE for calculating the physical properties of the ligands. The authors greatly appreciate Prof. Bruce Mueller and Dr. Jignesh Patel for their invaluable help with statistical analysis and the SAS program. Thanks are also due to many other colleagues at the University of Michigan who have provided valuable feedback.
Abbreviations
- MOAD
Mother of All Databases
- PDB
Protein Data Bank
- BSA
buried surface area
- ESA
exposed surface area
- MSA
molecular surface area
- SASA
solvent-accessible surface area
- ΔGbind
free energy of binding
- KNI-10033
(4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-[[(2R)-2-(2-isoquinolin-5-yloxyethanoylamino)-3-methylsulfanyl-propanoyl]amino]-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide
- KNI-10075
(4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-[[(2R)-2-(2-isoquinolin-5-yloxyethanoylamino)-3-methylsulfonylpropanoyl]amino]-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide
References
- 1.Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Prot. Struct. Func. Bioinfo. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 2.Smith RD, Hu L, Falkner JA, Benson ML, Nerothin JP, Carlson HA. Exploring protein-ligand recognition with Binding MOAD. J. Mol. Graph. Model. 2006;24:414–425. doi: 10.1016/j.jmgm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama Y. Druggability: Selecting optimized drug candidates. Drug Discov. Today. 2005;10:1577–1579. doi: 10.1016/S1359-6446(05)03675-5. [DOI] [PubMed] [Google Scholar]
- 6.Norvell JC, Machalek AZ. Structural genomics programs at the US National Institute of General Medical Sciences. Nat. Struct. Biol. 2000;(7 Suppl):931. doi: 10.1038/80694. [DOI] [PubMed] [Google Scholar]
- 7.Luque I, Freire E. Structure-based prediction of binding affinities and molecular design of peptide ligands. Methods Enzymol. 1998;295:100–127. doi: 10.1016/s0076-6879(98)95037-6. [DOI] [PubMed] [Google Scholar]
- 8.Williams DH, Stephens E, O’Brien DP, Zhou M. Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors. Angew. Chem. Int. Ed. 2004;43:6596–6616. doi: 10.1002/anie.200300644. [DOI] [PubMed] [Google Scholar]
- 9.Coleman RG, Salzberg AC, Cheng AC. Structure-based identification of small molecule binding sites using a free energy model. J. Chem. Inf. Model. 2006;46:2631–2637. doi: 10.1021/ci600229z. [DOI] [PubMed] [Google Scholar]
- 10.Hajduk PJ, Huth JR, Tse C. Predicting protein druggability. Drug Discov. Today. 2005;10:1675–1682. doi: 10.1016/S1359-6446(05)03624-X. [DOI] [PubMed] [Google Scholar]
- 11.Cele AZ, Metz JT. Ligand efficiency indices as guideposts for drug discovery. Drug Discov. Today. 2005;10:464–469. doi: 10.1016/S1359-6446(05)03386-6. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 13.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nat. Rev. Drug Discov. 2004;3:660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- 15.Molecular Operating Environment (MOE), 2007.08. Montreal, CN: Chemical Computing Group, Inc.; 2007. [Google Scholar]
- 16.Wildman SA, Crippen GM. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 1999;39:868–873. [Google Scholar]
- 17.SAS, release 9.1. Cary, N.C.: SAS Institute Inc.; 2002–2003. [Google Scholar]
- 18.JMP, release 7.01. Cary, N.C.: SAS Institute Inc.; 2007. [Google Scholar]
- 19.Coleman RG, Sharp KA. Travel depth, a new shape descriptor for macromolecules: Application to ligand binding. J. Mol. Biol. 2006;362:441–458. doi: 10.1016/j.jmb.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 20.Wang R, Fang X, Lu Y, Yang CY, Wang S. The PDBbind database: Methodologies and updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 21.Yang C-Y, Wang R, Wang S. M-Score: A knowledge-based potential scoring function accounting for protein atom mobility. J. Med. Chem. 2006;49:5903–5911. doi: 10.1021/jm050043w. [DOI] [PubMed] [Google Scholar]
- 22.Babaoglu K, Shoichet BK. Deconstructing fragment-based inhibitor discovery. Nat. Chem. Biol. 2006;2:720–723. doi: 10.1038/nchembio831. [DOI] [PubMed] [Google Scholar]
- 23.Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery: leads by design. Drug Discov. Today. 2005;10:987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 24.Hajduk PJ. Fragment-based drug design: how big is too big? J. Med. Chem. 2006;49:6972–6976. doi: 10.1021/jm060511h. [DOI] [PubMed] [Google Scholar]
- 25.Lafont V, Armstrong AA, Ohtaka H, Kiso Y, Amzel LM, Freire E. Compensating enthalpic and entropic changes hinder binding affinity optimization. Chem. Biol. Drug Des. 2007;69:413–422. doi: 10.1111/j.1747-0285.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 26.Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974;248:338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- 27.De Young LR, Dill KA. Partitioning of nonpolar solutes into bilayers and amorphous n-alkanes. J. Phys. Chem. 1990;94:801–809. [Google Scholar]
- 28.Sharp KA, Nicholls A, Fine RF, Honig B. Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science. 1991;252:106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- 29.An J, Totrov M, Abagyan R. Comprehensive identification of "druggable" protein ligand binding sites. Genome Inform. 2004;15:31–41. [PubMed] [Google Scholar]
- 30.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 31.Kubinyi H. Drug research: myths, hype and reality. Nat. Rev. Drug Discov. 2003;2:665–668. doi: 10.1038/nrd1156. [DOI] [PubMed] [Google Scholar]
- 32.Strachan RT, Ferrara G, Roth BL. Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discov. Today. 2006;11:708–716. doi: 10.1016/j.drudis.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Whitty A, Kumaravel G. Between a rock and a hard place? Nat. Chem. Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 34.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 35.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15422–15427. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russ AP, Lampel S. The druggable genome: an update. Drug Discov. Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 37.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J. Med. Chem. 2005;48:2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 38.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. Unraveling hot spots in binding interfaces: Progress and challenges. Curr. Opin. Struc. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 40.Soga S, Shirai H, Kobori M, Hirayama N. Use of amino acid composition to predict ligand-binding sites. J. Chem. Inf. Model. 2007;47:400–406. doi: 10.1021/ci6002202. [DOI] [PubMed] [Google Scholar]
- 41.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, Salzberg AC, Huang ES. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 42.Brooijmans N, Kuntz ID. Molecular recognition and docking algorithms. Annu. Rev. Biophys. Biomol Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 43.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Prot. Struct. Func. Genetics. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 44.Krovat EM, Steindl T, Langer T. Recent advances in docking and scoring. Curr. Comput.-Aided Drug Des. 2005;1:93–102. [Google Scholar]
- 45.Mohan V, Gibbs AC, Cummings MD, Jaeger EP, DesJarlais RL. Docking: successes and challenges. Curr. Pharm. Des. 2005;11:323–333. doi: 10.2174/1381612053382106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following information is available in the supporting information: box plots and distribution analysis from JMP; Tukey-Kramer HSD analysis; patterns obtained from the non-redundant dataset and the full dataset without protein-cofactor complexes; data for the three enzymes with a large range in affinities for a small range of ligand sizes; listing of the high-affinity complexes.