Abstract
Islet amyloid polypeptide (IAPP, also known as amylin) is responsible for pancreatic amyloid deposits in type 2 diabetes. The deposits, as well as intermediates in their assembly, are cytotoxic to pancreatic β-cells and contribute to the loss of β-cell mass associated with type 2 diabetes. The factors which trigger islet amyloid deposition in vivo are not well understood but peptide membrane interactions have been postulated to play an important role in islet amyloid formation. To better understand the role of membrane interactions in amyloid formation two-dimensional infrared spectroscopy was used to compare the kinetics of amyloid formation for human IAPP both in the presence and in the absence of negatively charged lipid vesicles. Comparison of spectral features and kinetic traces from the two sets of experiments provides evidence for the formation of an ordered intermediate during the membrane-mediated assembly of IAPP amyloid. A characteristic transient spectral feature is detected during amyloid formation in the presence of vesicles which is not observed in the absence of vesicles. The spectral feature associated with the intermediate raises in intensity during the self assembly process and subsequently decays in intensity in the classic manner of a kinetic intermediate. Studies with rat IAPP, a variant which is known to interact with membranes but does not form amyloid, confirm the presence of an intermediate. The analysis of 2D IR spectra in terms of specific structural features is discussed. The unique combination of time and secondary structure resolution of 2D IR spectroscopy has enabled the time-evolution of a hIAPP intermediate to be directly monitored for the first time. The data presented here demonstrates the utility of 2D IR spectroscopy for studying membrane-catalyzed amyloid formation.
Keywords: Amylin, IAPP, Peptide Membrane Interactions, Two-dimensional Infrared Spectroscopy
INTRODUCTION
The aggregation of proteins into amyloid plaques plays a role in more than 20 different human diseases, including type 2 diabetes, Alzheimer’s and Parkinson’s diseases. Each disease is associated with a different protein, although the plaques have many characteristics in common.1, 2 They consist of a dense meshwork of rigid fibrils typically 50 to 100 Å in diameter and of varying lengths. The fibrils are formed by proteins adopting a cross-β configuration3, 4 in which the individual β-strands are oriented perpendicular to the long axis of the fibrils.4, 7 Amyloid deposits can be destructive through direct obstruction of tissue, as for example in light chain deposition disease, or by disrupting normal islet architecture in type 2 diabetes,8 or by causing cell dysfunction and death,9-12 leading to decreased production of insulin. However, the mechanism by which amyloid plaques cause cell death is poorly understood and it is not clear that the deposits themselves are the major cause of the insulin loss. Increasing evidences highlight a potential role for intermediates in the assembly of amyloid as the key toxic species.13-16 Thus, while the presence of amyloid deposits is diagnostic, the plaques themselves are only one part of the disease mechanism since the symptoms of these diseases can occur with little or no amyloid formation.
Cell dysfunction might be caused by unfolded or partially aggregated proteins interacting with the cell membranes before amyloid deposits develop. In vitro studies have found that the polypeptide associated with type 2 diabetes, the human islet amyloid polypeptide (hIAPP also known as amylin), which is the focus of the present work, forms cation-permeable pores in membranes.17 Furthermore, amyloid formation causes vesicles to leak by extracting lipids15 and forming porous holes.13 In addition, prefibrillar assemblies of undefined hIAPP structure degrade vesicles.14 In vivo studies have found that transgenic mice expressing hIAPP require a high fat diet or altered lipid metabolism in order for amyloid formation to occur.12 Finally, amyloid fibrils form much faster in the presence of model membranes.18-20 Thus, there is strong evidence, at least for some amyloid diseases, that plaques are not the central feature of the pathogenic process and that the cytotoxic species may be a transient structure that somehow interacts with cellular membranes, according to the in vitro biophysical studies.
The realization that the cytotoxic species are likely to be intermediates in the aggregation processes has spurred many kinetic studies to follow fiber formation and elucidate the structures of intermediate species. However, most standard structural tools (such as NMR) are difficult or impossible to apply to studies of the kinetics of amyloid. As a result, the most commonly used techniques are fluorescence, using dyes such as thioflavin T (ThT) that bind to amyloid deposits and circular dichroism (CD) spectroscopy. ThT fluorescence is a good marker for fibril formation and can be used to monitor fibril growth kinetics, but it provides no information on intermediates since it does not generally bind to pre-fibril species.18, 19 CD can also be used to follow the kinetics of fibril assembly as well as provide information on secondary structure, although there can be technical and interpretive difficulties in membrane peptide studies because of light scattering artifacts and the fact that the CD spectra of amyloid fibrils are not well understood.19, 20 Indeed, our understanding of the CD spectra of β-sheets is, in general, much less developed than our understanding of the CD spectra of α-helical proteins and peptides. Nonetheless, CD is an important tool and there is evidence from CD studies that the formation of fibrils associated with type 2 diabetes proceeds through α-helical intermediate.21 Similar evidence of α-helical transients from CD spectra is also observed for hIAPP when the aggregation pathway is catalyzed by lipid vesicles.19, 20 Very recently, electron spin-resonance (ESR) studies of spin-labeled peptides found that residues 9 to 22 near the N-terminal do indeed form a helix upon binding to membranes.22 ESR studies have yielded important information however they necessarily involve perturbing the system by introducing a Cys mutation and then attaching a relatively large and fairly polar nitroxide spin label. Membranes appear to act as catalysts for fiber formation because the rate of fiber formation increases when vesicles are present and the final fiber structure is the same when grown with or without membranes,1, 18 although there is evidence that fiber formation disrupts the membranes themselves.15
In this paper, we use automated two dimensional infrared spectroscopy (2D IR) spectroscopy to probe the folding pathway of hIAPP in dilute solution and when catalyzed by membranes. We aim to address the differences in the aggregation pathway in the presence and absence of lipid vesicles. In addition, we wish to demonstrate the broad utility of automated 2D IR for these types of investigations. 2D IR is a relatively new spectroscopic technique but is beginning to be applied to a number of biophysical questions. Unfortunately problems associated with light scattering, a key difficulty in studies of membrane proteins and amyloid, plus other technical issues can make the method difficult to apply to heterogeneous systems. Automated 2D IR spectroscopy utilizes a new technology for generating mid-IR pulse trains that allows spectra to be collected in a rapid-scan mode.23 In this mode, 2D IR spectra are collected in a continuous fashion, about one spectrum per 0.5 second, and then post-processed to achieve the necessary signal-to-noise ratio. Importantly, the methodology also allows the use of phase cycling to reduce artifacts caused by scattering; a feature which is particularly important for studying heterogeneous systems such as peptide membrane samples. This rapid-scan version of 2D IR spectroscopy is very different from standard methods that take many minutes or hours for a single spectrum and often require repeated sample preparations for signal-to-noise averaging.24 Precisely repeating an experiment is problematic for aggregating systems like amyloids in which the aggregation process typically follows slightly different kinetics from experiment to experiment due to small differences in nucleation times. Thus, on-the-fly measurements are necessary. A significant advantage of this technique is that it gives unprecedented time resolution when compared to any other spectroscopic technique that has been applied to study the kinetics of amyloid formation. In addition, IR spectra are well known to be sensitive to secondary structure. We have used this new technology to monitor the folding kinetics of hIAPP aggregation in dilute solution.25, 26 In this paper, we compare the aggregation pathways in the presence and absence of membranes. We find significant differences in the spectra and kinetics, which provide evidence for the population of an intermediate during the membrane-catalyzed reaction.
EXPERIMENTAL METHODS
Peptide Preparation
hIAPP and rat IAPP (rIAPP) were purchased from Bachem and dissolved in deuterated hexafluoroisopropanol (d-HFIP) at a concentration of 0.5 mM. An aliquot of the d-HFIP stock solution was made into D2O solution after evaporating d-HFIP. The hIAPP or rIAPP D2O solution was lyophilized in 0.1 mM DCl multiple times and then one time in D2O to remove residual TFA. The sample was then redissolved in d-HFIP. The denaturant HFIP was then removed by evaporation under a stream of nitrogen. This protocol is similar to others in the literature and is commonly used in studies of IAPP.18-20
Large Unilamellar Vesicle (LUV) Preparation
1, 2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1, 2-dioleoyl-sn-glycero-3-phosphate (DOPA) chloroform solutions were obtained from Avanti. Appropriate amounts of the two lipid solutions were mixed at a molar ratio of 7: 3. The chloroform was evaporated under nitrogen gas flow and the samples were dried in a vacuum desiccator overnight. The dry lipid film was rehydrated in D2O at pH 6, subjected to five freeze-thaw cycles, and extruded with a mini-extruder fitted with a 100 nm polycarbonate membrane (Avanti) to produce LUVs with a diameter of 100 nm.
Sample Preparation
To initiate the control experiment of folding without vesicles, the peptide sample (hIAPP or rIAPP) was redissolved in 5 μL 4% dimethyl sulfoxide (DMSO) D2O at pH 6 to give a final peptide concentration of 0.5 mM. For vesicle catalysis experiments, 0.5 μL lipid D2O solution (about 80 mM lipid concentration) was added to the hIAPP or rIAPP sample at pH 6. The final concentration of peptide was approximately 0.45 mM and the final concentration of lipid was approximately 7 mM. The partition coefficient of IAPP between negatively charged vesicles and water is about 50000, so that only a tiny fraction of hIAPP does not associate with the membrane under these conditions.20 For the control experiment, no lipid was added to the peptide sample. The aggregation process was monitored in IR cell which consisted of two CaF2 windows separated by a 100 um Teflon spacer. Residual concentrations of DMSO (< 5% v/v) have no measurable effect on membrane integrity.18 This method of sample preparation leads to a dead time of about 2 minutes.
Automated 2D IR Spectroscopy
The aggregation was monitored using our rapid-scan 2D IR spectrometer in a pump-probe beam geometry.27 The pump has two pulses whose delays are set by a Ge acousto-optic modulator (AOM) based pulse shaper, as has been reported previously.23, 28 Briefly, femtosecond mid-IR pulses were initially generated by difference frequency mixing (DFM) of two femtosecond near-IR outputs of a β-barium borate (BBO)-based optical parametric amplifier (OPA). A ruled grating (150 g/mm) was then used to disperse the mid-IR pulses into the frequency domain. The Ge AOM was used to modulate the phase and intensity of the frequency profile so as to create the desired phase cycled pulse sequence. Rather than implementing a pump-probe experiment in the most traditional sense, so that
| (1) |
we instead implemented a phase-cycling scheme to improve the signal-to-noise and remove scatter from pump pulses. In this phase cycling scheme, we took the difference in transmission generated from four pump pulse trains with absolute phase shifted by π so that
| (2) |
The waveform generator for the pulse shaper can continuously cycle 426 pulse shapes at a time, which amounts to 106 time delays when using four phase cycles. To scan over >2 ps of the first coherence time with the 106 time delays, the relative phase of the pump pulse pair was cycled with a rotating frame frequency of 1225 cm-1, which enables regular sampling with a 22 fs step. It took 0.43 seconds to scan a single 2D IR spectrum at a 1 kHz repetition rate. The 2D IR spectra reported in this manuscript were generated by averaging 500 consecutive scans. For the kinetic traces, each data point is calculated from a running average over 300 scans. Thus each spectrum represents a running average over a time period of 5 minutes and each data point in the kinetic traces defines a time resolution of 3 min.
RESULTS
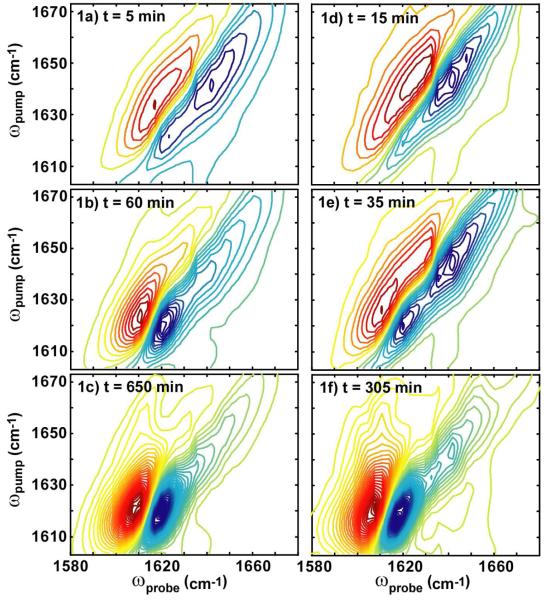
Shown in Fig. 1a-c are representative series of 2D IR spectra collected during the aggregation process of hIAPP in the absence of membranes. The data are comparable to previously published spectra. The first 2D IR spectrum shown in Fig. 1a is at t = 5 minutes. The most prominent features in these spectra are a pair of out-of-phase peaks at ωpump = 1642 cm-1. In 2D IR spectra, vibrational modes create doublets in which the negative peak is located on the diagonal and includes transitions between the vibrational ground state and its first excited state (e.g. ν=0→1).27 A diagonal slice through this peak can be interpreted much like a traditional infrared absorption peak, albeit with improved frequency resolution. The positive peak is created by pulse orderings that include transitions up to the overtone (e.g. ν=1→2) so that it is shifted along ωprobe by the anharmonic shift, which is about 17 cm-1. The large diagonal widths of these peaks and the anharmonic shift are typical of random coil peptide structures with large structural distributions, which is consistent with the unfolded nature of hIAPP. While this region is traditionally assigned to random coil structures, the so-called “random coil” region also contains well-defined secondary structures such as α-helix and small β-sheets.29 Thus, when referring to this spectral region, we use quotes to remind the reader that this region is not purely random coil, which is a topic we discuss in more depth below (see Discussion: Possible structural intermediates). As aggregation proceeds, a doublet appears near ωpump = 1617 cm-1. This feature is indicative of β-sheet formation and the doublet is caused by the anti-symmetric stretch motion of the amide I bands. The normal mode is caused by strong coupling between the amide I (carbonyl) vibrational modes to create exciton states. The anharmonic shift of this transition is smaller, which is another consequence of exciton coupling.30, 37 Thus, the doublet at 1617 cm-1 can be used to monitor the kinetics of β-sheet growth, while the doublet at 1642 cm-1 monitors the disappearance of “random coil” and the appearance of other secondary structures including α-helix.
Figure 1.
(a-c) Representative 2D IR spectra at folding time t = 5 minutes, 60 minutes and 650 minutes during uncatalyzed aggregation and (d-f) three spectra at folding times t = 15, 35 and 305 minutes during lipid vesicle (7: 3 DOPC/DOPA) catalyzed aggregation. Lipid vesicles were added at t = 14 min. Red contours are positive and correspond to the same intensity interval in all spectra.
Shown in Fig. 1d-f are a set of spectra recorded for hIAPP aggregation under identical conditions, except that hIAPP fibril formation was catalyzed by lipid vesicles made up of a mixture of 7: 3 DOPC: DOPA in D2O. This composition of lipids was chosen based on previous biophysical studies which characterized hIAPP fiber formation kinetics using Thioflavin T fluorescence and circular dichroism (CD) spectroscopy.19 The vesicles were added at 14 minutes into the folding kinetics. Fig. 1d shows the hIAPP spectrum immediately after lipid was added. Fig. 1e and 1f are spectra of hIAPP with lipid vesicles at later times. At first glance, the comparisons between the catalyzed and uncatalyzed reactions look superficially similar: the β-sheet features increase with time as amyloid fibrils are formed and the final 2D IR spectra have very similar features. These generic observations are consistent with what is already known about fibril formation: vesicles catalyze fibrils and that the fibrils have close to the same, if not identical structures, according to transmission electron microcopy (TEM) data.1, 18 However, closer inspection of the data reveals several interesting differences that can only be caused by large differences in the respective self-assembly pathways. In what follows, we illustrate the main three differences we found between experiments performed with and without vesicles, which are differences in (1) kinetics, (2) anharmonic shifts, and (3) line widths.
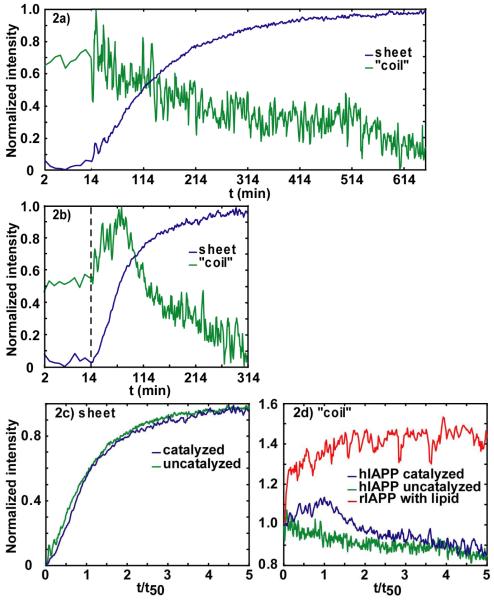
A comparison of the kinetics for the uncatalyzed and catalyzed processes is shown in Fig. 2a and 2b, respectively. The kinetics of β-sheet and “random coil” secondary structures are followed by monitoring their diagonal peak frequencies at 1617 and 1642 cm-1, respectively. For the uncatalyzed processes, we see the same trend in the kinetics that was reported previously.25, 26 Following a lag time of about 10 minutes, the β-sheet intensity increases while the random coil intensity decreases. Although the exponential-like changes in intensity of the random coil and β-sheet features are slightly different, which suggests a possible intermediate, the differences are subtle and there are no distinguishable spectroscopic signatures of intermediates found in the 2D IR spectra.25 However, when fibril formation is catalyzed by membranes, the differences are striking, which is the focus of this paper. One important difference is that β-sheet formation occurs more quickly, as expected for a catalyzed folding event, with a time-to-half-maximum (t50) of 60 minutes as compared to the uncatalyzed reaction which has t50 = 110 minutes. Fluorescence studies have demonstrated that when the time-axes are scaled, the kinetics of fibril formation with and without membrane catalysis exhibit identical progress curves,18 which is also the case here. Shown in Fig. 2c are the β-sheet kinetics with the x-axis scaled so that the time-to-half-maximum intensity (t50) is normalized to unity. In this plot, the two kinetics traces are nearly identical, which would suggest that the aggregation pathways are identical. Thus, other than by the difference in timescales, the aggregation pathway with and without vesicles cannot be differentiated by β-sheet kinetics alone.
Figure 2.
Intensities in anti-symmetric β-sheet and “random coil” regions versus aggregation time for (a) uncatalyzed and (b) membrane-catalyzed hIAPP folding. The dashed line indicates at t=14 minutes lipid vesicles were added. Intensities are normalized to minimum and maximum of individual kinetic traces. (c) Normalized intensities of anti-symmetric β-sheet feature for uncatalyzed (green) and catalyzed (blue). (d) Intensities in “random coil” region (1642 cm-1), for the (green) uncatalyzed, (blue) catalyzed hIAPP experiment and (red) rIAPP experiment. Intensities are normalized to starting values at t= 0.
While the kinetics of β-sheet formation is similar when scaled, there are striking differences in the kinetics of the “random coil” frequency range, shown in Fig. 2d. In the uncatalyzed reaction, the loss of “random coil” is well-fit to a single exponential.25 In the catalyzed reaction, the “random coil” intensity instead increases, coming to a maximum at t = 60 minutes (which corresponds closely to the t50 time of the β-sheet), before exponentially decreasing, as shown in Fig. 2b. This maximum is never observed without lipid vesicles present in the solution. A rise and fall in intensity is observed at all frequencies between 1635 and 1655 cm-1. Regardless of the precise structural origins of these intensities, the rise and fall of the catalyzed reaction proves that the aggregation process is occurring through an intermediate structure or structures that absorbs near the random coil frequency.
To investigate this intermediate further, we also measured the kinetics of rat IAPP (rIAPP) with vesicles. rIAPP is particularly useful in this regard since it is known to associate with membranes but does not form amyloid even in the presence of lipid vesicles.19, 20 The intensity of rat IAPP at 1642 cm-1 is plotted in Fig. 2d after addition of lipid vesicles (following the same protocols as for hIAPP. See Materials and Methods). A clear rise in intensity is observed which then levels off. Since it only takes a few hundred microseconds for rIAPP to diffuse to the vesicle surface, the rise in intensity at the beginning part of the rIAPP kinetics trace correlates with the formation of rIAPP membrane bound structure. Thus, by comparison to hIAPP, we conclude that the rise and fall of the catalyzed kinetics in hIAPP is created by a structural intermediate bound to the membrane.
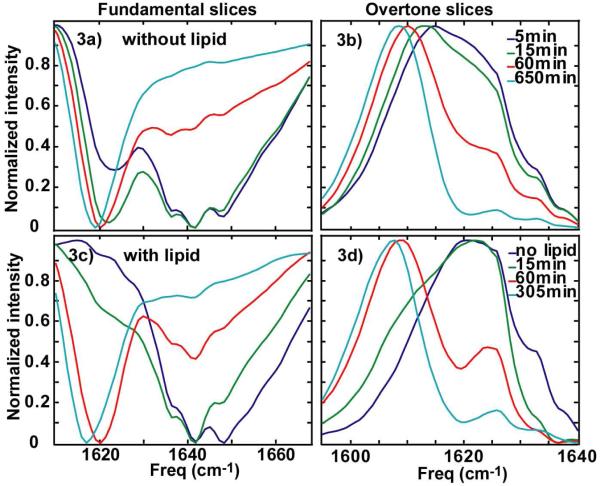
Membrane catalysis also narrows the structural distribution of hIAPP, which is observed in the diagonal line widths of the 2D IR spectra. Shown in Fig. 3a and 3c are slices along the diagonal for several 2D IR spectra. In the uncatalyzed case, the “random coil” feature exhibits a main peak at 1642 cm-1, as well as two shoulders that appear at 1636 and 1648 cm-1. Together, they have a full-width-at-half maximum (FWHM) of 30 cm-1. We cannot resolve the relative intensities of these three features, although they all disappear in tandem during the aggregation process, suggesting that they all arise from the same structural assembly. What is interesting is that the addition of lipids changes the relative intensity of these three features by significantly reducing the contribution from the highest frequency feature. The effect serves to decrease the FWHM of the “random coil” feature by 10 cm-1. The overtone states also narrow upon addition of lipids (Fig. 3b and 3d). Since the diagonal widths are a measure of the inhomogeneous distribution of the amide I modes in the peptide backbone, narrowing of the diagonal widths of the fundamental and overtone states indicates that there is a smaller structural distribution of peptides when folding occurs through a lipid catalyzed mechanism. Narrowing cannot be attributed to dehydration of the peptide by the membrane because dehydration causes a higher frequency shift. Rather, it is consistent with adoption of a secondary structure such as an α-helix, which has a lower amide I frequency and a narrower line width (see Discussion below).
Figure 3.
Diagonal slices along (a) fundamental peaks and (b) overtone peaks for uncatalyzed aggregation. Diagonal slices along (a) fundamental and (b) overtone peaks for catalyzed aggregation.
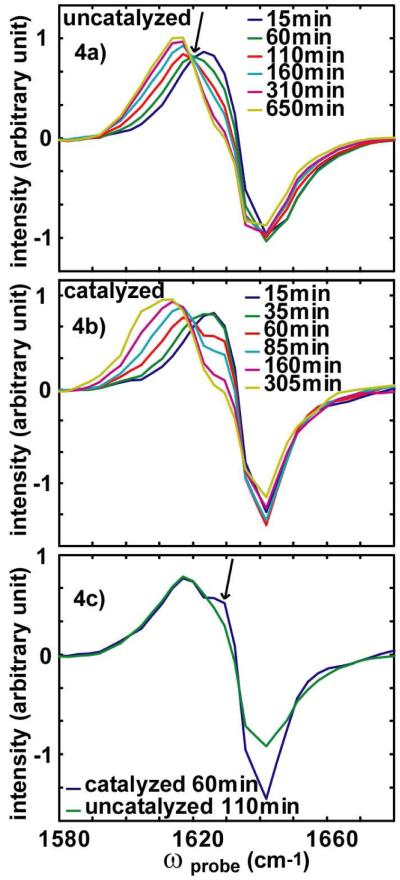
The final piece of evidence for the existence of an intermediate comes from the diagonal anharmonicity. Shown in Fig. 4a and 4b are slices along ωprobe for ωpump = 1642 cm-1. These slices cut through the “random coil” doublet as well as the tail of the β-sheet, so that both the disappearance of the “random coil” features and the appearance of the β-sheet can be monitored on the same graph. For the uncatalyzed case, these spectra can be fit to two components, one of which represents the “random coil” features and the other the β-sheet rich amyloid structure.25 This two-component fit describes the data remarkably well, which is consistent with the presence of an isobestic point at 1620 cm-1 (marked with an arrow in Fig. 4a). Isobestic points are often indicative of a two-state process and are a necessary but not a sufficient condition for a two state transition. In contrast; the membrane-catalyzed aggregation slices cannot be fit to two components and does not have a well-defined isobestic point. This proves that the reaction must involve more than two states. To better compare the two reactions, we overlap in Fig. 4c a slice from the catalyzed reaction at t = 60 minutes, which is at the maximum of the “random coil” absorption when the intermediate is most intense, and the uncatalyzed reaction at t = 110 minutes. On the scaled time-axis, these two time points are equivalent. At this point, the “random coil” feature, which exhibits a bleach at 1642 cm-1, has a narrower anti-diagonal linewidth, indicating that its homogeneous lifetime is longer. It also has a larger (negative) intensity. Also present is a shoulder near 1629 cm-1 (marked with arrow in Fig. 4c), that does not appear in the uncatalyzed reaction. In contrast it is present in all slices from 1635 cm-1 to 1648 cm-1 along ωpump in the catalyzed reaction at this time point. This shoulder scales with the intensity of the transient observed in the 1642 cm-1 kinetics (Fig. 2b); it is absent at t=0, maximum at t = 60 minutes and disappears again around t = 120 minutes. Since the shoulder appears at a higher frequency than the random coil overtone absorption, it must be created by a peptide structure with a smaller anharmonic shift. The only way that the anharmonic shift can be smaller than the random coil is if it derives from a strongly excitonically coupled peptide with an ordered structure.30 Secondary structures such as α-helices and β-sheets have anharmonic shifts smaller than random coils for these reasons. Thus, based on the observed kinetics, the lack of an isobestic point and a small anharmonic shift, we attribute this feature to a transiently populated intermediate structure as well.
Figure 4.
Slices along ωprobe for ωpump = 1642 cm-1 in the (a) uncatalyzed and (b) catalyzed spectra. (c) Comparison of slices at ωpump = 1642 cm-1 at t = t50 during uncatalyzed (green) and catalyzed (blue) aggregations.
DISCUSSION
We have presented three pieces of evidence that the assembly mechanism of hIAPP amyloid differs when catalyzed by lipid vesicles. This includes a difference in the kinetics whereby the “random coil” spectral features increases and then decreases in intensity while the fibrils grow in the presence of lipid vesicles rather than monotonically decreasing. A narrowing of the diagonal lineshape is also observed in the membrane catalyzed experiments indicating a smaller structural (inhomogeneous) distribution. Finally an isobestic point is observed in the uncatalyzed reaction, but in the catalyzed reaction a shoulder appears instead whose intensity scales with the kinetics of the intermediate. Together, these three features demonstrate that lipid catalyzed fiber formation occurs through a membrane-bound intermediate structure whose spectral features overlap with the “random coil” features and has a maximum population that occurs at the t50 of the β-sheet kinetics. Our data indicate that this intermediate has significant non-random secondary structure, which we discuss after first quantifying the kinetic timescales of the intermediate.
Modeling the kinetic traces
The simplest kinetic scheme that describes our observations is that the initial distribution of peptides (RC) converts to the amyloid fiber (AF) after passing through a membrane bound intermediate (I) via
| (S1) |
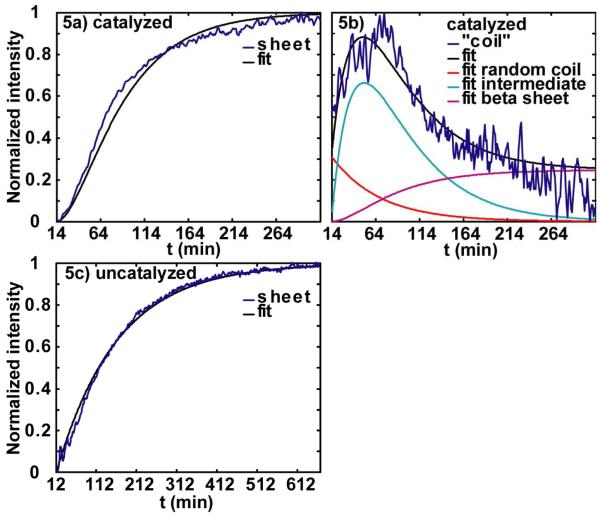
In this reaction, k1, k-1 and k2 are the time constants for each step. Using this reaction scheme, we then fit our data to extract these time constants. (Rate equations are given in the Supporting Information.) For the “random coil” region intensity, we fitted it with three components (random coil intensity, intermediate intensity and β-sheet intensity) because it contains contributions from all three. The fits are shown in Fig. 5a and 5b, for 2D IR data sets with scaled x-axes. The data could be fit with a range of k1 and k2, but the reverse rate k 1 was always at least 8 times smaller than k1. A small reverse reaction rate is consistent with the large binding constant (50000) of the peptide to the membrane20 and the fact that the reaction strongly favors the fibril products. Thus, in the final fits, we neglected k-1, which gave a unique fit with rate constants of and . By fixing one time constant while letting the other vary, we tested the covariance of k1 and k2, which we have used to estimate the error to our fits, which are for k1, and for k2. We report rate constants from scaled kinetics so that the results can be compared to other kinetic experiments, since the exact folding time depends on experimental conditions such as sample concentrations and the presence of small amounts of organic solvents, as well as hard-to-control parameters such as turbulence of mixing (which we suspect is the primary reason for different kinetics from run to run).30
Figure 5.
Kinetics and fits for lipid vesicle catalyzed aggregation, (a) β-sheet and (b) ‘random coil ’. (c) β-sheet kinetics and fit for aggregation without catalyzation.
Peptides that associate with the membrane are predicted to follow the above reaction scheme, but peptides that remain in the solution can aggregate into fibers via an uncatalyzed mechanism
| (S2) |
This simple kinetic scheme does not fully describe the nucleation kinetics of an aggregation mechanism, but because the lag time in these experiments is small, the rate constant adequately describes the time scale. Fits to the uncatalyzed kinetics (Fig. 5c) put the scaled reaction constant at with an error bar of . Thus, in principle, a portion of the ensemble that is not associated with the membrane will be folding on a timescale that competes with the catalyzed reaction pathway. However, the flux through the uncatalyzed pathway is negligible, because the partition coefficient of hIAPP between negatively charged vesicles and water is about 50000 and the nucleation rate constant is 25 times larger in lipid phase than in water phase, so that only a tiny amount of the peptide is unbound.20 Furthermore, the association of peptide with the membrane occurs within the first millisecond upon mixing, due to the rapid diffusion time of the peptides and the currents created upon stirring the sample. Thus, we can safely neglect the contribution of uncatalyzed peptide aggregation in our analysis of the catalyzed aggregation.
We also tested whether the data can be used to determine if the intermediate is on-pathway or off-pathway to forming the final fibril structure by fitting the data to the reaction scheme:
| (S3) |
The data can also be reasonably fit with this reaction scheme as well, which gives rate constants of with an error bar of , with an error bar of and . Thus it is formally not possible to distinguish between an on- or an off-pathway model. In fact this is not surprising and a similar situation is consistently encountered in kinetic studies of the folding of soluble globular proteins. Evidence for well-structured intermediates in amyloid aggregation is scarce, although many studies have suggested that the cytotoxic nature of amyloids arises from partially folded intermediates that permeabilize the cell membrane.15, 17 Whether or not the intermediate is on- or off-pathway may have little bearing on the cytotoxic properties of amyloid.
Possible structural features of the intermediates
What is the structure of the intermediate? At this time, we do not know the structure of the intermediate, but it appears to be linked to the two shoulders observed in the spectra of hIAPP at early times. Interpretation of spectral features in the ‘random coil’ region is complicated because random coil as well as helical feature and small β-sheets can all give signals in the same region. At early times in the folding process, three peaks appear in the “random coil” region, at 1636, 1642, and 1648 cm-1, regardless of whether folding is catalyzed or not. The 1642 cm-1 is traditionally assigned to random coil structures.29 We do not know what secondary structures the two shoulders correspond to, but the presence of vesicles changes their relative populations (Fig. 3a and 3c).
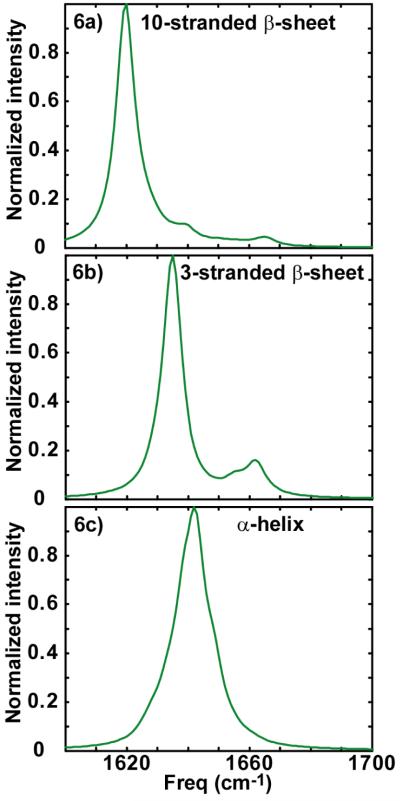
Infrared spectroscopy is sensitive to the secondary structures of peptides because of hydrogen bonding, environmental electrostatics, solvation, and vibrational coupling.32-34 Of these, couplings are the strongest indicator of structure because they alter the vibrational eigenstates frequencies, intensities, and anharmonicities. The reason that random coil structures typically absorb near 1642 cm-1 and are very broad is because the couplings between the local amide I modes of the residues are irregular due to structural disorder. As a result, the eigenstates are broadly distributed in frequency and intensity. However, for a β-sheet, the couplings between the local modes give rise to a periodic Hamiltonian. As a result, there is a well-defined frequency distribution of the eigenstates.35-37 Furthermore, due to the symmetry of the 2D sheet, only two of these eigenstates carry oscillator strength and appear in the infrared spectra (in the limit of a perfectly ordered and infinitely large β-sheet), which is the origin of the anti-symmetric and symmetric stretches around 1620 and 1670 cm-1, respectively. Shown in Fig. 6a is a linear absorption spectrum calculated for a β-sheet of 10 strands, each composed of 10 residues, assuming that the coupling between amide I local modes is described by transition dipole coupling.38 Each local mode is assigned a frequency of 1642 cm-1 except those amide groups at the edges which are assigned frequency of 1652 cm-1 to account for difference in hydrogen bonding.36 The strongest mode of the calculated β-sheet is the anti-symmetric stretch, which appears at 1619 cm-1, while the symmetric stretch is weaker at 1665 cm-1. The other weaker modes are caused by end-effects, which arise for finite sized β-sheets. Simulations by Hahn et al. show that the frequency of the anti-symmetric stretch mode is very sensitive to the number of strands in the β-sheet.36 To demonstrate this effect, we also show in Fig. 6b, a simulated absorption spectrum of a 3 stranded β-sheets, which we find has a strong absorption at 1635 cm-1. α-helices also absorb in this frequency range. Shown in Fig. 6c is the infrared spectrum predicted for a 14 residue α-helix. An α-helix also has two allowed modes, but because the amide I transition dipoles are oriented nearly parallel to the helix axis, almost all of the oscillator strength lies in a single eigenstate.39 α-helices in water absorb around 1635 cm-1 due to the strong positive coupling between neighboring amide I local modes. Trans-membrane helices absorb near 1655 cm-1, due to desolvation of the peptide backbone.24, 33 Thus, a structure like the membrane bound α-helix observed using ESR,22 could explain one of the three features in the 2D IR spectra, but it appears that other secondary structures are also present such as smaller β-sheet aggregates.
Figure 6.
(a) Linear absorption spectrum calculated for a β-sheet of 10 strands, each composed of 10 residues. (b) Simulated absorption spectrum of 3 stranded β-sheet with each strand consisting 10 residues. (c) Infrared spectrum predicted for α-helix with 14 residues.
Another signature which indicates that the intermediate is structurally ordered comes from the anharmonic shift. Coupled vibrational modes exhibit a decrease in the anharmonic shift (e.g. the frequency separation between the fundamental (blue) and overtone (red) transitions). For instance, this effect causes the anharmonic shift of the β-sheet at 1617 cm-1 to be smaller than the random coil at 1642 cm-1 (8 vs. 17 cm-1). A similar effect is seen for the intermediate. In Fig. 3c we pointed out the presence of a shoulder in slices through the catalyzed “random coil” peak that we assigned to the presence of an intermediate. This shoulder appears about 4 cm-1 higher in frequency, a smaller anharmonic shift than the random coil (the exact frequency is hard to determine because fits to the slices are not unique without knowing the spectral shape of the features in more detail). This smaller anharmonic shift is consistent with the vibrational coupling in a periodic secondary structure.
We present these simulations and the anharmonic shift data to make the point that the broad “random coil” frequency range also reports on a variety of secondary structures, including α-helices and small β-sheets. Thus, the presence of shoulders on the 1642 cm-1 random coil peaks is highly suggestive of peptide structures with periodic coupling constants e.g. well-defined secondary structures. Moreover, the distribution of these secondary structures must change when in contact with lipid membranes, since the relative contribution of the high frequency shoulder decreases. These secondary structures form immediately after dissolving the peptides, since they are present in both the catalyzed and uncatalyzed spectra at the earliest times we can monitor. These shoulders also appear in the spectra of samples that have long lag times before the onset of folding.26 Similar features (1642 cm-1 random coil peak with two shoulders) and changes in the distribution (intensity decrease of the higher frequency shoulder) were also seen in rIAPP spectra after lipid vesicles were added. At this time we do not attempt a more precise structural characterization. Possible structures might be tested with more precise coupling models or from isotope labeling studies. Both of these approaches are underway.
Comment on simplicity of the kinetic schemes
We conclude with a comment about the simplicity of the kinetic schemes used to model the 2D IR kinetics in this paper. The kinetic scheme proposed here is the simplest scheme that describes the observed data. However, we know that this scheme is too crude. We have recently published an analysis of the uncatalyzed aggregation kinetics of isotope labeled samples of hIAPP using 2D IR spectroscopy.26 Isotope labeling allowed us to resolve the aggregation kinetics of individual residues, in addition to the eigenstates of the coupled amide I local modes monitored in this article. With isotope labeling, we found the kinetics on a residue-by-residue basis differ dramatically from one other, indicating that aggregation does not proceed in a two-step reaction as suggested here, but evolves through a series of primarily β-sheet intermediates. The series of intermediates is obscured in unlabeled peptide kinetics, because the unlabeled eigenstates, while sensitive to global secondary structures, are insensitive to specific details. For instance, if a β-sheet forms near the C-terminus, it has a spectroscopic feature nearly identical to a β-sheet forming near the N-terminus, since unlabeled residues are indistinguishable. With regards to the data in this paper, the simple observation that membrane-catalyzed folding is obviously different than uncatalyzed folding, even for unlabeled peptides, indicates that the folding pathways are dramatically different. We expect that when experiments have been performed using isotopically labeled peptides, the differences will be even more pronounced and give rise to a much more detailed mechanism as to how lipid membranes catalyze fibril formation.
CONCLUSIONS
In conclusion, we have used automated 2D IR spectroscopy to monitor the secondary structures and aggregation kinetics of the polypeptide associated with type 2 diabetes, hIAPP, in the presence and absence of lipid membranes. We find that the kinetics of aggregation differ dramatically when catalyzed by lipid membranes and we have identified several spectral features that point to the presence of excitonically coupled membrane bound secondary structure(s). Membranes shift the relative populations of these structures. A very simple kinetic scheme involving an intermediate has been postulated. The fact that the spectral features and kinetics measured by 2D IR spectroscopy differs when membrane catalyzed, points to a significantly different pathway. Whether this pathway differs in relative energies (and hence lifetimes) of intermediates or whether structurally different intermediates are populated is a topic left for the future. Intermediates have been postulated as being responsible for the cytotoxic nature of partially formed amyloids.13-16 This is the first study which has directly probed the nature of intermediates in real time. We anticipate that isotope labeling studies will provide more detailed informative about the aggregation pathway for catalyzed reactions, so that a step-by-step mechanism can be obtained with residue-specific structural resolution. The studies presented here pave the way for future isotope labeling studies.
We end with a note about the advantages of using 2D IR spectroscopy over FTIR to follow amyloid kinetics. Some, but not all, of the information reported here could potentially have been obtained using FTIR spectroscopy. However, the non-linear nature of 2D IR spectroscopy has the clear advantage that it enhances strong absorbers over weaker ones, which reduces the background from solvent modes. Another major advantage is that the anharmonic shifts measured by off-diagonal peaks in 2D IR spectroscopy provides strong evidence for excitonically coupled secondary structures, which cannot be determined by FTIR. A third advantage of 2D IR spectroscopy when implemented with our pulse shaping technology is that it can remove scattered light from the spectra using phase cycling.40 Heterogeneous samples like amyloids and membranes also strongly scatter light, which is a major problem in FTIR and 2D IR spectroscopy. But the real advantages of 2D IR spectroscopy will be utilized in future experiments that focus on 2D lineshapes and cross peaks, which have the potential to detect in a time-resolved fashion solvent expulsion and identify the secondary structures of intermediates, among other properties. The work presented here sets the stage for such future studies and demonstrates the utility of 2D IR methods for the study of amyloid formation in challenging heterogeneous environments.
Supplementary Material
ACKNOWLEDGEMENT
This work is supported by grants from the NIH DK79895 to MTZ and GM078114 to DPR. We also thank for comments from reviewers.
REFERENCES
- 1.Sunde M, Blake C. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv. Protein. Chem. 1997;50:123–159. doi: 10.1016/s0065-3233(08)60320-4. [DOI] [PubMed] [Google Scholar]
- 2.Tycko R. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr. Opin. Struc. Biol. 2004;14:96–103. doi: 10.1016/j.sbi.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Mclean LR, Balasubramanism A. Promotion of beta-structure by interaction of diabetes associated polypeptide (amylin) with phosphatidylcholine. Biochim. Biophys. Acta. 1992;1122:317–320. doi: 10.1016/0167-4838(92)90411-6. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths JM, Ashburn TT, Auger M, Costa PR, Griffin RG, Lansbury PT. Rotational resonance solid-state NMR elucidates a structural model of pancreatic amyloid. J. Am. Chem. Soc. 1995;117:3539–3546. [Google Scholar]
- 5.Makin OS, Serpell LC. Structural characterization of islet amyloid polypeptide fibrils. J. Mol. Biol. 2004;355:1279–1288. doi: 10.1016/j.jmb.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 6.Jack E, Newsome M, Stockley PG, Radford SE, Middleton DA. The organization of aromatic side groups in an amyloid fibril probed by solid-state H-2 and F-19 NMR spectroscopy. J. Am. Chem. Soc. 2006;128:8089–8099. doi: 10.1021/ja0581898. [DOI] [PubMed] [Google Scholar]
- 7.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supermolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemisty. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark A, Edwards CA, Ostle LR, Sutton R, Rothbard JB, Morris JF, Turner RC. Localization of islet amyloid peptide in lipofuscin bodies and secretory granules of human β-cells and in islets of type-2 diabetic subjects. Cell Tissue Res. 1989;257:179–185. doi: 10.1007/BF00221649. [DOI] [PubMed] [Google Scholar]
- 9.Porte D, Kahn SE. β-cell dysfunction and failure in type 2 diabetes - potential mechanisms. Diabetes. 2001;50:S160–S163. doi: 10.2337/diabetes.50.2007.s160. [DOI] [PubMed] [Google Scholar]
- 10.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Pancreatic-islet cell toxicity of amylin associated with type-2 diabetes-mellitus. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 11.Janson J, Ashley RH, Harrison D, Mclntyre S, Butler PC. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- 12.Kahn SE, Andrikopoulos S, Verchere CB. Islet amyloid: a long-recognized but underappreciated pathological feature of type 2 diabetes. Diabetes. 1999;48:241–253. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 13.Anguiano M, Nowak RJ, Lansbury PT. Protofibrillar islet amyloid polypeptide permeabilized synthetic vesicles by a pore-like mechanism that may be relevant to type II diabetes. Biochemistry. 2002;41:11338–11343. doi: 10.1021/bi020314u. [DOI] [PubMed] [Google Scholar]
- 14.Porat Y, Kolusheva S, Jelinek R, Gazit E. The human islet amyloid polypeptide forms transient membrane-active prefibrillar assemblies. Biochemistry. 2003;42:10971–10977. doi: 10.1021/bi034889i. [DOI] [PubMed] [Google Scholar]
- 15.Sparr E, Engel MFM, Sakharov DV, Sprong M, Jacobs J, de Kruijff B, Hoppener JWM, Killian JA. Islet amyloid polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. Febs. Lett. 2004;577:117–120. doi: 10.1016/j.febslet.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 16.Kayed R, Head E, Thompson JL, Mclntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;302:814–818. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 17.Mirzabekov TA, Lin MC, Kagan BL. Pore formation by the cytotoxic islet amyloid peptide amylin. J. Boil. Chem. 1996;271:1988–1992. doi: 10.1074/jbc.271.4.1988. [DOI] [PubMed] [Google Scholar]
- 18.Knight JD, Miranker AD. Phospholipid catalysis of diabetic amyloid assembly. J. Mol. Biol. 2004;341:1175–1187. doi: 10.1016/j.jmb.2004.06.086. [DOI] [PubMed] [Google Scholar]
- 19.Jayasinghe SA, Langen R. Lipid membranes modulate the structure of islet amyloid polypeptide. Biochemistry. 2005;44:12113–12119. doi: 10.1021/bi050840w. [DOI] [PubMed] [Google Scholar]
- 20.Knight JD, Hebda JA, Miranker AD. Conserved and cooperative assembly of membrane-bound α-helical states of islet amyloid polypeptide. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- 21.Goldsbury C, Goldie K, Pellaud J, Seelig J, Frey P, Muller SA, Kistler J, Cooper GJS, Aebi U. Amyloid fibril formation from full-length and fragments of amylin. J. Struc. Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 22.Apostolidou M, Jayasinghe SA, Langen R. Structure of α-helical membrane-bound human islet amyloid polypeptide and its implications for membrane-mediated misfolding. J. Biol. Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim S, Strasfeld DB, Ling YL, Zanni MT. Automated 2D IR spectroscopy using a mid-IR pulse shaper and application of this technology to the human islet amyloid polypeptide. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14197–14202. doi: 10.1073/pnas.0700804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee P, Krummel AT, Fulmer EC, Kass I, Arkin IT, Zanni MT. Site-specific vibrational dynamics of the CD3 ζ membrane peptide using heterodyned two dimensional infrared photon echo spectroscopy. J. Chem. Phys. 2004;120:10215–10224. doi: 10.1063/1.1718332. [DOI] [PubMed] [Google Scholar]
- 25.Strasfeld DB, Ling YL, Shim S, Zanni MT. Tracking fiber formation in human islet amyloid polypeptide with automated 2D IR spectroscopy. J. Am. Chem. Soc. 2008;130:6698–6699. doi: 10.1021/ja801483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shim S, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. 2D IR spectroscopy defines the pathway of amyloid formation with residue specific resolution. Submitted. (Note to Referees: This paper is currently under review but has not been accepted for publication yet.) [Google Scholar]
- 27.Hamm P, Lim M, DeGrado WF, Hochstrasser RM. The two dimensional IR nonlinear spectroscopy of a cyclic penta-peptide in relation to its three-dimensional structure. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2036–2041. doi: 10.1073/pnas.96.5.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim S, Strasfeld DB, Fulmer EC, Zanni MT. Fentosecond pulse shaping directly in the mid-IR using acousto-optic modulation. Opt. Lett. 2006;31:838–840. doi: 10.1364/ol.31.000838. [DOI] [PubMed] [Google Scholar]
- 29.Mantsch HH, Chapman D. Infrared spectroscopy of biomolecules. Wiley-Liss, Inc.; New York: 1995. [Google Scholar]
- 30.Wang J, Hochstrasser RM. Characteristics of the two dimensional infrared spectroscopy of helices from approximate simulation and analytic models. Chem. Phys. 2004;297:195–219. [Google Scholar]
- 31.Padrick SB, Miranker AD. Islet amyloid: phase partitioning and secondary nucleation are central to the mechanism of fibrillogenesis. Biochemistry. 2002;41:4694–4703. doi: 10.1021/bi0160462. [DOI] [PubMed] [Google Scholar]
- 32.Barth A, Zscherp C. What vibrations tell us about proteins. Quarterly Reviews of Biophysics. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 33.Decatur SM. Elucidation of residue-level structure and dynamics of polypeptides via isotope-edited infrared spectroscopy. Acc. Chem. Res. 2006;39:169–175. doi: 10.1021/ar050135f. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee P, Kass I, Arkin I, Zanni MT. Picosecond dynamics of a membrane protein revealed by 2D IR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran A, Mukamel S. The origin of vibrational mode couplings in various secondary structural motifs of polypeptides. Proc. Natl. Acad. Sci. U. S. A. 2004;101:506–510. doi: 10.1073/pnas.2533089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn S, Kim S, Lee C, Cho M. Characteristic two-dimensional IR spectroscopic features of antiparallel and parallel β-sheets polypeptides: simulation studies. J. Chem. Phys. 2005;123:084905–084915. doi: 10.1063/1.1997151. [DOI] [PubMed] [Google Scholar]
- 37.Cheatum CM, Tokmakoff A. Signatures of β-sheet secondary structures in linear and two-dimensional infrared spectroscopy. J. Chem. Phys. 2004;120:8201–8215. doi: 10.1063/1.1689637. [DOI] [PubMed] [Google Scholar]
- 38.Hamm P, Woutersen S. Coupling of the amide I modes of the glycine dipeptide. Bull. Chem. Soc. Jpn. 2002;75:985–988. [Google Scholar]
- 39.Miyazawa T. Perturbation treatment of the characteristic vibrations of polypeptide chains in various configurations. J. Chem. Phys. 1960;32:1647–1652. [Google Scholar]
- 40.Shim S, Zanni MT. How to turn your pump-probe instrument into a multidimensional spectrometer: 2DIR and Vis spectroscopies via pulse shaping. PCCP. doi: 10.1039/b813817f. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.