Abstract
Trim5α from primates (including humans), cows, and rabbits has been shown to be an active antiviral host gene that acts against a range of retroviruses. Although this suggests that Trim5α may be a common antiviral restriction factor among mammals, the status of Trim5 genes in rodents has been unclear. Using genomic and phylogenetic analyses, we describe an expanded paralogous cluster of at least eight Trim5-like genes in mice (including the previously described Trim12 and Trim30 genes), and three Trim5-like genes in rats. Our characterization of the rodent Trim5 locus, and comparison to the Trim5 locus in humans, cows, and rabbits, indicates that Trim5 has undergone independent evolutionary expansions within species. Evolutionary analysis shows that rodent Trim5 genes have evolved under positive selection, suggesting evolutionary conflicts consistent with important antiviral function. Sampling six rodent Trim5 genes failed to reveal antiviral activities against a set of eight retroviral challenges, although we predict that such activities exist.
Keywords: Retrovirus, rodent, Trim5, adaptive evolution, restriction factor, paralog
INTRODUCTION
In order to counteract pathogens, host organisms evolved multiple strategies within the broad context of adaptive and innate immunity. One such innate immune strategy involves the host factor known as Trim5α (Bieniasz, 2004; Nisole et al., 2005). Trim5α was identified as the factor responsible for preventing the infection of rhesus macaque cells with HIV-1 (Keckesova et al., 2004; Stremlau et al., 2004). Members of the Trim gene family are characterized by the presence of a TRI-partite Motif (TRIM) consisting of RING, B-Box and Coiled-Coil domains (Nisole et al., 2005; Reymond et al., 2001). Some TRIM proteins, such as Trim5α, contain a fourth domain called the PRYSPRY domain, also known as the B30.2 domain (James et al., 2007; Reymond et al., 2001). In humans there are more than 70 members of the TRIM gene family, some of which appear to have vital roles in restricting viral infections (Nisole et al., 2005).
Trim5α specifically recognizes viral capsids in the cytoplasm via its PRYSPRY domain (Sebastian and Luban, 2005), imposing a restriction before reverse transcription takes place. Under some circumstances restriction occurs after reverse transcription (Perron et al., 2007; Stremlau et al., 2006). The mechanism of viral inhibition by Trim5α appears to occur in multiple steps that include premature uncoating and/or capsid sequestration and destruction (Anderson et al., 2006; Stremlau et al., 2006; Wu et al., 2006; Sawyer et al., 2005). Although other members of the TRIM multi-gene family have been shown to exhibit antiviral activity (Nisole et al., 2005), none are as dramatic as the restriction activity of Trim5α against the retroviruses it recognizes.
In primates, Trim5α evolves at an extremely rapid rate (Liu et al., 2005; Sawyer et al., 2006; Sawyer et al., 2007). Such rapid evolution can be a hallmark of proteins involved in virus-host interactions because of evolutionary dynamics at the protein-protein interaction interface. Signatures of adaptive evolution (also known as positive selection) can be identified in comparisons of nucleotide sequences as a significant abundance of non-synonymous changes versus synonymous ones. Studying these signatures of positive selection can provide important information about the evolutionary history of genes and critical residues involved in host-virus interactions. For example, the region of the PRYSPRY of Trim5α that serves as a major determinant for the specificity of Trim5α towards capsid (Nakayama et al., 2005; Stremlau et al., 2005; Yap et al., 2005) was accurately predicted independently by identification of a group of amino acids under strong positive selection (Liu et al., 2005; Sawyer et al., 2005).
Trim5α-mediated restriction appears to be widespread in mammals. After its identification in rhesus macaques (Stremlau et al., 2004), Trim5α was found to be present in diverse species of primates (Brennan et al., 2007; Keckesova et al., 2004; Nisole et al., 2004; Sawyer et al., 2005) and other mammals such as cows (Si et al., 2006; Ylinen et al., 2006) and rabbits (Schaller et al., 2007). Cows have an expanded clade of five paralogous Trim5 genes, one of which (Trim5-3) functions as an antiviral factor (Sawyer et al., 2007; Si et al., 2006; Ylinen et al., 2006). In contrast, dogs lack Trim5α due to disruption of its open reading frame (Sawyer et al., 2007). Other studies have investigated whether rodents have orthologs of the mammalian Trim5 gene, but have failed to reach consensus (Schaller et al., 2007; Si et al., 2006). Part of this confusion arose from the fact that the two mouse TRIM genes with closest sequence similarity to Trim5 are named Trim12 and Trim30 (the human genome does not encode genes with these designations). A mouse TRIM gene known by its RIKEN cDNA number (9230105E10Rik) was recently referred to as Trim5 (Schaller et al., 2007). However, due to a lack of rigorous examination of the syntenic mouse chromosomal region, the presence of Trim5 in rodents remains inconclusive.
Here, we use genomic and phylogenetic analyses to rigorously analyze the Trim5 locus in the genomes of mouse and rat. We find an expanded paralogous cluster of at least eight Trim5-like genes in mice and three Trim5-like genes in rats. Of the mouse Trim5-like genes, at least five contain all four canonical domains of Trim5α. Further analysis suggests that most of these mouse Trim5 genes are expressed. Arguing against the notion that these are redundant or decaying gene duplications, we show that mouse Trim5 homologs have evolved under positive selection. In some cases the signature of selection is especially strong, suggesting a role in immunity similar to antiviral mammalian Trim5α orthologs. Murine Trim5 genes did not restrict any of the extant retroviruses we tested. Based on our studies, we predict that the viruses rodent Trim5 orthologs restrict (or may have restricted in the past, (Kaiser et al., 2007)) remain to be identified.
RESULTS
Identification of a rodent Trim5 locus
The UCSC genome browser BLAT tool (Kent, 2002) was used to query the mouse genome (Mouse Genome Sequencing Consortium et al., 2002) July 2007 assembly with human Trim5α, which revealed a locus of multiple, adjacent Trim5-like genes on the syntenic mouse chromosome 7 (7qE3). This cluster contains the genes 9230105E10Rik, Trim12, Trim30, as well as several unnamed gene predictions. Similarly, our search of the rat genome assembly (Gibbs et al., 2004) with human Trim5α revealed a locus of three unnamed, predicted rat Trim5-like genes on the syntenic chromosome 1 (1q32). Although genome databases were helpful in identifying this locus, the presence of tandem gene duplication events can potentially result in assembly errors due to repeat-rich regions. Indeed, the assembly and gene annotation of the mouse Trim5 locus changed during the course of preparation of this study. Therefore, we identified single or multiple overlapping bacterial artificial chromosome (BAC) clone sequences that encompass each locus. By manually examining each BAC for individual exons of Trim5α, we confirmed the gene content of the locus, and collated the predicted gene sequences for each gene (Figure 1A). We performed a similar analysis for Trim6, Trim34, and Trim22 because these represent proximal genes to Trim5 in all mammals tested so far (Sawyer et al., 2007).
Figure 1. A paralogous expansion of rodent Trim5 genes.
(A) The mouse and rat Trim5 loci are compared to the human locus. All three loci sit in an olfactory receptor gene expansion, here shown with only two of the genes on each side (white arrows). Distances between genes are drawn roughly to scale, with numbers on the top (for mouse) and bottom (for rat) indicating distance between genes, based on BAC sequences. The Trim5 homologs are shown in green arrows, with dark green color for a human Trim5-like pseudogene (P1). The asterisk (*) denotes partial Trim5-like sequences found in the mouse genome. Trim34 homologs are shown in orange, Trim6 homologs in blue, and Trim22 is shown in magenta. Mouse Trim12-X is not included because it is on the X chromosome. 30-L1 and 30-L2 refer to rat Trim30-like1 and Trim30-like2, respectively.
(B) The DNA sequence for RBCC domains were used to construct a maximum likelihood tree with 100 replicates for estimating bootstrap support. Mouse Trim12-X and Trim30-4 are not included because they do not contain the RBCC domains. Color profiles match those used in (A). Scale for branch length is shown below the tree. (C) Phylogeny of the species used in (B), based on reference (Hedges, 2002).
Using these mouse and rat sequences, as well as previously published sequences from the Trim5 loci in humans, rhesus macaques, cows, dogs, and rabbits, we constructed a phylogenetic tree using maximum likelihood methods to infer the evolutionary histories of the rodent Trim5-like genes. We excluded exon 8 (which encodes the PRYSPRY domain) from our phylogenetic analysis because this domain has evolved rapidly in some cases, and therefore might bias phylogenetic analysis. As an outgroup, we included human Trim21, a closely related Trim gene that does not belong to the Trim5 locus (Figure 1B). Our phylogenetic analysis revealed four major clades whose members cluster tightly together with strong bootstrap support: a Trim6 clade, a Trim34 clade, a Trim22 clade, and a Trim5 clade. Strong bootstrap support (97%) for this multi-species Trim5 clade distinguishes it from the next closest relatives of Trim5, which are Trim22, Trim6 and Trim34. Finally, when present, these genes form branches in an order consistent with the divergence of species, as predicted for orthologous genes (Figure 1C) (Hedges, 2002).
Similar to the previously published human, cow, and dog loci (Sawyer et al., 2007), the rodent Trim5 locus sits within an expansion of olfactory receptor genes that includes Trim6 and Trim34 (Figure 1A). Mice are unique compared to rats, humans and cows, in that they contain two Trim34 genes that we have named Trim34-1 and Trim34-2. While the human locus contains a single Trim5 gene, tandem gene duplications have resulted in eight mouse Trim5 genes and three rat Trim5 genes. The cow locus contains a similar expansion of Trim5 genes. However, unlike the cow locus, the Trim5 genes in rodents are in antisense orientation with respect to Trim6 and Trim34. Notably, Trim22, which is present in the human and dog loci, is absent from both rodent and cow loci that have undergone independent expansions of the Trim5 genes. Taken together, these findings confirm that the Trim5 loci identified in rodents is in synteny with the Trim5 loci characterized in other mammals.
We find that all of the rodent Trim5 genes are orthologous to Trim5 from humans, rhesus macaques, cows, and rabbits. Interestingly, there are two distinct monophyletic groups within the rodent Trim5 gene cluster: Those related to Trim12 or to Trim30. Unnamed rodent genes were given names to reflect their presence in either the Trim12 or Trim30 phylogenetic grouping, a system that also results in grouping genes by their chromosomal positions. Mouse genes phylogenetically associated with Trim12 were named Trim12-1 and Trim12-2 (formerly mouse 9230105E10Rik). Similarly, genes associated with Trim30 were named Trim30-1, Trim30-2 and Trim30-3. The rat gene most closely related to mouse Trim12 was named rat Trim12, and those associated with mouse Trim30 were named rat Trim30-like1 and Trim30-like2. The latter two genes appear to result from independent duplication events after the mouse and rat species split. Therefore, they were tagged ‘-like,’ so as not to imply orthology to mouse Trim30-1 and Trim30-2.
Domain organization and expression of rodent Trim5 homologs
The primate restriction factor Trim5α contains four canonical domains: RING, B-box, coiled-coil, and PRYSPRY. Our domain analysis revealed that five out of eight mouse Trim5 genes, and each of the three rat Trim5 genes, contain all four canonical domains of primate Trim5α (Figure 2). Also considering their phylogenetic arrangement, we refer to these collectively as rodent Trim5α. Two of the three non-canonical mouse Trim5 genes lack the PRYSPRY domain; these are mouse Trim12 and Trim30-1. The presence of a PRYSPRY domain downstream of Trim12 is not detectable in the BAC sequence. On the other hand, Trim30-1 contains a cryptic PRYSPRY domain that is present in the BAC sequence at approximately the expected distance from the upstream exon, but this domain has been pseudogenized due to numerous stop codons. The third non-canonically structured mouse Trim5 gene (Trim30-4) lacks the RING, B-Box and coiled-coil domains, and consists of only a partial PRYSPRY domain downstream of a start codon. In addition to these mouse Trim5 genes on chromosome 7, we located a Trim12-like gene outside the mouse Trim5 locus on chromosome X, which we named Trim12-X. It encodes a partial coiled-coil domain together with a PRYSPRY domain, all downstream of a start codon. Interestingly, in addition to lacking introns, Trim12-X also contains a poly-A sequence downstream of its stop codon, suggesting that it may have arisen from a retrotransposition event. A similar Trim5 gene was observed external to the main Trim5 gene locus in the cow genome (Sawyer et al., 2007). While screening the mouse BAC sequence corresponding to the Trim5 locus, we also discovered short RING domain like sequences (30–45 nucleotides in length) that appear to be byproducts of recombination events that reorganized this gene cluster. These findings demonstrate that the mouse Trim5 locus has undergone dynamic evolutionary events that are quite distinct from those in the primate lineages.
Figure 2. Domain organization and expression of rodent Trim5 loci genes.
Domains of rodent Trim5, Trim6 and Trim34 genes are compared to those of human Trim5. Five of the mouse and three of the rat Trim5 paralogs contain all four of the canonical domains of Trim5α. Previously existing (if available) RefSeq or Genbank accession numbers are shown below each gene name and are gathered from the NCBI and UCSC genome browser online servers. The expression files for mouse Trim5 genes containing the four canonical domains of Trim5α are shown for NIH3T3 cells. Expressed indicates that we were able to amplify cDNA from NIH3T3 RNA extracts, using reversetranscriptase-PCR. Not expressed indicates that we were not able to amplify cDNA from NIH3T3 RNA extracts, even though were designed primers according to predicted sequences. Not applicable indicates that the NIH3T3 cell line was not suitable to amplify that gene.
To investigate which rodent Trim5 homologs are expressed, we first checked the NCBI expressed sequence tags (ESTs) online database for ESTs corresponding to each gene (Figure 2). Most rodent Trim5 homologs contained ESTs from several tissues suggesting that they are expressed. We also screened RNA from mouse cell lines to confirm the expression of each Trim5α homolog containing all canonical domains. Using primers specific to each mouse Trim5 gene (see Methods for primer sequences) we performed reverse transcriptase PCR (RT-PCR) on cytoplasmic RNA from NIH3T3 cells (from Mus musculus). We found that the full length Trim5 homologs, with one exception (Trim12-1), are expressed (summarized in Figure 2). It is possible that Trim12-1 is also expressed based on a solitary EST from a 16 day old neonate mouse (Figure 2). These data indicate that most rodent Trim5 α homologs are indeed transcribed.
Evidence of recombination among rodent Trim5 homologs between RBCC and PRYSPRY domains
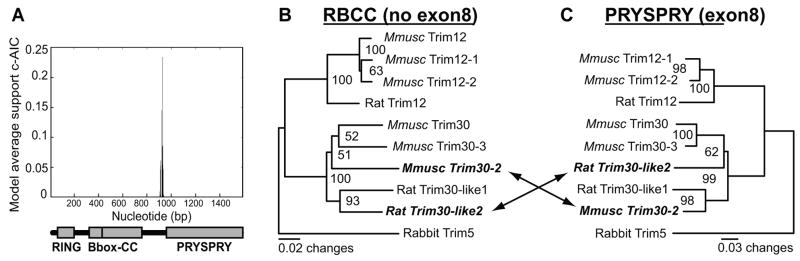
Given the tandem arrangement of rodent Trim5 genes, we looked for evidence of recombination among mouse and rat Trim5 homologs before performing evolutionary analysis. This is because recombination can result in signatures that can resemble adaptive evolution and may hinder the reliability of evolutionary analyses for detecting positive selection (Anisimova et al., 2003). We analyzed rodent Trim5 sequences for evidence of recombination using the Genetic Algorithm for Recombination Detection (GARD) tool (Kosakovsky Pond et al., 2006, Pond and Frost, 2005). Using the single breakpoint analysis, GARD identified a recombination breakpoint with high confidence among the rodent Trim5 homologs that mapped to the start of exon 8, which ecodes the PRYSPRY domain (Fig. 3A). Next, using rabbit Trim5 as an outgroup, we compared two phylogenetic trees constructed using either the PRYSPRY domain (encoded by exon8) or the RBCC domains (excluding exon8; Figure 3B, C) to see if we could confirm the recombination breakpoint identified by GARD. The trees yielded different branching patterns for mouse Trim30-2 and rat Trim30-like2, suggesting that an ancestral recombination event had taken place for these genes. To see if these two topologies were significantly different from one another, we performed the Kishino-Hasegawa (Kishino and Hasegawa, 1989) and Shimodaira-Hasegawa (Shimodaira and Hasegawa, 1999) tests which forces both topologies onto the sample set and assesses likelihood. Both tests confirmed the two trees to be significantly different (p<0.05) (data not shown). These results imply that recombination has shuffled the histories of the RBCC and PRYSPRY domains in the rodent Trim5 genes, but we obtained no evidence of recombination within either the RBCC or PRYSPRY domains themselves.
Figure 3. Recombination among rodent Trim5 genes.
(A) The online GARD tool identifies a breakpoint upstream of the PRYSPRY (exon8) domain for the full-length canonical Trim5α sequences from mice, rats and rabbits. The second-order Akaike Information Criterion (c-AIC) value is shown on the y axis as a measure to show the goodness of fit of this location of the breakpoint compared to other locations identified by GARD (not shown). The model average support value (numbers on the y axis) by itself has no meaning; they are used by GARD to identify the best single breakpoint among a series of possible breakpoints. Nucleotide position is indicated on the x axis. DNA sequences of the (B) RBCC domains (excluding exon8), and of the (C) PRYSPRY (exon8) domain for rodent Trim5 homologs were used to construct a maximum likelihood tree. Rabbit Trim5α was used as an outgroup. Sequences that showed evidence of recombination are shown in bold text. Mmusc stands for Mus musculus. Mmusc Trim12 was excluded because it lacks a PRYSPRY domain (exon8). Scale for branch length is shown below the tree.
Positive selection among rodent Trim5 paralogs
We next asked whether any of the rodent Trim5 homologs show signs of adaptive evolution. Due to the recombination breakpoint between the RBCC and PRYSPRY domains, we analyzed RBCC and PRYSPRY domain phylogenies separately for positive selection. To strengthen our analysis we also included Trim5 homologs from the mouse species, Mus dunni. Since the genome sequence database for Mus dunni is very limited, we amplified Mus dunni Trim5α paralogs using RT-PCR on RNA from MDTF cells. We amplified Trim12-2, Trim30, and Trim30-3 from MDTF cells, but could not recover Trim12-1 and Trim30-2 (data not shown). Next, using rabbit, rhesus and human Trim5 as an outgroup, we again constructed two phylogenetic trees using the maximum likelihood method for rodent Trim5 homologs from mice (Mus musculus and Mus dunni) and rats based on the RBCC (all but exon8) or PRYSPRY (exon8 only) sequences (Figure 4) to account for the potential recombination breakpoint.
Figure 4. Rodent Trim5 homologs are rapidly evolving.
Maximum likelihood trees for DNA sequences of (A) RBCC domains (excluding exon8) and (B) PRYSPRY domain (exon8) are shown for rodent Trim5 homologs, with bootstrap values at each node out of 100 replicates. Rabbit, rhesus and human Trim5α are used as out-groups. Branch dN/dS values calculated by PAML are shown above each branch. The number of non-synonymous changes (N) and synonymous changes (S) are shown below each branch in parentheses as (N,S). Branch dN/dS values significantly greater than 1 are highlighted as a bold line. Lineages with the highest dN/dS on each tree are highlighted in bold text (Mmusc Trim12 for RBCC, Mdunn Trim30 for PRYSPRY). MMusc stands for Mus musculus, and Musdunn for Mus dunni. Mmusc Trim12 was excluded from the PRYSPRY tree (B) because it lacks a PRYSPRY domain.
We analyzed our sample set for evidence of positive selection using the codeml application from the phylogenetic software package PAML (Yang, 2007). By using input sequences and phylogenetic topology, PAML compares the substitution rates of non-synonymous (dN) and synonymous (dS) codon changes for each sequence. The ratio of dN to dS (dN/dS) provides information on the nature of selective pressures acting on each gene (Holmes, 2004; Yang and Nielsen, 1998). A higher than expected rate of non-synonymous changes (dN/dS greater than 1) indicates positive selection. Using likelihood ratio tests (LRT) to compare Nssites models (M1, M7) in which positive selection is prohibited (null model), to models (M2, M8) in which positive selection is allowed, we find that allowing positive selection provides a much better fit to the data (Table 1) for both the RBCC domains (p<0.001) as well as the PRYSPRY domains (p=0.002) of Trim5 genes from mice, rats and rabbits. In the RBCC domain, we estimate that 7% of codons have evolved with an average dN/dS ratio of 3, whereas in the PRYSPRY domain, 6% are estimated to have evolved with an average dN/dS ratio of 2.5. Similar results were also obtained on excluding the rabbit Trim5 gene, a known antiviral gene. In contrast, no evidence of positive selection is found in the Trim6 and Trim34 genes (data not shown).
Table 1. Likelihood ratio tests for positive selection among rodent and rabbit Trim5 homologs.
(A) Maximum likelihood trees of DNA sequences for RBCC (excluding exon8) and PRYSPRY (exon 8) are used for Trim5 paralogs from mice, rats and for rabbit Trim5α (see figure 4 for tree). The analysis for RBCC domains was repeated by excluding the Trim with the highest branch dN/dS value (Mmusc Trim12, see figure 4A). Similarly, the analysis for the PRYSPRY domain was repeated by excluding the Trim with the highest branch dN/dS value (Mdunni Tim30, see figure 4B) and also excluding rabbit Trim5 α.
(B) Mus species phylogeny (Tucker 2006) is used for Trim30 orthologs from mice and rats (see figure 5A for PRYSPRY tree). The analysis for exon8 domains was repeated by excluding Mus dunni Trim30 (the highest branch dN/dS value).
| A RBCC (no exon8) of Trim5 paralogs from Mus musculus, Mus dunni, Rat, Rabbit. | |||
|---|---|---|---|
| Codon model1 | M7 vs M82 | Tree length3 | |
| f3×4 | p<0.001 | 3.85 | |
| f61 | p<0.001 | 4.1 | |
| Excluding Mus musculus Trim12: | f61 | p<0.001 | 4.06 |
| PRYSPRY (exon8) of Trim5 paralogs from Mus musculus, Mus dunni, Rat, Rabbit. | |||
| Codon model | M7 vs M8 | Tree length | |
| f3×4 | p=0.0021 | 4.39 | |
| f61 | p=0.006 | 4.42 | |
| Excluding Mus dunni Trim30: | f61 | p=0.019 | 4.38 |
| Excluding Rabbit Trim5: | f61 | p=0.019 | 4.38 |
| B SPRY (exon8) of Trim30 orthologs from mouse species and rat. | |||
| Codon model | M7 vs M8 | Tree length | |
| f3×4 | p=0.003 | 1.63 | |
| f61 | p=0.0075 | 1.63 | |
| Excluding Mus dunni Trim30: | f61 | p=0.014 | 1.52 |
Two different codon models are used (f3×4 and f61) to ensure that codon frequency models yield similar results.
Likelihood ratio tests were done using log likelihood values obtained from PAML, comparing a null model (M7) to a positive selection model (M8). p values were obtained running a chi square test of log likelihood values, with 2 degrees of freedom.
Tree length values obtained by PAML based on the input tree, used to assess evolutionary depth of sample input.
We also calculated the dN/dS values for each branch on the Trim5 phylogenetic tree (figure 4), separately for the RBCC and PRYSPRY domains. Because these dN/dS values are averaged over the entire domain, most lineages exhibited a dN/dS less than 1 with wide variation among different lineages. Indeed, when we compared the likelihood of a single dN/dS fit over the entire tree to one that allowed different dN/dS values for each branch, the latter provided a much better fit for both the RBCC (p<0.01) and PRYSPRY (p<0.02) domains, indicating that position selection on both domains has been quite episodic, similar to previous observations of other TRIM genes (Sawyer et al., 2005; Sawyer et al., 2007). In particular, the lineage leading to Trim12 from Mus musculus in the RBCC tree shows the highest dN/dS value of 6.9, with 19 non-synonymous changes compared to 1 synonymous change (Figure 4A). Surprisingly, Trim12 is one of the mouse Trim5 homologs that lacks a PRYSPRY domain. Because this domain interacts with viral capsid, it frequently evolves the most rapidly among antiviral Trim5 genes. To rule out that the overall signature of positive selection for Trim12 might be the sole contributor to the significant LRT for the entire RBCC tree, we excluded Mus musculus Trim12 from our analysis. The null model was still rejected favorably (p<0.001, Table 1) indicating that positive selection is widespread among the RBCC domains for rodent Trim5 homologs, albeit particularly dramatic in the case of Trim12. Similarly, in the PRYSPRY (exon8) tree, Trim30 from Mus dunni shows the highest dN/dS value, with 11 non-synonymous changes compared to zero synonymous changes (Figure 4B). This rate is very similar to that seen among PRYSPRY domains of antiviral Trim5 genes from primates and cows (Liu et al., 2005; Sawyer et al., 2007), suggesting that Mus dunni Trim30 may have recently been subject to similar evolutionary pressures. Again, Mus dunni Trim30 does not solely contribute to the observed positive selection in the PRYSPRY domain, as excluding it still resulted in a robust signature of positive selection (p<0.02). Nevertheless, our results suggest that whereas positive selection is present among the PRYSPRY domain for rodent Trim5 homologs, Mus dunni Trim30 is probably contributing most of the non-synonymous changes among the PRYSPRY sequences. Taken together, these findings show that positive selection is common, episodic and, in some cases, quite strong among rodent Trim5 homologs.
Positive selection among mouse Trim30 orthologs localized to the variable loops of the PRYSPRY domain
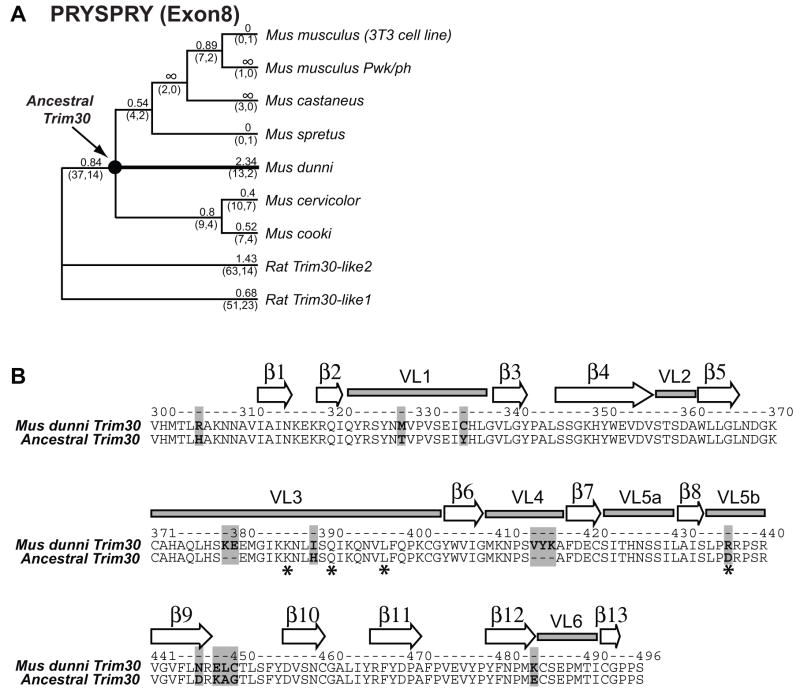
Because of the observed rapid evolution of the Mus dunni Trim30 PRYSPRY domain (Figure 4), we sequenced orthologous Trim30 sequences genomic DNA from the following additional mouse species: Mus castaneus, Mus spretus, Mus cookii, Mus cervicolor, Mus musculus, Mus dunni and Mus Pahari. Using these samples, we sequenced exon 2 and 8 of Trim30, corresponding to the RING-Bbox and the PRYSPRY domains, respectively. Phylogenetic analysis confirmed that the genomic sequences we obtained are indeed orthologs of Trim30 (data not shown). The sequences obtained from Mus pahari seemed orthologous to other Trim30-like genes rather than Trim30 and were therefore excluded from further analysis. Next, using the PRYSPRY (exon 8) sequences and a published tree for these mouse species (Tucker, 2006) we analyzed the substitution rates of each Trim30 ortholog since these species diverged. Likelihood ratio tests corroborated evidence for positive selection among mouse and rat Trim30 orthologs by rejecting the null model (p=0.0015) (Table 1). Analyzing the branch values revealed once again that Mus dunni Trim30 has the highest dN/dS value of 2.34 with 13 non-synonymous changes compared to 2 synonymous changes (Figure 5A), once again confirming the degree of rapid evolution unique to Mus dunni Trim30.
Figure 5. Codons of mouse Trim30 orthologs under positive selection are locate in the variable loops of the PRYSPRY domain.
(A) Phylogeny of Mus species (Tucker 2006) used in our study are shown for Trim30 orthologs, with an out-group to rat Trim30 paralogs. Branch dN/dS values calculated by PAML are shown above each branch. The number of non-synonymous changes (N) and synonymous changes (S) are shown below each branch in parentheses as (N,S). Branch dN/dS values significantly greater than 1 are highlighted as a bold line. The black circle indicates the node for the ancestral Trim30 sequence for Mus dunni.
(B) Protein sequence alignment of the PRYSPRY (exon8) domain for Mus dunni and its ancestral Trim30 is shown. Differences are highlighted with a gray box. The position of the beta-sheets and variable loops are based on structural work from reference (James et al., 2007). Codons under positive selection identified by PAML based on Bayes-Empirical-Bayes analysis (with posterior probability greater than 95%) are shown in *. White arrows and gray rectangles indicate beta-sheets and variable loops in the tertiary structure, respectively.
We next used Naïve Empirical Bayes and Bayes Empirical Bayes analyses to analyze which codons in the PRYSPRY domain of mouse Trim30 orthologs have accumulated most of the non-synonymous changes throughout mouse evolution. The codons that determine the specificity of human or rhesus Trim5α towards HIV-1 are located in variable loop 1 (VL1) of its PRYSPRY domain (Sawyer et al., 2005; Stremlau et al., 2005; Yap et al., 2005). Similarly, codons subject to positive selection in cow Trim5 genes and even primate Trim22 genes also are concentrated in this loop, which is exposed on the tertiary structure of PRYSPRY domains and interacts with factors in the cellular environment (James et al., 2007). We asked if the codons of Mus dunni Trim30 under positive selection also map to these variable loops of the PRYSPRY domain. We mapped residues predicted to be evolving under positive selection (posterior probability >95%) on an alignment of the Mus dunni Trim30 and a reconstructed ‘ancestral’ PRYSPRY sequence (see node on Figure 5A). We found that almost all of the changes in Mus dunni Trim30 PRYSPRY domain, compared to the constructed ancestral node, are located in the variable loops (Figure 5B, highlighted in gray). Three residue under positive selection with >95% posterior probability, all map to loop 3. These indicate that surface-exposed loops of the PRYSPRY domain of mouse Trim30 orthologs have rapidly evolved throughout mouse evolution, consistent with their potential role in antiviral defense.
Screening mouse Trim5α homologs for anti-viral activity
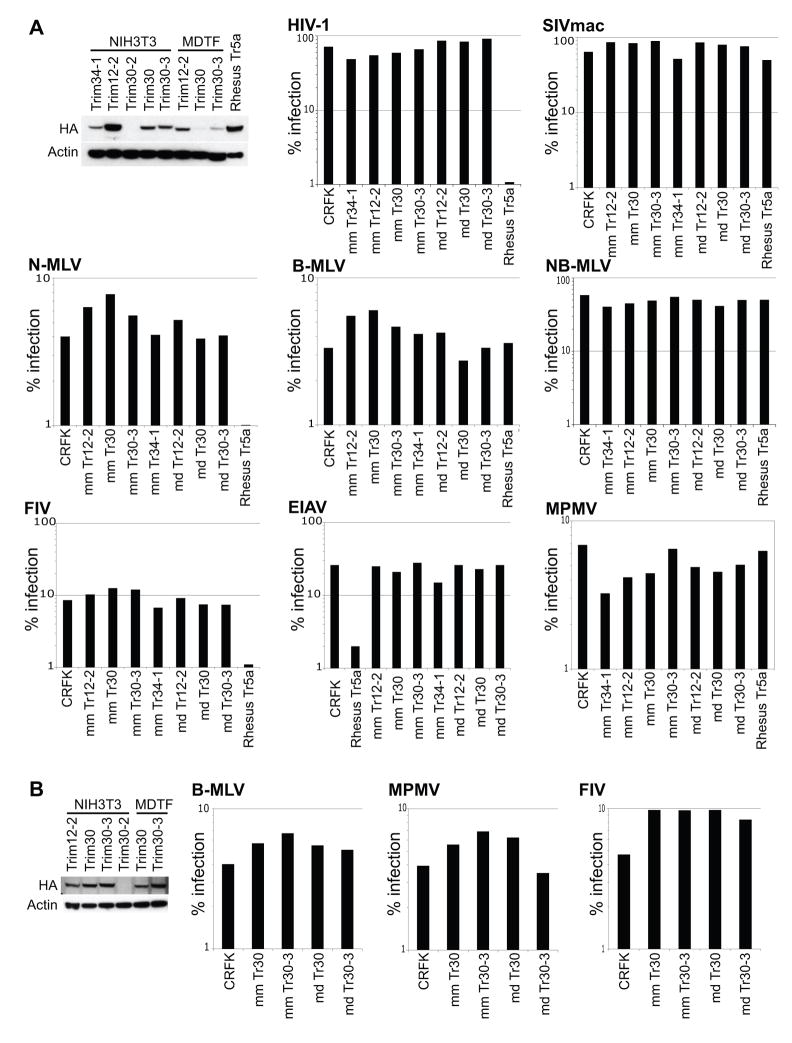
The rapid evolution of mouse canonical Trim5α homologs, particularly of Mus dunni Trim30, suggested that these genes could participate in antiviral defense. We set up an assay to test these Trim5 homologs against a diverse group of retroviruses. Since all four canonical domains are required for Trim5α activity, we focused only on the mouse Trim5 genes that contain these domains and that are expressed in mouse cell lines (Trim12-2, Trim30, Trim30-2, Trim30-3). We cloned a cDNA corresponding to each of these genes (with an epitope tag at the N-terminus) and generated CRFK (Crandel Feline Kidney) cells stably expressing each mouse Trim5α homolog. With the exception of Trim30-2, we were able to generate CRFK cells that stably express each gene from both NIH3T3 and MDTF cells (Mus musculus and Mus dunni, respectively). Initial low expression yields for MDTF Trim30 and Trim30-3 (Figure 6A) prompted us to re-transduce these clones for better expression (Figure 6B). Next, we challenged each mouse Trim5α with a diverse range of retroviruses including lentiviruses (HIV-1, SIVmac, FIV, EIAV), gammaretroviruses (N-MLV, B-MLV), and a betaretrovirus (MPMV) (Figure 6). These viruses are known to be restricted by at least one of the existing Trim5 genes in one or more primates (Brennan et al., 2007; Keckesova et al., 2004; Nisole et al., 2004; Perron et al., 2007; Saenz et al., 2005; Stremlau et al., 2004). Moreover, HIV-1 and SIVmac are restricted by rabbit Trim5α (Schaller et al., 2007), while N-MLV, SIVmac, HIV-1 and EIAV are restricted by cow Trim5α (Si et al., 2006; Ylinen et al., 2006). Thus, although this is a limited set of retroviruses, there is a precedent for restriction by one or more Trim5α proteins. Although our positive control, Trim5α from rhesus macaques, restricted the expected retroviruses, we did not find any virus that Trim12-2, Trim30, and Trim30-3 restricted in our assay. These results do not rule out the hypothesis that mouse Trim5 homologs can inhibit other viruses however.
Figure 6. Retroviruses used in our assays were not restricted by mouse Trim5 homologs.
CRFK cell lines were transduced to stably express Trim5 homologs containing all four of the canonical domains of Trim5α, cloned from NIH3T3 and MDTF cells, representing Mus musculus and Mus dunni respectively. Stable cell lines for Trim34-1 from NIH3T3 cells and Rhesus Trim5α were also generated. Each Trim gene was cloned with an N-terminal HA tag for detection via immunoblotting. (A and B) 50 ug of lysates per well, immunoblotted with anti-HA to check for expression of each Trim5 homolog. Anti-actin was used as a loading control. A second attempt at stable cell line generation was needed to boost the levels of MDTF Trim30 and Trim30-3 (B). We were unable to generate CRFK cells stably expressing Trim30-2 from either NIH3T3 or MDTF cells.
(A and B) Each stable cell line expressing one of the listed Trim genes was challenged with the list of retroviruses at sufficient amounts to give non-saturating levels of infection. Each virus is generated to express GFP upon infection. These retroviruses were used because they represent a broad range of viruses restricted by antiviral Trim5 orthologs from primates, cows and rabbits. HIV-1 (human immunodeficiency virus-1), SIVmac (simian immunodeficiency virus macaque strain), MPMV (mason-pfizer monkey virus), N-MLV (N-tropic murine leukemia virus), B-MLV (B-tropic murine leukemia virus), NB-MLV (N- and B-tropic murine leukemia virus), FIV (feline immunodeficiency virus), EIAV (equine infectious anemia virus). Percentage of cells that are infected are plotted on the y-axis as a log-scale.
DISCUSSION
Here, we describe the organization and evolutionary history of multiple orthologs of the antiviral restriction factor Trim5α found in both mouse and rat genomes. We show that Trim5 has undergone a paralogous gene expansion in both rat and mice relative to the primate Trim5 locus. Similar to old world primate and bovine Trim5α, murine Trim5 genes show evidence for positive selection with most of these signatures found in the variable loops of the PRYSPRY domain. Although we were unable to find antiviral activity for any of the full-length murine Trim5 genes (using a set of eight retroviral challenges), their signatures of positive selection indicate that selective pressures (possibly in the form of viral capsids) have acted upon these genes throughout rodent evolution. Moreover, we find that Trim5 has undergone independent evolutionary fates within each species.
Paralogous gene expansions are subject to several possible outcomes. For instance, gene families can functionally diversify and take on functional duties under unique temporal or spatial expression profiles (Nei and Rooney, 2005). In the case of Trim5α, the expansions might allow for broader specificities towards incoming viruses, thus expanding the defensive shield against these pathogens. Together with other known mouse restriction factors such as Fv1 (Best et al., 1996), mouse Trim5 paralogs may further broaden the range of defense. Of note, the Trim5 gene expansion is not unique to rodents; cows have had a similar but independent expansion that has resulted in at least six Trim5 genes (Sawyer et al., 2007). This suggests that paralogous expansions of Trim5 genes have occurred in at least two distinct lineages among mammals (rodents and cows), and possibly more species-specific Trim5α expansions will become apparent as genome projects continue to expand. The fact that the mouse expansion is not as drastic in rats suggests that the mouse expansion occurred after the mouse-rat species split, dating to between 41 million years ago (Hedges, 2002) and 10 million years ago (Guenet and Bonhomme, 2003). Tracing the history of the Trim5 locus during rodent evolution could reveal interesting dynamics. Interestingly, Mus pahari, the earliest diverged mouse in our sample set, contained Trim30-like sequences that were not direct Trim30 orthologs (data not shown). In particular, screening diverse mouse species’ genomes for Trim5 paralogs will provide a more accurate history for the mouse Trim5 locus.
The murine Trim5 expansion serves as an interesting contrast to the APOBEC3 locus which contains only one gene in mice, and seven genes in primates. It is not clear what selection pressures may have driven the expansion of one antiviral locus over another, although existing evidence suggests that the APOBEC3 genes have a broader specificity against multiple different viruses as well as against different retroelements (Strebel and Khan, 2008), while the Trim5 genes have a much more limited specificity against particular retroviruses. Nevertheless, it appears that primates have more heavily ‘invested’ in an Apobec3-based defense strategy whereas other mammals have favored a more Trim-based strategy. Having multiple Trim5 genes may seem beneficial at first, but that could come at a cost, which may explain why Trim5 expansions have not occurred in primates. In some cases, a loss-of-antiviral-function even seems to be tolerated in certain human populations with Trim5 alleles containing the H43Y mutation (Sawyer et al., 2006).
Some members of the TRIM multigene family appear to have arisen as a result of paralogous expansions before mammalian speciation (Reymond et al., 2001). This possibility is further strengthened by the fact that the Trim5 locus is in synteny among humans, mice, rats, dogs and cows ((Sawyer et al., 2007), and work presented here). The fact that the Trim5 loci among these species sits in an expansion of class 1 olfactory receptor genes ((Taylor et al., 2006), and work presented here) suggests the possibility that these and other Trim genes may have expanded due to this association with olfactory receptor gene expansions. Olfactory receptor gene clusters have had a history of dramatic expansions in mammalian genomes (Young and Trask, 2002). It is therefore possible that recombination events triggered by olfactory receptor gene expansions could have facilitated the expansion of Trim5 paralogs in both mouse and cow genomes.
The fact that most of the mouse Trim5 paralogs have not become pseudogenized after their expansion suggests that they have either retained functions of their parental genes, or have acquired novel functions. While work described here predicts antiviral roles for these paralogs, it appears that some have unique cellular roles as well. Previous reports have shown Trim12 and Trim30 transcripts to be down-regulated in NOD mice, which are susceptible to autoimmunity (Liston et al., 2007). Recently, Trim30 was shown to function as an anti-inflammatory factor that is able to down regulate NF-kB signaling by degrading signaling intermediates, TAB2 and TAB3 (Shi et al., 2008). An older report that referred to Trim30 as Rpt-1 showed that it was able to inhibit HIV-1 LTR-driven transcription (Patarca et al., 1988). Whether these functions are unique to these mouse paralogs, or are ancestral functions of Trim5-like proteins remains to be investigated.
Trim5 genes in primates are known to encode splice variants. Trim5α is one of these splice variants that harbors all four of the domains. Another primate splice variant, Trim5delta for example (Xu et al., 2003), does not encode the PRYSPRY domain and was shown to be a dominant inhibitor of Trim5α restriction (Passerini et al., 2006). During the work described here we did not investigate whether any of the rodent Trim5 genes encode alternative splice variants. It is possible that mouse Trim5 genes that do not encode all four domains (such as Trim12, Trim12-X, Trim30-1, Trim30-4) may obviate the need for alternative splice variants. However, the high dN/dS values for mouse Trim12 demonstrates, to our knowledge, the first evidence of a RBCC domain-only Trim5 homolog that contains such high rates of adaptive evolution. Since Trim5 proteins are known to multimerize, this finding may indicate a functional significance for mouse Trim12 in heteromultimerizing with other paralogs that contain the PRYSPRY domain. It is possible that these truncated mouse Trim5 genes could encode for unique cellular functions or could act as dominant inhibitors of mouse Trim restriction factors.
Retroviruses that we used in our assays were not restricted by mouse Trim5α homologs. However, the signatures of positive selection seen among these homologs, especially in Mus dunni Trim30, strongly suggest that these genes have an evolutionary history similar to their mammalian antiviral orthologs. The viruses they restrict remain to be identified. However, it is also possible that the viruses that led to the selective events were long in the past and are now either extinct, or exist as endogenous elements in the mouse genome. Almost 40% of the mouse genome consists of endogenous transposable elements, 10% of which are endogenous retroviruses (ERVs) while the rest are mostly retrotransposons (Mouse Genome Sequencing Consortium et al., 2002). Many of the mouse ERVs are still active and are capable of replicating and introducing mutations into the mouse germ line (Maksakova et al., 2006; Stocking and Kozak, 2008). In contrast, most ERVs in humans are extinct, with the exception of HERV-K (Turner et al., 2001). Therefore, it is possible that murine Trim5 proteins serve to keep in check one or more of the many active endogenous retroviruses encoded by their genome. The retroviruses we used in our antiviral assays do have endogenous relatives in the mouse genome. For example, N-MLV and B-MLV (genus Gammaretrovirus) are related to class I mouse ERVs, while MPMV (genus Betaretrovirus) is related to class II mouse ERVs. However, the list of retroviruses used in our assay, albeit phylogenetically diverse, does not even come close to represent the broad diversity of endogenous and exogenous retroviruses that mice encounter (Boeke and Stoye, 1997; Rosenber and Jolicoeur, 1997; Stocking and Kozak, 2008). It is therefore not unlikely that selective pressures from such broad pathogenic diversity may have selected for a rich repertoire of antiviral genes, resulting in the cluster of paralogous mouse Trim5 genes described here. The present studies will make it possible to probe the function of Trim5 proteins in an in vivo model using targeted knock-outs in mice.
MATERIALS & METHODS
Gathering sequences from the rodent Trim5 gene cluster
The UCSC genome browser BLAT tool (Kent, 2002) was used to search the mouse and rat genome databases using human sequences of Trim5α, Trim22, Trim6 and Trim34 as queries. Mouse BAC sequences were used to confirm the genome assembly at this locus: AC123830, AC142110, AC122400, AC135109. Rat BAC sequences used were AC112864 and AC113720. The olfactory receptor genes that flank each side of the rodent Trim5 loci were also identified through annotations available on the UCSC genome browser, and were confirmed on BAC clones. To exhaustively identify partial Trim genes within the rodent Trim5 loci, we queried BAC sequences with individual exons from Trim5α, Trim22, Trim6 and Trim34.
Phylogenetic analysis
All TRIM-like rodent sequences were aligned with known Trim5, 22, 6, and 34 orthologs from human, macaque, cow, dog, and rabbit using the multiple alignment program Clustal X (Jeanmougin et al., 1998). PAUP*4.0b10 (Swofford, 2003) was used to infer phylogenies and create maximum likelihood (ML) trees from this alignment. General phylogeny of the Trim homologs was established using the DNA sequences corresponding to the RBCC domains (approximately 750bp in length), as this tree was consistently supported with high confidence. The ML tree was generated as follows: First, a maximum parsimony tree was created using a heuristic search. Next, using likelihood analysis, the substitution model and rate variation were empirically determined from the dataset. Finally, using the determined parameters, a ML tree was constructed using a heuristic search. Bootstrap analysis was performed using a full heuristic approach with 100 replicates. Nodes with less than 50% bootstrap support were collapsed or shown where appropriate.
ML trees were also obtained for data sets consisting of only Trim5 homologs (as in figures 3 and 4). In these cases, two trees were constructed using either full length DNA sequences but exon 8 (approximately 910bp), or using only exon 8 DNA sequences (approximately 700bp).
For Trim30 orthologs from different mouse species (as in figure 5A), previously published phylogeny for these species was used (Tucker, 2006).
Recombination analysis
In order to detect potential recombination events, the online GARD tool (Kosakovsky Pond et al., 2006) on the DataMonkey server (Pond and Frost, 2005) was utilized. GARD was asked to identify a single breakpoint in our data set, which it then uses the second-order Akaike Information Criterion (c-AIC) to assess the goodness of fit for possible recombination breakpoints. Next, two ML trees were constructed as described above, consisting of either exon8 only (which corresponds to the PRYSPRY domain) or the full sequence except exon8 (which corresponds to the RING, B-box, and coiled-coil domains). The resulting trees were compared to one another for consistency of branching topology. To see if the two topologies are significantly different, we used the KH and SH tests (Kishino and Hasegawa 1989, Shimodaira and Hasegawa 1999) using the RELL distribution, where we forced the two topologies onto the data set.
Cloning and sequencing of mouse Trim5α homologs
Mouse Trim5 homologs were cloned and sequenced using reverse transcribed RNA from NIH3T3 and MDTF cell lines, which are from Mus musculus and Mus dunni, respectively. Briefly, primers designed specifically to 5′ and 3′ ends of cDNA were used to amplify each mouse Trim5 homolog. Primers for mouse Trim12-1 and Trim12-2 are CACAGCAACTATGGCTTCACAATTCATG (forward), TTAAGAGTCTGGCCAGCAAATTGTCATGG (reverse). Primers for mouse Trim30 are ATGGCCTCATCAGTCCTGGAGATG (forward), CTAGGAGGGTGGCCCGCATATAG (reverse). Primers for mouse Trim30-2 are ATGGCCTCCTCAGCTCTGGC (forward), TCATTCTTTTGACTGTGTTTCCACAG (reverse). Primers for mouse Trim30-3 are ATGGCCTCATCAGTCCTGGAGATG (forward), CTAGGATGGTGGTCCGCATACTGTC (reverse). PCR products were first cloned into TOPO TA cloning vector (Invitrogen) for sequence verification, followed by cloning into a retroviral expression vector pLNCXm1 together with a N-terminal HA tag. Orthologs of Trim30 from mouse species were sequenced from genomic DNA corresponding to Mus musculus, Mus Spretus, Mus Cervicolor, Mus Cooki, Mus Caroli, and Mus Pahari. These samples were provided generously by Priscilla K. Tucker. Nested primers were designed to bind flanking introns and amplify exon2 and exon8 of Trim30 directly from genomic DNA. Exon2 primer pairs are GTGGTCCTTCCTTGACCTCTGC (forward), ATGTAAAAGAGTATATAGAATGATATATCAACTTCC (reverse), and their nested pairs TTCCACTTCTTACTTGGTCCTTTTCC (forward), AAGCCTATAATGCTCAGGTAAGTACC (reverse). Exon8 primers are TCTCTGTTAAGTCTCAGGTTTCATGG (forward), TTAGAATGATGCAAGGGGTTTCTTGC (reverse), and their nested pairs CCACTATGTGCAGCCCTCTGC (forward), GCATAAGATACACCAGACATGGGG (reverse).
PAML, positive selection and statistical analyses
DNA sequences for either the RBCC domain (excluding exon 8) or the PRYSPRY domain (exon8) were aligned using ClustalX and input files for PAML were prepared using the online protein to nucleic acid alignment converter PAL2NAL (Suyama et al., 2006). The codeml program in PAML was used to examine positive selection in two analyses: Codon models and branch models.
In codon model analysis, each codon site was analyzed for signatures of positive selection. First, likelihood ratio tests were performed to compare the null model (M1 and M7) to the positive selection model (M2 and M8). Both M1 and M2 assume that all codons fall into a few discrete categories of dN/dS. M1 assumes that no codons have dN/dS greater than 1, while M2 allows this. M7 and M8 assume a beta-distribution of codon dN/dS values. M7 assumes that no codon has a dN/dS greater than 1, while M8 allows this. The maximum likelihood estimates obtained were then used to calculate the p values using the chi square test, with 2 degrees of freedom. These analyses were performed using two separate models of codon frequencies: the f3×4 codon frequency table, and the f61 codon frequency table (where the frequency of each of the 61 non-stop codons is empirically derived from the dataset). In cases where codon models accommodating positive selection were supported, codon positions subject to positive selection was identified with codeml Naive Empirical Bayes and Base Empirical Bayes analysis. The percentage of codons with average dN/dS values are reported from the M7 vs M8 comparison.
In branch model analysis, branch-specific values of dN/dS were assessed in codeml using the free ratio model allowing one dN/dS per branch. Likelihood ratio tests were performed to compare the universal model (M0) where dN/dS is forced to be same on all branches, to the unique branch model (M1) where a unique dN/dS is allowed for each branch. The maximum likelihood estimates obtained were then used to calculate the p values using the chi square test. Degrees of freedom for branch analysis was calculated by the formula 2(n-1)-1, where n equals the number of taxa.
Generation of stable cell lines
Mouse Trim5 homologs were cloned into the retroviral expression vector pLNCXm1 as described above. To generate virus stocks for transductions, 293T cells were co-transfected with each mouse Trim5 retroviral vector construct, together with vectors that encode Gag-Pol, and vectors that encode the glycoprotein of Vesicular Stomatitis Virus (VSV-G). Trans-IT transfection reagent was used (Mirus Bio). Viral supernatant were collected and filtered 3 days post transfection. CRFK (Crandel Feline Kidney) cells were transduced with virus containing each of the mouse Trim5α homologs and stable transductions were selected for using G418 at 0.4mg/ml. Expression of constructs were confirmed using immunoblotting against the HA epitope tag.
Restriction assays
Infectious virus stocks were prepared by transient transfection of 293T cells using the Trans-IT transfection reagent (Mirus Bio). DNA constructs for virus stocks include the following: pL-VSV-G (an expression vector of VSV-G protein for pseudotyping) (Bartz and Vodicka, 1997), pCMV-tat (to allow efficient expression of VSV-G, which is under the HIV-1 LTR promoter) (Bartz and Vodicka, 1997), Gag-Pol expression vector for each virus, and GFP-ecoding MLV-based transfer vectors (Miller and Rosman, 1989). CRFK cells stably expressing each mouse Trim5α homolog were infected with GFP-expressing retroviruses at non-saturating levels. 2 days post infection, flow cytometry was used to measure the percentage of cells expressing GFP upon infection.
Acknowledgments
We thank Priscilla K. Tucker for her generous gift of mouse genomic DNA samples; Nels Elde, Nisha Duggal, and Yegor Voronin for comments on the manuscript. This work was supported by NIH grant R37 AI30937 (M.E.), Searle Scholar and Burroughs-Wellcome Investigator Awards (H.S.M.), and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and NIH F32 GM074299 (S.L.S).
Footnotes
DECLARATION OF COMPETING INTERESTS
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz SR, Vodicka MA. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Intrinsic immunity: A front-line defense against viral attack. Nat immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Stoye JP. Retroviruses. In: Coffin JM, Hughes SH, Varmus HE, editors. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. Cold Spring Harbor Laboratory Press; New York: 1997. p. 343. [PubMed] [Google Scholar]
- Brennan G, Kozyrev Y, Kodama T, Hu SL. Novel TRIM5 isoforms expressed by macaca nemestrina. J virol. 2007;81:12210–12217. doi: 10.1128/JVI.02499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al. Genome sequence of the brown norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Guenet JL, Bonhomme F. Wild mice: An ever-increasing contribution to a popular mammalian model. Trends genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat rev genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Adaptation and immunity. PLoS biol. 2004;2:E307. doi: 10.1371/journal.pbio.0020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc natl acad sci USA. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with clustal X. Trends biochem sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human Trim5αlpha antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ. The human and african green monkey Trim5αlpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc natl acad sci USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J mol evol. 1989;29:170–9. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: A genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Liston A, Hardy K, Pittelkow Y, Wilson SR, Makaroff LE, Fahrer AM, et al. Impairment of organ-specific T cell negative selection by diabetes susceptibility genes: Genomic analysis by mRNA profiling. Genome biol. 2007;8:R12. doi: 10.1186/gb-2007-8-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Wang YQ, Liao CH, Kuang YQ, Zheng YT, Su B. Adaptive evolution of primate Trim5αlpha, a gene restricting HIV-1 infection. Gene. 2005;362:109–116. doi: 10.1016/j.gene.2005.06.045. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–2. 984–6, 989–90. [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nakayama EE, Miyoshi H, Nagai Y, Shioda T. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of african green monkey Trim5αlpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J virol. 2005;79:8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu rev genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc natl acad sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat rev microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- Passerini LD, Keckesova Z, Towers GJ. Retroviral restriction factors Fv1 and TRIM5alpha act independently and can compete for incoming virus before reverse transcription. J virol. 2006;80:2100–5. doi: 10.1128/JVI.80.5.2100-2105.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patarca R, Freeman GJ, Schwartz J, Singh RP, Kong QT, Murphy E, et al. Rpt-1, an intracellular protein from helper/inducer T cells that regulates gene expression of interleukin 2 receptor and human immunodeficiency virus type 1. Proc natl acad sci USA. 1988;85:2733–2737. doi: 10.1073/pnas.85.8.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. The human Trim5αlpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J virol. 2007;81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenber N, Jolicoeur P. Retroviruses. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviral pathogenesis. Cold Spring Harbor Laboratory Press; New York: 1997. p. 475. [PubMed] [Google Scholar]
- Saenz DT, Teo W, Olsen JC, Poeschla EM. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate Trim5αlpha proteins. J virol. 2005;79:15175–15188. doi: 10.1128/JVI.79.24.15175-15188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Akey JM, Emerman M, Malik HS. High-frequency persistence of an impaired allele of the retroviral defense gene Trim5αlpha in humans. Curr biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate Trim5αlpha identifies a critical species-specific retroviral restriction domain. Proc natl acad sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Hue S, Towers GJ. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J virol. 2007;81:11713–11721. doi: 10.1128/JVI.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Luban J. Trim5αlpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol biol evol. 1999;16:1114–6. [Google Scholar]
- Si Z, Vandegraaff N, O’huigin C, Song B, Yuan W, Xu C, et al. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc natl acad sci USA. 2006;103:7454–7459. doi: 10.1073/pnas.0600771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocking C, Kozak CA. Murine endogenous retroviruses. Cell mol life sci. 2008 doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Khan MA. APOBEC3G encapsidation into HIV-1 virions: Which RNA is it? Retrovirology. 2008;5:55. doi: 10.1186/1742-4690-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component Trim5αlpha restricts HIV-1 infection in old world monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, et al. Specific recognition and accelerated uncoating of retroviral capsids by the Trim5αlpha restriction factor. Proc natl acad sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of Trim5αlpha determines the potency of human immunodeficiency virus restriction. J virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic acids res. 2006;34:W609–12. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of eurasian mus based on mitochondrial and nuclear gene sequences. Mol phylogenet evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- Taylor TD, Noguchi H, Totoki Y, Toyoda A, Kuroki Y, Dewar K, et al. Human chromosome 11 DNA sequence and analysis including novel gene identification. Nature. 2006;440:497–500. doi: 10.1038/nature04632. [DOI] [PubMed] [Google Scholar]
- Tucker PK. Mouse in biomedical research: History, genetics and wild mice. In: Fox JG, Newcomer C, Smith A, Barthold S, Quimby F, Davisson M, editors. Systematics of the genus mus. Academic Press; 2006. pp. 13-13–23. [Google Scholar]
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus Trim5αlpha restriction of HIV-1 reverse transcription and infection. Proc natl acad sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yang L, Moitra PK, Hashimoto K, Rallabhandi P, Kaul S, et al. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp cell res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol biol evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J mol evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5αlpha leads to HIV-1 restriction. Curr biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Ylinen LM, Keckesova Z, Webb BL, Gifford RJ, Smith TP, Towers GJ. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J virol. 2006;80:7332–7338. doi: 10.1128/JVI.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Trask BJ. The sense of smell: genomics of vertebrate odorant receptors. Hum Mol Genet. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]