Abstract
RNase A (bovine pancreatic ribonuclease) is the founding member an extensive family of divergent proteins that share specific elements of sequence homology, a unique disulfide-bonded tertiary structure, and the ability to hydrolyze polymeric RNA. Among the more intriguing and perhaps counterintuitive findings, at current state of the art, the connection between ribonuclease activity and characterized host defense functions is quite weak. Whether this is a scientific reality or more a reflection of what has been chosen for study remains to be determined. Several of the RNase A family ribonucleases are highly cationic and have cytotoxic and bactericidal properties that are clearly (eosinophil cationic protein, leukocyte RNase A-2) or are probably (RNase 7) unrelated to their enzymatic activity. Interestingly, peptides derived from the leukocyte RNase A-2 sequence are nearly as bactericidal as the entire protein, suggesting that, among other functions, the RNase A superfamily may be serving as a source of gene-scaffolds for the generation of novel cytotoxic peptides. Other RNase A ribonucleases are somewhat less cationic (mouse angiogenin 4, zebrafish RNases) and have moderate bactericidal activities that have not yet been explored mechanistically. Additional host defense functions characterized specifically for the RNase, eosinophil-derived neurotoxin include reducing infectivity of RNA viruses for target cells in culture, which does require ribonuclease activity, chemoattraction of immature human dendritic cells via a G-protein coupled receptor-dependent mechanism, and activation of toll-like receptor 2. The properties of individual RNase A ribonucleases, recent experimental findings, and important questions for the near and distant future will be reviewed.
Keywords: leukocyte, cationic, eosinophil, evolution, bactericidal, virus
Introduction
RNase A (also known as bovine pancreatic RNase) is perhaps the best-characterized of all known mammalian proteins, and much of what has been learned about its chemistry and catalytic mechanism has had a direct impact on our understanding of the nature and evolution of the gene superfamily that bears its name [1]. However, it is not at all evident mechanistically how RNase A ribonucleases, whose members share specific elements of sequence homology, a unique three-dimensional disulfide-bonded structure, and the ability to hydrolyze polymeric RNA, might function to promote host defense. Given the timing of experimental discoveries, many of the better-known connections to host defense, including bactericidal, helminthotoxic and cytotoxic functions, preceded discovery of the gene family, and (perhaps as a result) the vast majority of host defense functions currently characterized for RNase A ribonucleases are not related at all to their enzymatic activity. This observation may relate more closely to the experimental directions taken by individual researchers rather than the nature of the biology itself, as there may be important functions that do rely on active enzymatic activity that simply have not yet been considered and explored. A summary of our current knowledge vis à vis RNase A ribonucleases and host defense is included in Table 1.
Table 1.
Summary of host defense functions examined for RNase A family ribonucleases.
| RNase A Ribonuclease | Species | PI | Host Defense Activities |
|---|---|---|---|
| Eosinophil-derived neurotoxin (RNase 2) | Human | 9.0 | Chemoattractant for immature human dendritic cells [2]. |
| Enhances maturation of dendritic cells [3]1 | |||
| Reduces infectivity for RNA viruses (HRSV, HIV) in tissue culture assays [4 - 6]2 | |||
| Produced in macrophages in response to LPS and TNF-α [3] | |||
| Activates TLR2 and augments Th2 biased immune responses [7] | |||
| Eosinophil cationic protein (RNase 3) | Human | 10.8 | Cytotoxin for helminthes [8 -14], strong bactericidal activity [15] |
| Reduces infectivity for HRSV in tissue culture assay [16] | |||
| Eosinophil-associated RNase 2 | Mouse | 9.70 | Chemoattractant for immature human dendritic cells [2] |
| Eosinophil-associated RNase 11 | Mouse | 9.10 | Produced by alveolar macrophages in response to Th2 cytokine stimulation [17] |
| Produced in response to respiratory virus infection, up-regulated in the absence of type I interferon signaling [18] | |||
| Eosinophil-associated RNases 1 and 2 | Rat | 8.6 | Moderate bactericidal activity against E. coli and S. aureus [19, 20] |
| Angiogenin (RNase 5) | Human | 10.1 | Moderate bactericidal activity against S. pneumoniae [21]; conflicting report [22] |
| Angiogenin-4 | Mouse | 9.0 | Moderate bactericidal activity against L. monocytogenes and E. faecalis [21] |
| Produced in response to bacterial LPS [21]. | |||
| RNase 7 | Human | 10.3 | Broad spectrum strong bactericidal activity, very strong against E.faecium [23] |
| RNase 8 | Human | 8.4 | Broad spectrum moderate bactericidal activity [24] |
| Leukocyte RNase A-2 | Chicken | 10.9 | Bactericidal against E. coli, S. aureus [25] and Salmonella sps. (unpublished data) |
| Dr-RNases 1, 2, and 3 | zebrafish | 9.0, 9.2, 8.8 | Broad spectrum moderate bactericidal activity against E. coli [26] |
Values for isoelectric point (pI) are calculated based on primary amino acid sequences. Moderate bactericidal activity, low micromolar concentrations in standard overnight incubation assay reduce colony count 10 - 100 fold; strong bactericidal activity, low micromolar concentrations reduce colony count 104 to 107 fold.
Activity also shared with human pancreatic ribonuclease, or RNase 1,
Activity dependent on enzymatic (ribonuclease) activity.
The RNase A Superfamily
As noted earlier, the protein RNase A, initially isolated from bovine pancreatic tissue, was a favorite subject for protein structure and functional studies during the 1950s and 1960s, due to its thermostability and the relative abundance of its source tissue [27 - 29]. However, during the late 1980s, several seemingly unrelated proteins from other source tissues were isolated, their genes cloned and sequenced, and these proteins were ultimately found to have significant sequence homology to pancreatic ribonuclease. The founding members of what was to become the RNase A superfamily include angiogenin (RNase 5) [30], the eosinophil cationic protein (ECP / RNase 3) and eosinophil-derived neurotoxin (EDN / RNase 2) [31 - 34] and several of the bullfrog oocyte cytotoxins [35, 36]. The biochemical nature of RNase A ribonucleases and the emergence of the RNase A gene superfamily has been reviewed extensively [37, 38]; a phylogenetic tree documenting the relationships among representative RNase A ribonucleases is shown in Figure 1. Upon completion of the human genome, eight fully catalytically active RNase A ribonucleases (numbered 1 - 8) were identified together with five divergent sequences that encode proteins that are structurally incapable of degrading polymeric RNA [40 - 42]. Each RNase A ribonuclease displays some degree of nucleotide preference within RNA substrates, but they do not have rigidly selective recognition or cleavage sites. Among the signature features of this gene family, all active RNase A ribonuclease coding sequences are initiated with a signal sequence, all mature proteins include 6 - 8 appropriately spaced cysteines that form distinct disulfide bonds, and all include two catalytic histidines (H) and a single lysine (K), the latter within an invariant sequence motif (CKXXNTF). As a rule, RNase A ribonuclease coding sequences are found on a single exon, and, in the human genome, are found on chromosome 14 [42].
Figure 1. Neighbor-joining phylogenetic tree featuring the relationships among the major mammalian and non-mammalian RNase A lineages.
Amino acid sequences were aligned using ClustalW (http://clustalw.genome.jp/) and an unrooted tree was created using Mega 4.0 Evolutionary Genetics software [39; http://www.megasoftware.net/]. Bootstrap values over 50 (1000 replicates) are shown. Sequences are from GenBank and NCBI Protein databases (http://www.ncbi.nlm.nih.gov/sites/entrez); accession numbers are available from the author on request.
Host defense functions have been attributed to three specific lineages of the mammalian branch of the RNase A superfamily, including the aforementioned eosinophil secretory ribonucleases, which are rapidly diverging proteins with prominent bactericidal, helminthotoxic, and antiviral activities; RNase 7, a bactericidal cationic protein initially isolated from human skin and the closely-related RNase 8; and the angiogenins, proteins originally characterized as promoting blood vessel growth, and just recently shown to have distinct, species-specific anti-pathogen functions. Many fewer non-mammalian RNase A lineages have been studied, however host defense functions have recently been identified for the highly cationic chicken leukocyte RNase A-2, and for three RNase A ribonucleases from the zebrafish Danio rerio.
The Eosinophil Ribonucleases
The concept that RNase A ribonucleases had a role to play in promoting host defense initially emerged from studies on the eosinophil granule proteins, eosinophil cationic protein (ECP / RNase 3) and eosinophil-derived neurotoxin (EDN / RNase 2). Although the results of in vitro experiments suggest that these mediators interact with and destroy target pathogens (parasites, bacteria), it is important to understand that the role of their source cells, the eosinophils themselves, in promoting host defense remains somewhat enigmatic and highly controversial. Although eosinophils are recruited in large numbers in response to parasitic infection and allergic provocation, there is no consensus as to their overall roles, functions, or contributions to these disease states [43 - 45]. Most recently, even the issue of whether or not eosinophils function via degranulation has come under significant scrutiny [46].
To review this history of these proteins: ECP was first identified and isolated as an arginine rich cationic protein secreted from eosinophils [8, 47] that was toxic to human parasites when evaluated in vitro [9 - 11]. Likewise, ECP has documented bactericidal activity in vitro [15] which has been exploited as a useful and general model of cytotoxicity, although there is in fact no evidence that eosinophils themselves have any role to play in combating bacterial infection in vivo. Similarly, EDN was identified as a component of eosinophils that induced a cerebellar syndrome (the Gordon phenomenon) when injected intrathecally into rabbits [48], although there is likewise no evidence to suggest that this represents a physiologic phenomenon. Interestingly, although EDN and ECP have similar biochemical properties, EDN is relatively neutral and significantly less effective when evaluated in vitro against helminthes and bacterial pathogens [10 - 15].
Gleich and colleagues were the first to report the amino terminal sequences of EDN and ECP, to observe that the sequences were similar to one another as well as to that of pancreatic RNase, and to demonstrate that each had ribonuclease activity [49, 50]. Molecular cloning confirmed membership of both these proteins into what was then the emerging RNase A gene superfamily [31-34].
Among the more interesting features of these proteins, primate EDN and ECP and the expanded group of rodent orthologs, the eosinophil-associated ribonucleases, or Ears, are known for their unusual degree of interspecies divergence, and the evolutionary constraints to which they appear to be responding have been hypothesized as directly related to their roles in promoting host defense [16]. Specifically, we have shown that EDN and ECP arose as a gene pair as a result of a duplication event that occurred shortly after the divergence of the Old World from the New World monkeys, an event dated at ~ 50 million years ago, and, since that point in time, they have been accumulating non-silent mutations at a very rapid rate [51]. Also interesting, despite the rapid rate of change, all evolutionary variants maintain the features necessary for ribonuclease activity [52, 53]. Lee and colleagues [54] identified the first mouse eosinophil-associated ribonucleases (mEARs) 1 and 2, which are highly divergent orthologs of EDN and ECP (only ~50% amino acid sequence homology). Zhang and colleagues [55] subsequently provided evidence demonstrating that the rodent Ears underwent divergence via rapid-birth-death and gene sorting, a pattern observed only among gene families characterized by functional diversity (eg.…T cell receptor, MHC), while not proof, again suggestive of a role for these proteins in promoting host defense.
Erickson and colleagues [56] have explored the role of genetic diversity within human species, focusing on ECP polymorphisms [57] and their correlations with the course of infection with the helminth, S. mansoni. Interestingly, the authors demonstrated a lower prevalence of schistosomiasis among individuals in a Ugandan population who were homozygous for the sequence polymorphism encoding a more cytotoxic variant of ECP (434GG, including an arginine at position 97 as opposed to a threonine). Of note, while this polymorphism has an impact on ECP vis à vis its cytotoxicity, it does not alter its ribonuclease activity to any significant extent [58]. This is consistent with studies performed with ribonucleolytically-inactivated ECP, which demonstrated that the cytotoxic properties of ECP were not at all dependent on ribonuclease activity [59].
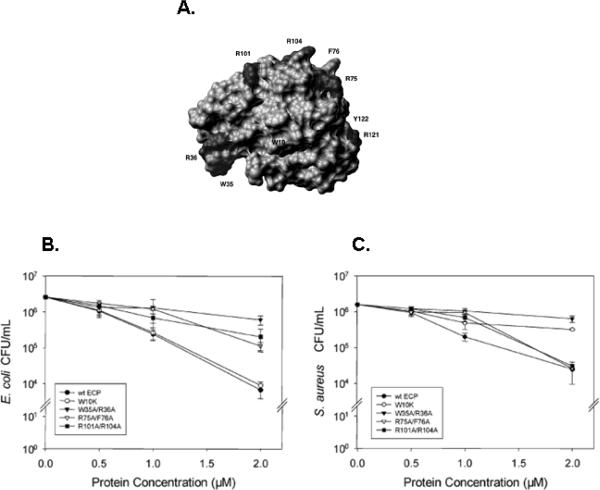
This last finding is worthy of further comment. Despite the persistence of ribonuclease activity in the face of rapid sequence change, ECP and ribonucleolytically-inactivated ECP can both function equally effectively in cytotoxicity assays. Certainly, a ribonuclease-activity-independent mechanism is consistent with the earlier findings of Young and colleagues [60] who reported that ECP functioned by destabilizing lipid membranes. This work was extended by Nogués and colleagues [61] who demonstrated that specific cationic and aromatic amino acids that map to the surface of ECP, as opposed to catalytic components, were crucial for bactericidal activity [Figure 2], and by Boix and colleagues [62] who explored the distinct roles of individual tryptophan molecules, and found the membrane destabilization properties of ECP to be consistent with a “carpet-like” mechanism. Another intriguing and yet-to-be-explored avenue of research is the possibility that ECP (and other cationic RNases) interact with and activate bacterial autolysins [63]. The molecular mechanism of ECP's cytotoxicity has been reviewed extensively [64]. However, it would seem unusual if not counterintuitive for a lineage to retain an enzymatic activity in the face of such otherwise rapid divergence for which it maintained no use whatsoever.
Figure 2. Structure-function analysis of the bactericidal activity of ECP.
(A) Spacefilling diagram documenting the location of various amino acids evaluated in a mutational analysis by Carreras et al. [61], reprinted with permission, followed by a demonstration of the importance of specific cationic arginines (R) and aromatic tryptophans (W) to the overall activity of ECP against (B) E. coli and (C) S. aureus. Overall, data suggest distinct mechanisms of action against gram negative vs. gram positive bacterial species.
Toward this end, we hypothesized that EDN and ECP might have also have activity against pathogens that were likewise subjected to evolutionary stress while remaining ribonucleolytically vulnerable; the literature linking eosinophils to the pathogenesis of disease caused by the identified target, single-stranded RNA respiratory virus pathogens, has been reviewed previously [16]. As such, we have shown that eosinophils and recombinant EDN can reduce the infectivity of the respiratory virus pathogen, human respiratory syncytial virus (hRSV) for target epithelial cells in culture [4, 16, Figure 3]. Addition of ribonuclease inhibitor eliminated the eosinophil-mediated antiviral effect; likewise, antiviral activity is not observed with the ribonucleolytically-inactivated recombinant EDN (rhEDNΔK38). We have recently expanded on this work by demonstrating a role for eosinophils in RSV clearance in vivo [65] and will proceed to explore this question with natural respiratory pathogens [66]. However, other than the existence of ribonucleolytic involvement at some level, the precise mechanism via which EDN reduces virus infectivity remains unclear. Of note, EDN has been shown to have activity against other viruses in similar in vitro settings, including human immunodeficiency virus [HIV; 5, 6].
Figure 3. (A) Isolated human eosinophils and (B) recombinant, ribonucleolytically active EDN reduce the infectivity of the pathogen, respiratory syncytial virus (RSV) for target epithelial cells in vitro.
As shown, ribonucleolytically inactivated EDN (catalytic lysine K38 converted to arginine) has no antiviral activity, although the mechanism via which EDN reduces virus infectivity remains otherwise unclear. Reprinted with permission from Domachowske et al. [4].
Another intriguing and original feature of EDN and its role in promoting host defense is its ability to promote maturation and chemotaxis of dendritic cells [2, 3]. EDN can promote chemotaxis of immature human dendritic cells nearly as effectively as the CXC chemokine, stromal-derived growth factor (SDF)-1α, and does so via a direct or indirect interaction with an otherwise unidentified pertussis (PTX) sensitive G protein coupled receptor [Figure 4]. Most recently, EDN was shown to activate TLR2 and to enhance antigen-specific Th2-biased immune responses in vivo [7]. Yang and colleagues have likewise not determined whether or not any of these features are directly related to ribonuclease activity per se, but have classified EDN as an alarmin, which are potent immunostimulants, including defensins, cathelicidin, and high-mobility group box protein 1, which serve as early warning signals to activate both innate and adaptive immune systems [67].
Figure 4. EDN as a chemoattractant.
(A) Migration of immature human dendritic cells to EDN was similar in magnitude to that seen to the chemokine SDF-1α and (B) was sensitive to inhibition by pertussis toxin (PTX). Reprinted with permission from Yang et al., [2].
The Angiogenins
Similar to the eosinophil RNases, angiogenin had been identified and substantially characterized prior to its identification as a member of the RNase A gene family [68]. Once its amino acid sequence was determined, it was clear that angiogenin (also known as RNase 5) shared the two histidine, one lysine catalytic triad, signature CKXXNTF motif, and paired cysteines typical of members of the enlarging RNase A gene superfamily. The angiogenin lineage has also been the subject of rapid diversification [69], and, recently, based on evidence from lower vertebrate sequences, Cho and colleagues [42] have suggested that angiogenin, with three as opposed to four paired cysteines, represents the more ancient of the RNase A ribonuclease lineages.
There is one functional angiogenin gene in the human genome, and six in the mouse. Of the six mouse angiogenin (Ang) genes, Hooper and colleagues [21] found that mouse Ang 4 was expressed in Paneth cells and had the unexpected property of having bactericidal activity against specific intestinal microbes [Figure 5]. Upon further evaluation, the authors found that mouse Ang 1 and human angiogenin also displayed antimicrobial activity, with 100-fold reductions of colony counts of S. pneumoniae and C. albicans observed in response to low micromolar concentrations of recombinant protein. Interestingly, Avdeeva and colleagues [22] recently reported that a commercial preparation of recombinant human angiogenin was no more effective than an albumin control at inhibiting the growth of S. pneumoniae or C. albicans. The reason for this remarkable discrepancy is not immediately apparent and will require further experimental clarification, although it might be noted that the pathogen in question, S. pneumoniae, is routinely identified by its sensitivity to detergent-mediated lysis (bile solubility test, or 2% sodium deoxycholate) and its growth in culture can be inhibited by remarkably small amounts of detergent contaminants.
Figure 5. Selective anti-pathogen activity of mouse angiogenin 4.
Although the basis for this activity remains unclear, mouse angiogenin 4 displays activity against E. faecalis and L. monocytogenes, but not against the other pathogens shown. Reprinted with permission from Hooper et al., [21].
RNases 7 and 8
Human RNase 7 was identified by Harder and Schröder [23] as part of a broad screening protocol aimed at identifying antimicrobial agents in human skin. Keratinocytes are a major source of RNase 7, which has strong bactericidal activity (colony reductions of >104 fold in response to low to mid micromolar concentrations) against a number of pathogens, with most profound activity against a clinical isolate of vancomycin-resistant Enterococcus (LD90 < 0.03 μM). RNase 7 is expressed in response to several characterized inflammatory stimuli, specifically interferon-gamma (IFN-γ), TNF-α, and IL-1β, as well as in direct response to bacteria which suggests the possibility of a TLR2 or TLR4-mediated mechanism. A parallel study by Zhang and colleagues [70] documented prominent liver expression, and commented on the unusual nature of the sequence, dominated by cationic lysines as opposed to arginines. Indeed, Huang and colleagues [71] have recently demonstrated that four specific clustered lysines (K1, K3, K111 and K112) are crucial for the membrane lytic activity of RNase 7. Although ribonuclease dependence has not been addressed directly, the correlation of lytic activity with bactericidal function suggests that, similar to ECP, ribonuclease activity per se is not a crucial feature of RNase 7-mediated cytotoxicity.
Initially identified by Zhang and colleagues [72], human RNase 8 is closely related to RNase 7, but is expressed only in the placenta. Although only a limited number of samples from each species have been evaluated, RNase 8 appears to have been pseudogenized in about half the known primate genomes. Rudolph and colleagues [24] have recently shown that human RNase 8 has antimicrobial activity against a variety of pathogens. Unfortunately (from an experimentalist's perspective), the mouse genome does not contain orthologs of either RNase 7 or RNase 8, thus the ways in which we can further our understanding of the roles of these proteins in promoting host defense in vivo are somewhat limited.
Non-Mammalian RNase A Ribonucleases
As might be expected, the vast majority of work has been focused on the characterization of RNase A ribonucleases in mammals, primarily in humans and human cell lines. Recently, the properties of several non-mammalian RNase A ribonuclease lineages have been revealed, and the potential role of these proteins in promoting endogenous host defense will be reviewed here.
The Zebrafish RNases
Two groups have recently reported their findings resulting from searches of the zebrafish (Danio rerio) sequence databases [26, 73, 74]. The zebrafish RNase A sequences are typical of the ancestral RNases, with appropriate catalytic histidines and lysine within the conserved RNase A superfamily motif, and 6 (as opposed to 8) conserved cysteines. The open reading frames encode relatively small proteins (MW 13 - 15 kDa) with a relatively low isoelectric points for this gene superfamily (pI = 8.8 - 9.2). Interestingly, despite their relatively low cationicity (comparable to human EDN, which has no detectable bactericidal activity in a similar assay [53]), recombinant proteins had some bactericidal activity, particularly against gram-negative E. coli, although not of the same magnitude as observed for the highly cationic RNases (e.g…RNase 7, or leukocyte RNase A-2, below). Given their limited cationicity, it would be most interesting to know whether the bactericidal activity observed for these proteins occurred via a distinct, perhaps ribonuclease-dependent mechanism. However, mucosal localization [Figure 6] and bactericidal activity among these distant RNase A ribonucleases stand in strong support of the authors' conclusions that host defense represents a primordial function of this gene lineage [26].
Figure 6. Zebrafish RNase A ribonucleases.
(A) Tissue distribution determined by RT-PCR including (E) eye, (H) heart, (B) brain, (L) liver, (G) gut, (T) testis, (O) ovary, (S) skin, and (M) skeletal muscle. (B) Comparative ribonuclease activity against yeast tRNA substrate. Reprinted with permission from Cho and Zhang, [26].
The Bullfrog RNases
The RNase A ribonuclease isolated from oocytes from the bullfrog, Rana pipiens, was one of the original members of the emerging RNase A gene superfamily [36], although for many years this sequence and the related sequences from Rana catesbeiana remained alone among the characterized non-mammalian members of this group. The focus of these frog RNases has been their biotherapeutic, as opposed to biologic potential; the Rana pipiens sequence, also known as onconase, (trade name Ranpirnase) is currently undergoing clinical trials for unresectable mesothelioma and non small cell lung cancer [75]. The biotherapeutic potential of this ribonuclease lineage is based all or in part on their structural resistance to mammalian cellular ribonuclease inhibitor, and thus, once taken up by the cell, its apparently unrestricted capacity to degrade cellular RNA [76]. Associated with this property, Ardelt and colleagues [77] have recently shown that onconase can also suppress intracellular oxidative stress. Remaining unexplored, however, is the role of these proteins in their natural setting. Of interest, the Rana ribonucleases have diverged into multigene clusters under positive selection [78], as have the rodent eosinophil ribonucleases, although the overall significance of this vis à vis host defense function remains speculative.
Chicken Leukocyte RNases A-1 and A-2
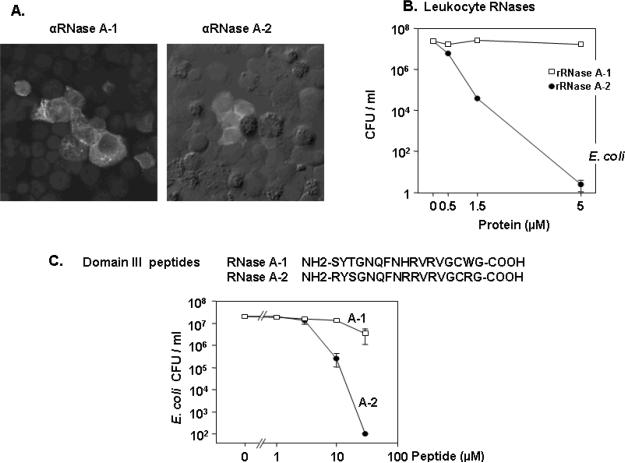
Nitto and colleagues [25] characterized two RNase A ribonucleases originally isolated from chicken leukocyte-derived source material [79]. Renamed Leukocyte RNases A-1 and A-2, both related proteins (diverging under positive selection pressure) were found in chicken peripheral blood heterophils as would be appropriate for prominent host defense proteins [Figure 7]. Although RNase A-1 is the more active ribonuclease, RNase A-2 is more cationic (pI = 11.0), angiogenic and profoundly bactericidal against both grampositive and gram-negative species, with micromolar quantities reducing the colony count of E. coli from 107 CFU/ml to near zero. Most intriguing, not only was ribonuclease activity irrelevant to bactericidal activity (similar to earlier findings with ECP [59]), but independent linear domains within the RNase A-2 sequence were nearly as effective at reducing the colony count as the full RNase A-2, suggesting that, not only is RNase activity unimportant, that the tertiary structure limited by the cysteines may also be dispensible. Taken one step further, these results suggest that, at least among duplicate genes, the ribonuclease gene structure may be serving merely as a scaffold to support the evolution of novel, non-ribonucleolytic host defense (and other?) peptides.
Figure 7. Bactericidal activities of chicken leukocyte RNase A-2 and domain III peptide.
(A) Leukocyte RNases A-1 and A-2 are found in chicken heterophils and bone marrow progenitors. (B) RNase A-2 is profoundly bactericidal, reducing the colony count of E. coli from 107 to near zero and (C) the 17- amino acid domain III peptide, taken from sequence near the C-terminus, is nearly equally effective. Reprinted with permission from Nitto et al., [22].
Conclusions
Our enlarging understanding of the RNase A gene superfamily has provided us with a greater appreciation of the role of these proteins in promoting host defense. Although the most prominent activity explored is ribonuclease-independent bactericidal activity, more recent explorations have focused on the role of these mediators, particularly EDN, in antiviral host defense, as chemoattractant agents and as endogenous ligands for toll-like receptors. Future work will permit us to focus on specific, unique and perhaps more subtle roles played by individual RNase A ribonuclease lineages.
Acknowledgements
I am deeply indebted to my colleagues who share my fascination with RNase A ribonucleases, and particularly to those who have permitted me to reprint their published work here. Work in my lab is funded by the NIAID Division of Intramural Research.
References
- 1.Beintema JJ, Kleineidam RG. The ribonuclease A superfamily: general discussion. Cell. Mol. Life Sci. 1998;54:825–832. doi: 10.1007/s000180050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang D, Rosenberg HF, Chen Q, Dyer KD, Kurosaka K, Oppenheim JJ. Eosinophil-derived neurotoxin (EDN), an antimicrobial protein with chemotactic activities for dendritic cells. Blood. 2003;102:3396–3403. doi: 10.1182/blood-2003-01-0151. [DOI] [PubMed] [Google Scholar]
- 3.Yang D, Chen Q, Rosenberg HF, Rybak SM, Newton DL, Wang ZY, Fu Q, Tchernev VT, Wang M, Schweitzer B, Kingsmore SF, Patel DD, Oppenheim JJ, Howard OM. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J. Immunol. 2004;173:6134–6142. doi: 10.4049/jimmunol.173.10.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domachowske JB, Dyer KD, Bonville CA, Rosenberg HF. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis. 1998;177:1458–1464. doi: 10.1086/515322. [DOI] [PubMed] [Google Scholar]
- 5.Lee-Huang S, Huang PL, Sun Y, Huang PL, Kung HF, Blithe DL, Chen HC. Lysozyme and RNases as anti-HIV components in beta-core preparations of human chorionic gonadotropin. Proc. Natl. Acad. Sci. USA. 1999;96:2678–2681. doi: 10.1073/pnas.96.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rugeles MT, Trubey CM, Bedoya VI, Pinto LA, Oppenheim JJ, Rybak SM, Shearer GM. Ribonuclease is partly responsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. AIDS. 2003;17:481–486. doi: 10.1097/00002030-200303070-00002. [DOI] [PubMed] [Google Scholar]
- 7.Yang D, Chen Q, Su SB, Zhang P, Kurosaka K, Caspi RR, Michalek SM, Rosenberg HF, Zhang N, Oppenheim JJ. Eosinophil-derived neurotoxin acts as an alarmin to activate TLR2-MyD88 signal pathway in dendritic cells and enhances Th2 immune responses. J. Exp. Med. 2007 doi: 10.1084/jem.20062027. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackerman SJ, Loegering DA, Venge P, Olsson I, Harley JB, Fauci AS, Gleich GJ. Distinctive cationic proteins of the human eosinophil granule: major basic protein, eosinophil cationic protein, and eosinophil-derived neurotoxin. J. Immunol. 1983;131:2977–2982. [PubMed] [Google Scholar]
- 9.Hamann KJ, Gleich GJ, Checkel JL, Loegering DA, McCall JW, Barker RL. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J. Immunol. 1990;144:3166–3173. [PubMed] [Google Scholar]
- 10.Molina HA, Kierszenbaum F, Hamann KJ, Gleich GJ. Toxic effects produced or mediated by human eosinophil granule components on Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 1988;38:327–334. doi: 10.4269/ajtmh.1988.38.327. [DOI] [PubMed] [Google Scholar]
- 11.Hamann KJ, Barker RL, Loegering DA, Gleich GJ. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis. J. Parasitol. 1987;73:523–529. [PubMed] [Google Scholar]
- 12.Ackerman SJ, Gleich GJ, Loegering DA, Richardson BA, Butterworth AE. Comparative toxicity of purified human eosinophil granule cationic proteins for schistosomula of Schistosoma mansoni. Am. J. Trop. Med. Hyg. 1985;34:735–745. doi: 10.4269/ajtmh.1985.34.735. [DOI] [PubMed] [Google Scholar]
- 13.Hamann KJ, Barker RL, Loegering DA, Gleich GJ. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis. J. Parasitol. 1987;73:523–529. [PubMed] [Google Scholar]
- 14.Hamann KJ, Gleich GJ, Checkel JL, Loegering DA, McCall JW, Barker RL. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J. Immunol. 1990;144:3166–3173. [PubMed] [Google Scholar]
- 15.Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol. 1989;142:4428–4434. [PubMed] [Google Scholar]
- 16.Rosenberg HF, Domachowske JB. Eosinophils, eosinophil ribonucleases, and their role in host defense against respiratory virus pathogens. J. Leukoc. Biol. 2001;70:691–698. [PubMed] [Google Scholar]
- 17.Cormier SA, Yuan S, Crosby JR, Protheroe CA, Dimina DM, Hines EM, Lee NA, Lee JJ. T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 2002;27:678–687. doi: 10.1165/rcmb.4882. [DOI] [PubMed] [Google Scholar]
- 18.Garvey TL, Dyer KD, Ellis JA, Bonville CA, Foster B, Prussin C, Easton AJ, Domachowske JB, Rosenberg HF. Inflammatory responses to pneumovirus infection in IFN-αβR gene-deleted mice. J. Immunol. 2005;175:4735–4744. doi: 10.4049/jimmunol.175.7.4735. [DOI] [PubMed] [Google Scholar]
- 19.Nakajima M, Hirakata M, Nittoh T, Ishihara K, Ohuchi K. Expression and purification of recombinant rat eosinophil-associated ribonucleases, homologues of human eosinophil cationic protein and eosinophil-derived neurotoxin, and their characterization. Int. Arch. Allergy Immunol. 2001;125:241–249. doi: 10.1159/000053822. [DOI] [PubMed] [Google Scholar]
- 20.Ishihara K, Asai K, Nakajima M, Mue S, Ohuchi K. Preparation of recombinant rat eosinophil-associated ribonuclease-1 and -2 and analysis of their biological activities. Biochim Biophys Acta. 2003;1638:164–172. doi: 10.1016/s0925-4439(03)00077-2. [DOI] [PubMed] [Google Scholar]
- 21.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 22.Avdeeva SV, Chernukha MU, Shaginyan IA, Tarantul VZ, Naroditsky BS. Human angiogenin lacks specific antimicrobial activity. Curr. Microbiol. 2006;53:477–478. doi: 10.1007/s00284-006-0033-6. [DOI] [PubMed] [Google Scholar]
- 23.Harder J, Schröder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 24.Rudolph B, Podschun R, Sahly H, Schubert S, Schröder JM, Harder J. Identification of RNase 8 as a novel human antimicrobial protein. Antimicrob. Agents Chemotherap. 2006;50:3194–3196. doi: 10.1128/AAC.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitto T, Dyer KD, Czapiga M, Rosenberg HF. Evolution and function of leukocyte RNase A ribonucleases of the avian species, Gallus gallus. J. Biol. Chem. 2006;281:25622–25634. doi: 10.1074/jbc.M604313200. [DOI] [PubMed] [Google Scholar]
- 26.Cho S, Zhang J. Zebrafish ribonucleases are bactericidal: implications for the origin of vertebrate RNase A superfamily. Mol. Biol. Evol. 2007;24:1259–1268. doi: 10.1093/molbev/msm047. [DOI] [PubMed] [Google Scholar]
- 27.Hirs CH, Moore S, Stein WH. Studies on structure of ribonuclease. Fed. Proc. 1956;15:840–848. [PubMed] [Google Scholar]
- 28.Hirs CH, Stein WH, Moore S. The amino acid composition of ribonuclease. J. Biol. Chem. 1954;211:941–950. [PubMed] [Google Scholar]
- 29.Aqvist SE, Anfinsen CB. The isolation and characterization of ribonucleases from sheep pancreas. J. Biol. Chem. 1959;234:1112–1117. [PubMed] [Google Scholar]
- 30.Kurachi K, Davie EW, Strydom DJ, Riordan JF, Vallee BL. Sequence of the cDNA and gene for angiogenin, a human angiogenesis factor. Biochemistry. 1985;24:5494–5459. doi: 10.1021/bi00341a032. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg HF, Tenen DG, Ackerman SJ. Molecular cloning of the human eosinophil-derived neurotoxin: a member of the ribonuclease gene family. Proc. Natl. Acad. Sci. U S A. 1989;86:4460–4464. doi: 10.1073/pnas.86.12.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamann KJ, Barker RL, Loegering DA, Pease LR, Gleich GJ. Sequence of human eosinophil-derived neurotoxin cDNA: identity of deduced amino acid sequence with human nonsecretory ribonucleases. Gene. 1989;83:161–167. doi: 10.1016/0378-1119(89)90414-9. [DOI] [PubMed] [Google Scholar]
- 33.Barker RL, Loegering DA, Ten RM, Hamann KJ, Pease LR, Gleich GJ. Eosinophil cationic protein cDNA. Comparison with other toxic cationic proteins and ribonucleases. J. Immunol. 1989;143:952–955. [PubMed] [Google Scholar]
- 34.Rosenberg HF, Ackerman SJ, Tenen DG. Human eosinophil cationic protein. Molecular cloning of a cytotoxin and helminthotoxin with ribonuclease activity. J. Exp. Med. 1989;170:163–176. doi: 10.1084/jem.170.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nitta R, Katayama N, Okabe Y, Iwama M, Watanabe H, Abe Y, Okazaki T, Ohgi K, Irie M. Primary structure of a ribonuclease from bullfrog (Rana catesbeiana) liver. J. Biochem. (Tokyo) 1989;106:729–735. doi: 10.1093/oxfordjournals.jbchem.a122924. [DOI] [PubMed] [Google Scholar]
- 36.Ardelt W, Mikulski SM, Shogen K. Amino acid sequence of an antitumor protein from Rana pipiens oocytes and early embryos. Homology to pancreatic ribonucleases. J. Biol. Chem. 1991;266:245–251. [PubMed] [Google Scholar]
- 37.Beintema JJ. Introduction: the ribonuclease A superfamily. Cell. Mol. Life Sci. 1998;54:763–765. doi: 10.1007/s000180050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyer KD, Rosenberg HF. The RNase A superfamily: generation of diversity and innate host defense. Mol. Divers. 2006;10:585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Dudley J, Nei M, Kumar S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 40.Penttinen J, Pujianto DA, Sipila P, Huhtaniemi I, Poutanen M. Discovery in silico and characterization in vitro of novel genes exclusively expressed in the mouse epididymis. Mol. Endocrinol. 2003;17:2138–2151. doi: 10.1210/me.2003-0008. [DOI] [PubMed] [Google Scholar]
- 41.Castella S, Fouchecourt S, Teixeira-Gomes AP, Vinh J, Belghazi M, Dacheux F, Dacheux JL. Identification of a member of a new RNase a family specifically secreted by epididymal caput epithelium. Biol. Reprod. 2004;70:319–328. doi: 10.1095/biolreprod.103.022459. [DOI] [PubMed] [Google Scholar]
- 42.Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics. 2005;85:208–220. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 43.Rothenberg ME, Hogan SP. The eosinophil. Ann. Rev. Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 44.Jacobsen EA, Taranova AG, Lee NA, Lee JJ. Eosinophils: singularly destructive effector cells or purveyors of immunoregulation? J. Allergy Clin. Immunol. 2007;119:1313–1320. doi: 10.1016/j.jaci.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 45.Klion AD, Nutman TB. The role of eosinophils in host defense against helminth parasites. J. Allergy Clin. Immunol. 2004;113:30–37. doi: 10.1016/j.jaci.2003.10.050. [DOI] [PubMed] [Google Scholar]
- 46.Lee JJ, Lee NA. Eosinophil degranulation: an evolutionary vestige or a universally destructive effector function? Clin. Exp. Allergy. 2005;35:986–994. doi: 10.1111/j.1365-2222.2005.02302.x. [DOI] [PubMed] [Google Scholar]
- 47.Olsson I, Persson AM, Winqvist I. Biochemical properties of the eosinophil cationic protein and demonstration of its biosynthesis in vitro in marrow cells from patients with an eosinophilia. Blood. 1986;67:498–503. [PubMed] [Google Scholar]
- 48.Durack DT, Ackerman SJ, Loegering DA, Gleich GJ. Purification of human eosinophil-derived neurotoxin. Proc. Natl. Acad. Sci., USA. 1981;78:5165–5169. doi: 10.1073/pnas.78.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gleich GJ, Loegering DA, Bell MP, Checkel JL, Ackerman SJ, McKean DJ. Biochemical and functional similarities between human eosinophil-derived neurotoxin and eosinophil cationic protein: homology with ribonuclease. Proc. Natl. Acad. Sci. USA. 1986;83:3146–3150. doi: 10.1073/pnas.83.10.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slifman NR, Loegering DA, McKean DJ, Gleich GJ. Ribonuclease activity associated with human eosinophil-derived neurotoxin and eosinophil cationic protein. J. Immunol. 1986;137:2913–2917. [PubMed] [Google Scholar]
- 51.Rosenberg HF, Dyer KD, Tiffany HL, Gonzalez M. Rapid evolution of a unique family of primate ribonuclease genes. Nature Genet. 1995;10:219–223. doi: 10.1038/ng0695-219. [DOI] [PubMed] [Google Scholar]
- 52.Rosenberg HF, Domachowske JB. Eosinophils, ribonucleases and host defense: solving the puzzle. Immunol Res. 1999;20:261–74. doi: 10.1007/BF02790409. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberg HF, Dyer KD. Eosinophil cationic protein and eosinophilderived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem. 1995;270:21539–21544. doi: 10.1074/jbc.270.37.21539. [DOI] [PubMed] [Google Scholar]
- 54.Larson KA, Olson EV, Madden BJ, Gleich GJ, Lee NA, Lee JJ. Two highly homologous ribonuclease genes expressed in mouse eosinophils identify a larger subgroup of the mammalian ribonuclease superfamily. Proc. Natl. Acad. Sci. USA. 1996;93:12370–12375. doi: 10.1073/pnas.93.22.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Dyer KD, Rosenberg HF. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl. Acad. Sci. USA. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eriksson J, Reimert CM, Kabatereine NB, Kazibwe F, Ireri E, Kadzo H, Eltahir HB, Mohamed AO, Vennervald BJ, Venge P. The 434(G>C) polymorphism within the coding sequence of Eosinophil Cationic Protein (ECP) correlates with the natural course of Schistosoma mansoni infection. Int. J. Parasitol. 2007;37:1359–1366. doi: 10.1016/j.ijpara.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, Rosenberg HF. Sequence variation at two eosinophilassociated ribonuclease loci in humans. Genetics. 2000;156:1949–1958. doi: 10.1093/genetics/156.4.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trulson A, Byström J, Engstrom A, Larsson R, Venge P. The functional heterogeneity of eosinophil cationic protein is determined by a gene polymorphism and post-translational modifications. Clin. Exp. Allergy. 2007;37:208–218. doi: 10.1111/j.1365-2222.2007.02644.x. [DOI] [PubMed] [Google Scholar]
- 59.Rosenberg HF. Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem. 1995;270:7876–7881. doi: 10.1074/jbc.270.14.7876. [DOI] [PubMed] [Google Scholar]
- 60.Young JD, Peterson CG, Venge P, Cohn ZA. Mechanism of membrane damage mediated by human eosinophil cationic protein. Nature. 1986;321:613–616. doi: 10.1038/321613a0. [DOI] [PubMed] [Google Scholar]
- 61.Carreras E, Boix E, Rosenberg HF, Cuchillo CM, Nogués MV. Both aromatic and cationic residues contribute to the membrane-lytic and bactericidal activity of eosinophil cationic protein. Biochemistry. 2003;42:6636–6644. doi: 10.1021/bi0273011. [DOI] [PubMed] [Google Scholar]
- 62.Torrent M, Cuyás E, Carreras E, Navarro S, López O, de la Maza A, Nogués MV, Reshetnyak YK, Boix E. Topography studies on the membrane interaction mechanism of the eosinophil cationic protein. Biochemistry. 2007;46:720–733. doi: 10.1021/bi061190e. [DOI] [PubMed] [Google Scholar]
- 63.Ginsburg I. Bactericidal cationic peptides can also function as bacteriolysis-inducing agents mimicking beta-lactam antibiotics?; it is enigmatic why this concept is consistently disregarded. Med. Hypotheses. 2004;62:367–374. doi: 10.1016/j.mehy.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 64.Boix E, Nogués MV. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defense. Mol Biosyst. 2007;3:317–335. doi: 10.1039/b617527a. [DOI] [PubMed] [Google Scholar]
- 65.Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, Foster PS, Matthaei KI. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood. 2007;110:1578–1586. doi: 10.1182/blood-2007-01-071340. [DOI] [PubMed] [Google Scholar]
- 66.Rosenberg HF, Bonville CA, Easton AJ, Domachowske JB. The pneumonia virus of mice infection model for severe respiratory syncytial virus infection: identifying novel targets for therapeutic intervention. Pharmacol. Therap. 2005;105:1–6. doi: 10.1016/j.pharmthera.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 67.Oppenheim JJ, Tewary P, de la Rosa G, Yang D. Alarmins initiate host defense. Adv. Med. Biol. 2007;601:185–194. doi: 10.1007/978-0-387-72005-0_19. [DOI] [PubMed] [Google Scholar]
- 68.Tello-Montoliu A, Patel JV, Lip GY. Angiogenin: a review of the pathophysiology and potential clinical applications. J. Thromb. Haemost. 2006;4:1864–1874. doi: 10.1111/j.1538-7836.2006.01995.x. [DOI] [PubMed] [Google Scholar]
- 69.Zhang J, Rosenberg HF. Diversifying selection of the tumor-growth promoter angiogenin in primate evolution. Mol. Biol. Evol. 2002;19:438–445. doi: 10.1093/oxfordjournals.molbev.a004099. [DOI] [PubMed] [Google Scholar]
- 70.Zhang J, Dyer KD, Rosenberg HF. Human RNase 7: a new cationic ribonuclease of the RNase A superfamily. Nucl. Acids Res. 2003;31:602–607. doi: 10.1093/nar/gkg157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang YC, Lin YM, Chang TW, Wu SJ, Lee YS, Chang MD, Chen C, Wu SH, Liao YD. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. J. Biol. Chem. 2007;282:4626–4633. doi: 10.1074/jbc.M607321200. [DOI] [PubMed] [Google Scholar]
- 72.Zhang J, Dyer KD, Rosenberg HF. RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucl. Acids Res. 2002;30:1169–1175. doi: 10.1093/nar/30.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pizzo E, Buonanno P, Di Maro A, Ponticelli S, De Falco S, Quarto N, Cubellis MV, D'Alessio G. Ribonucleases and angiogenins from fish. J. Biol. Chem. 2006;281:27454–27460. doi: 10.1074/jbc.M605505200. [DOI] [PubMed] [Google Scholar]
- 74.Pizzo E, D'Alessio G. The success of the RNase scaffold in the advance of biosciences and in evolution. Gene. 2007 doi: 10.1016/j.gene.2007.05.006. In press. [DOI] [PubMed] [Google Scholar]
- 75.Pavlakis N, Vogelzang NJ. Ranpirnase--an antitumour ribonuclease: its potential role in malignant mesothelioma Exper. Opin. Biol. Therap. 2006;6:391–399. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- 76.Costanzi J, Sidransky D, Navon A, Goldsweig H. Ribonucleases as a novel pro-apoptotic anticancer strategy: review of the preclinical and clinical data for ranpirnase. Cancer Invest. 2005;23:643–650. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- 77.Ardelt B, Juan G, Burfeind P, Salomon T, Wu JM, Hsieh TC, Li X, Sperry R, Pozarowski P, Shogen K, Ardelt W, Darzynkiewicz Z. Onconase, an anti-tumor ribonuclease suppresses intracellular oxidative stress. Int. J. Oncol. 2007;31:663–669. doi: 10.3892/ijo.31.3.663. [DOI] [PubMed] [Google Scholar]
- 78.Rosenberg HF, Zhang J, Liao YD, Dyer KD. Rapid diversification of RNase A superfamily ribonucleases from the bullfrog, Rana catesbeiana. J. Mol. Evol. 2001;53:31–38. doi: 10.1007/s002390010188. [DOI] [PubMed] [Google Scholar]
- 79.Klenova EM, Botezato I, Laudet V, Goodwin GH, Wallace JC, Lobanenkov VV. Isolation of a cDNA clone encoding the RNasesuperfamily-related gene highly expressed in chicken bone marrow cells. Biochem. Biophys. Res. Commun. 1992;185:231–239. doi: 10.1016/s0006-291x(05)80980-5. [DOI] [PubMed] [Google Scholar]