Introduction
In 2006 alone nearly 234,460 men in the United States will be diagnosed with prostate cancer. In addition, about 27,000 will die from, rather than with, their disease (1). Although there is considerable variability in disease incidence by age, race, and family history, about 70% of all cases are ≥65 years (yrs) at diagnosis, and the median age at diagnosis in the United States is 68 yrs (2). The disease is more frequent among African American than Caucasians; Asian men have the lowest reported incidence (3, 4).
Segregation analysis were the first to suggest a strong hereditary component for prostate cancer, particularly among younger men. Two similar studies based on ascertainment of family history through probands treated with radical prostatectomy showed evidence for the dominant transmission of a rare high-risk allele/s (population prevalence of 0.3%–0.6%), with carriers having an 88%–89% risk of getting prostate cancer by age 85, compared to 3%–5% in non-carriers (5, 6). Carter et al., have suggested that the cumulative proportion of prostate cancer cases within the U.S. population that is attributable to high-risk susceptibility alleles is 43% for men diagnosed ≤55 years, 34% for men ≤70 years, and 9% for men ≤85 years (5). By comparison, population-based studies from Sweden (6) and Australia (7, 8), estimate a higher population prevalence of carriers (1.1%–1.67%) and a lower lifetime incidence (63%–79%). The latter studies also suggest that 23% of all prostate cancer cases diagnosed <65 years may be due to inherited mutations in susceptibility genes (6).
In addition to evidence for autosomal dominant Mendelian inheritance, several studies also support an X-linked or recessive model with higher relative risks for prostate cancer in men with affected siblings compared to affected fathers. A segregation analysis from Australia found that the best fitting models included a dominantly inherited increased risk that was greater at younger ages (penetrance of 70% by age 80) and a recessively inherited or X-linked increased risk that was greater at older ages of diagnosis (8). The latter study also found that all two-locus models gave better fit than single-locus models, suggesting that multiple loci are responsible for the disease.
Several types of epidemiological studies also present compelling evidence for the existence of prostate cancer susceptibility loci. Both case-control and cohort studies show that having a first-degree relative with prostate cancer increases a man’s risk of being diagnosed with the disease by two- to three-fold relative to those without a family history (9). If the relative is diagnosed before age 65 (RR=5.9) or if there are ≥3 affected first-degree relatives (RR=10.9), the risk is increases significantly (9–11). Twin studies report higher concordance rates for monozygotic (19%–27%) than dizygotic (4%–7%) twins (7, 12, 13), with the largest study reporting a relative risk (R.R.) for prostate cancer of 12.3 (95% CI 8.4–18.1) in monozygotic twins (13).
Efforts to identify susceptibility loci for hereditary prostate cancer (HPC) have been ongoing for several years (14–30). At the heart of the problem is the extreme locus and disease heterogeneity that are hallmarks of the disease (31–35). With over a dozen genome scans completed to date, suggestive evidence for loci has been described on nearly every chromosome, and efforts to replicate results using seemingly similar data sets has been challenging (31–35). Yet, in recent years, it appears that progress is finally being made. Strategies for overcoming the problem of locus heterogeneity and ultimately identifying prostate cancer loci are the focus of this discussion.
Genome Wide Scans for Prostate Cancer Susceptibility
In 1993, Carter et al. (36) provided a definition for hereditary prostate cancer (HPC) based on families meeting at least one of the following criteria: 1) prostate cancer in ≥3 first-degree relatives; 2) prostate cancer in three successive generations of the maternal or paternal lineages; or 3) two first-degree relatives affected at age ≤55 years. We, like other researchers, have used these criteria to ascertain families for genome-wide scans aimed at finding prostate cancer susceptibility loci. In aggregate, the community has published the results from over a dozen genome wide scans (37–39). This has allowed the community to make several predictions regarding the genetic nature of prostate cancer in high-risk families. These are as follows:
Prostate cancer susceptibility is caused by mutations in multiple genes
Different genes are likely associated with distinct population frequencies in various ethnic groups; and
Distinct models of Mendelian inheritance and varying levels of penetrance are associated with different loci.
Loci Mapped to Date
Among data sets of hereditary families, several genes have been suggested as causative. However there has been extreme difficulty in both confirming linkage results, as well as results derived from candidate genes analysis of selected populations. Among the most frequently debated genes are HPC2/ELAC2 (40), HPC1/RNASEL (41), (42), and BRCA2 (43). For each, both confirmatory and lack of replication studies are reported. In some cases, such as that of HPC1/RNASEL, replication studies have focused exclusively on isolated populations such as Ashkenazi Jewish patients or Finnish men, in an effort to reduce problems associated with heterogeneity (44–46).
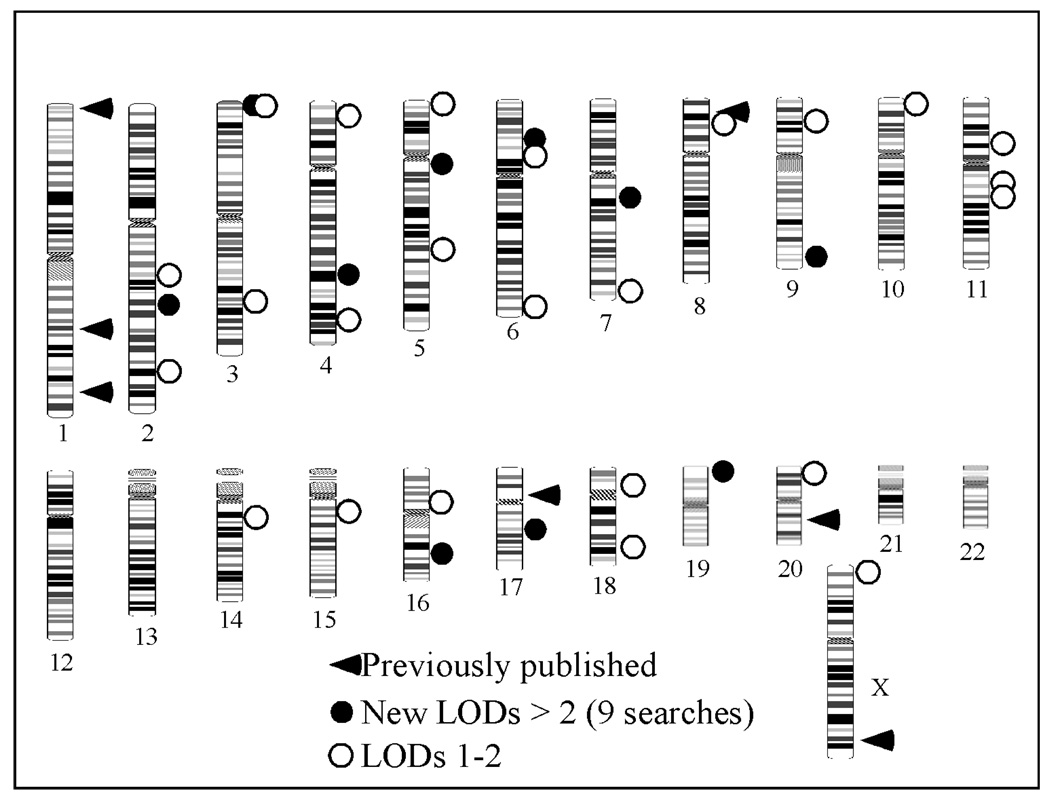
In late 2003, eight genome-wide scans for prostate cancer susceptibility, including our own of 255 families were published (21). The aggregate results are summarized in a review by Easton and colleagues (33, 38) and shown in Figure 1. The eight scans include 1293 families with multiple cases of prostate cancer. Across all studies, eleven peaks with Lod scores > 2.0 were observed, identifying regions on chromosomes 2, 3, 4, 5, 6, 7, 9, 16, 17, 19, and 20 as containing putative susceptibility loci. However other than a replication by the group describing the initial results on chromosome 20, none of these peaks coincided precisely with any of the previously reported linkage peaks. The most promising were the results on chromosome 19 by Wiklund et al. (24) which is close to the peak on chromosome 19 originally reported by reported by Hsieh et al. (27).
Figure 1.
Prostate Cancer Linkage Peaks
Of even greater surprise was the fact that no chromosomal region was reported with a significance level of 2.0 or greater by more than one study. In addition, only one study demonstrated a Lod score ≥3.0—that of Cunningham et al. (19), which reported a Lod score of 4.77 on chromosome 20q13. The same group had originally mapped HPC20 and the data in the replication study included a subset of the original mapping families. The highest reported Lod score by any other group on chromosome 20, was Lod = 1.02.
The results from other candidate loci were similarly discouraging. A total of 24 additional Lod scores >1.0 were reported, identifying every chromosome as carrying a putative locus except 12, 13, 21 and 22. No Lod scores >1.0 were observed at the HPC2 locus on 17p or the HPCX locus on the X chromosome. Indeed, Lod scores in excess of 1.0 were noted only by Xu et al. (25) and Cunningham et al. (19). Again, both data sets contained families from the original study. Scores in excess of 1.0 were observed on chromosome 16q only by Witte et al., (47) which was the hypothesis generating group, and one other group. In addition to Xu et al. (25) who originally reported linkage to 8p (25), only one other group noted a Lod score >1.0. In summary, none of the “candidate loci” received true statistical confirmation by an independent group and only the locus at 19p proposed by Hsieh et al. (27) was replicated by an independent group with a Lod score of ≥2.0.
These facts speak to the enormous level of both phenotypic heterogeneity and locus heterogeneity observed with prostate cancer. Clearly there are many genes contributing to the disease with varying levels of penetrance. The introduction of the prostate specific antigen blood test (PSA) contributes to the overall phenotypic variability, as man are now diagnosed earlier in life then they might have been previously. Indeed, most data sets in the literature today are mixed and reflect a subset of men diagnosed before and after PSA came into common use, which is estimated to have occurred in the late 1980s. To try and overcome these difficulties and find prostate cancer loci, the prostate cancer research community at large has employed four strategies, each of which are discussed in turn below.
Meta Analysis and the International Consortium of Prostate Cancer Genetics
The prostate cancer mapping community has formed an international working group termed the International Consortium of Prostate Cancer Genetics (ICPCG). The goal of the ICPCG is to work together, often sharing data prior to publication, to generate large meta data sets with increased power for tackling problems related to prostate cancer susceptibility. The advantage of this strategy is that is allows extensive sub classification and stratification, while still retaining sufficient numbers in each strata such that statistically meaningful analysis can be done. Appropriate corrections for multiple corrections can be made, and accurate results still achieved.
Thus far the group has focused on replication studies on chromosome 1 and 20 (48, 49). In the case of HPC1, the community has shown that only very large families with no evidence for linkage to the X chromosome likely can attribute their disease to mutations at HPC1. On chromosome 20 the consortium finds little evidence for replication in a dataset of over 1200 families, calling into question the original finding (49). More recently, the ICPCG has also undertaken studies focused on families with an excess of aggressive disease (50).
Clinical Features of Disease
Several studies have focused exclusively on aggressive disease (Reviewed in (51)). While many men will die with prostate cancer, it is those that die of prostate cancer that are the most clinically interesting and the ones that the research community most wishes to study. Initial studies focused on using Gleason score as a measure of aggressiveness. Gleason score is an assessment of tumor grade (52). To obtain a Gleason score, several regions of the tumor are independently scored by a pathologist and assigned a number of 1–5, representing well to poorly differentiated patterns. The higher the score the worst the prognosis (53). The two predominant scores are then added together to give a summary score between 2 and 10, with most tumors falling in the range of Gleason 5–7.
Some studies have treated Gleason score as a quantitative trait for outcome of disease aggressiveness, since it is reported to be a good predictor of survival (53). Others have treated the Gleason score as a covariate, using it to explain locus heterogeneity. For example, Witte et al. analyzed grade as a quantitative trait on 513 men from concordant sibships. They reported evidence for linkage on chromosomes 5q31-33, 7q32, and 19q12-13.11 (54). Using the same data set as Witte (54), Goddard et al. (18) used a variety of factors as covariates including sum of the sib-pair Gleason scores, mean family age at diagnosis, existence of male-to-male transmission (which argues against X linkage), and the number of affected first-degree relatives. In doing so, they detected linkage at three previously reported loci (1q24-25, 1q42.2-43, and 4q) and found linkage at Xq12-13 (LOD score 3.06, P= 0.00053), adjacent to the androgen receptor. In addition, they identified five other loci with Lod scores ≥2.5.
Interestingly, the loci at 1q24, Xq12 and chromosome 5 were evident only when Gleason score was considered as one of the covariates. In the absence of the covariates, results were weak to non existence. Others have found this approach to be similarly informative (55–57). Some of the strongest data using Gleason score as a measure of tumor aggressiveness have come from Slager et al. in a genome scan of 161 sibpairs (58). They not only strongly confirm the linkage results for chromosome 19q (P< 0.00001), but report evidence for linkage on chromosome 4 (P = 0.00012). In a subsequent and independent study, involving 175 brother pairs from 103 families the same group found evidence for linkage at 6q23 (P = 0.0009), 1p13-q21 and 5p13-q11 (59).
As more studies have been undertaken, it has become clear that a precise definition of aggressiveness is needed. Most recent studies have used a definition of aggressive disease (60–62) that includes families in which at least two genotyped men had at least one of the following disease features: regional- or distant-stage disease (based on pathology if a radical prostatectomy has been done, including T3, T4, N1 or M1, otherwise data from clinical staging is accepted); a Gleason score at diagnosis of ≥7 (poorly differentiated grade if no Gleason score is available); a pre-treatment prostate-specific antigen (PSA) score of 20 ng/ml or higher; and if deceased, death from metastatic prostate cancer at <65 years.
Using the above criteria, we reported suggestive evidence for linkage on chromosome 22 (dominant HLOD = 2.18) (62). We utilized clinical data from 784 affected men from 248 HPC families for whom a genomic screen had been previously performed (21). Disease characteristics described above were used to classify affected men into categories of clinically insignificant, moderate, or aggressive prostate cancer. Only men with aggressive disease were coded as affected in the linkage analysis. Suggestive linkage was observed at chromosome 22q11.1 (dominant HLOD = 2.75) and at 22q12.3-q13.1 using a recessive model of inheritance (HLOD = 1.90). Other studies have reported at least nominal evidence for linkage on chromosome 22. For instance, Lange et al. reported a Lod score of 1.87 at 45cM in a set of 16 African American families and a Lod = 1.87 at 51 cM in 79 families with four or more affected men (22). A subset of younger-age-at-onset families from Utah (HLOD = 2.42) also gave evidence for linkage in this region (29).
Other Cancers
While the above criteria have proven useful for dealing with the locus heterogeneity problem, they don’t fully solve the problem. One additional strategy is that of examining high-risk prostate cancer families for an excess of other diseases and then using the resulting group of families in an isolated analysis.
Johannesson et al. have done that with high risk prostate-kidney cancer families (63). An association between these prostate and kidney cancer has been suggested by at least two studies. In a Swedish study, Grönberg and colleagues examined 1,364 relatives of 62 HPC families for the incidence of other cancers and found a significant association for kidney cancer (Standardized Incidence Ratio, SIR = 2.51; 95% CI 1.15–4.77) (64). A second Swedish study utilizing the nation-wide Family Cancer Database reported a familial relative risk of 1.3 (95% CI 1.0–1.76) for kidney cancer in offspring of prostate cancer cases (65).
We selected a set of 15 prostate-kidney families from among the 154 families on which we had completed a genome wide scan using 441 microsatellite markers (21) The 15 families all reported both primary prostate and primary kidney cancer. All kidney cancer cases were confirmed by either death certificate or medical records. To be eligible for the study, the kidney case had to be either a prostate cancer case himself or a first-degree relative of a prostate cancer case. There are 191 (99 genotyped) individuals, and all families were of Caucasian ancestry. Ten of the kidney cancer cases also had prostate cancer, and the other five kidney cancer cases were first-degree relatives of prostate cancer cases.
While we found no statistically significant evidence for linkage in the initial analysis, we found two regions of suggestive linkage at 11q12 and 4q21, with HLOD scores of 2.59 and 2.10, respectively. The primary result on chromosome 11 was strengthened after excluding two families with members with transitional cell carcinoma (TCC). This was a valid strategy, as 11 TCC is a very different and much more rare disease then in renal cell carcinoma, which was reported by the majority of families.
The non parametric analysis revealed a Kong and Cox p-value of 0.004 for marker D11S1290 at 11p11.2. The 8 cM region between 11p11.2 and 11q12.2 was refined by the addition of 16 additional markers. The subset of HPC families with a median age of diagnosis >65 years demonstrated the strongest evidence for linkage (HLOD = 2.50). The p-values from non-parametric analysis ranged from 0.004 to 0.05 across five contiguous markers. There are some provocative candidate genes in the area, including the gene for Prostate-Specific Membrane Antigen (PSMA). However mutation screening of that gene is made difficult by the presence of another gene, termed PSMA-Like, that is located at 11q14.3 and shares 98% homology with the coding sequence of the PSMA gene itself. Studies pursing this interesting question continue in our lab and others.
Studies of Isolated Populations
Studies of isolated populations have frequently been undertaken in cancer genetics as a way to both deal with locus heterogeneity as well as identify founder mutations for specific tumor suppressor genes. In the case of prostate cancer, studies to date have focused on populations from Iceland (66–68), Finland (69) and Ashkenazi Jewish men (70–74).
In our collaborative group, Friedrichsen et al. have examined a genome wide scan in a set of 36 families of Ashkenazi Jewish origin (75). The 36 Jewish families represent a combined dataset of 17 Jewish families from the Fred Hutchinson Cancer Research Center (FHCRC)-based PROGRESS dataset, and 19 Ashkenazi Jewish families collected at Johns Hopkins University (JHU). All available family members, including 94 affected men, were genotyped using a set of microsatellite markers distributed across the genome at an average of about 8–10 cM density. To combine the two datasets, only markers present in the UCSC genome browser April 2003 assembly (http://genome.ucsc.edu/) were used (PROGRESS 421 markers, JHU 398 markers) and map order and distance between markers were taken from the UCSC map.
Since no segregation analysis has been done exclusively on Ashkenazi Jewish men, and models of inheritance were thus hard to predict, the data were analyzed primarily using nonparametric multipoint methods. The strongest signal in this data set was a significant linkage peak at 7q11-21, associated with a nonparametric linkage (NPL) score of 3.01 (p = 0.0013). Simulation analysis indicate that this corresponds to a genome-wide empirical p = 0.006. Empirical p values were calculated using the computer program Merlin (76), which was used to generate and analyze 1,000 replicates of the entire genome from the original dataset of 36 Jewish families.
After genotyping additional markers within the 7q11-21 peak, the NPL score increased to 3.35 (p = 0.0004) at marker D7S634 with an allele-sharing LOD of 3.12 (p = 0.00007). Detailed SNP analysis is underway in an attempt to find a shared haplotype that is over represented among affected versus unaffected men within the Jewish families. Within that haplotype should lie the susceptibility gene and variant of interest.
We had noted a minor signal on chromosome 7q in our initial genome scan (21). We were thus curious as to the degree to which Jewish families accounted for that result. In the 254 PROGRESS families we previously reported an HLOD of 2.25 (LOD = 1.55) at marker D7S2212 on 7q21 using a recessive parametric model. In a nonparametric analysis we reported an NPL score of 1.79 (P = 0.038) in the same region. Analysis of the 237 non-Jewish families from the PROGRESS dataset yielded an NPL score of 1.11 (P = 0.134), revealing no evidence for linkage. This suggested clearly that the majority of result in the original genome-wide scan for the PROGRESS families was due to the presence of a modest number of Jewish families.
Similar conclusions were reached by investigators at JHU. In the genome-wide study of 188 JHU families by Xu et al., the strongest result on 7q was an allele-sharing LOD of 1.63 with marker D7S486 which is adjacent to the region of interest (25). When 17 of the 19 JHU Ashkenazi Jewish families were analyzed using D7S486, the allele-sharing LOD was only 0.04, suggesting that the Johns Hopkins-collected Ashkenazi Jewish families do not contribute significantly to the results previously reported for 7q22.
Our analysis of 36 Jewish families also highlighted regions on chromosomes 1q31-32, 2p11, 3q27-28, 14q12, and 20q11 with p values of 0.02 to 0.06. The strongest of these was at 14q12 (P = 0.02). Other minor peaks with an NPL p value ≤ 0.05 included 3q27-28 (P = 0.03) and 20q11 (p = 0.04). While these may represent other loci that contribute to prostate cancer in Jewish, and perhaps non Jewish families, further investigation is clearly needed to draw definitive conclusions.
Summary
Studies to date suggest that prostate cancer is a genetically very heterogeneous disease. High-risk families, in which multiple men are affected likely reflect the contributions of a number of genes, some that are rare and highly penetrant, while others are more common and weakly penetrant. In this review we have discussed only the first type of loci, and found the identification of such genomic regions be a formidable problem. Replication between seemingly similar data sets is weak, likely reflecting the older age of onset associated with the disease, the inability to collect affected individuals from more the two generations in a family, and the variation seen in disease presentation, in addition to the underlying locus heterogeneity. Indeed, the definition of prostate cancer is ever changing, as diagnostic criteria and tools for pinpointing early lesions improve.
Are we making progress? Clearly the answer is yes. The ability to divide large data sets into homogenous subset of families likely to share common genetic underpinnings has improved power to identify loci and reproducibility between loci is now more common. Indeed, several groups report linkage to loci on chromosomes 1, 17, 19 and 22. Key to our continued success is our ever increasing ability to understand the disease. Identifying the subset of men who are likely to get clinically significant disease is the goal of genetic studies like these, and identifying the underlying loci is key for developing diagnostics. The willingness of the community to share data prior to publication, combine datasets, and to work together has been an important factor in the successes the community has enjoyed to date, and will likely be as important as we move forward to untangle the genetics of this complex and common disorder.
Acknowledgements
We and our many colleagues are grateful to the many men who continue to participate in prostate cancer genetic studies across the world. The authors thank the National Human Genome Research Institute of the National Institutes of Health for their continued support.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun M. Cancer Statistics, 2006. Vol. 56. CA: A Cancer Journal for Clinicians; 2006. pp. 106–130. [DOI] [PubMed] [Google Scholar]
- 2.Ries L, Harkins D, Krapcho M, Mariotto A, Miller B, Feuer E, Clegg L, Eisner M, Horner M, Howlader N, Hayat M, Hankey B, Edwards B. SEER Cancer Statistics Review, 1975–2003. Bethesda MD: National Cancer Institute; 2005. [Google Scholar]
- 3.Wingo PA, Bolden S, Tong T, Parker SL, Martin LM, Heath CW. Cancer statistics for African Americans, 1996. CA Cancer J Clin. 1996;46:113–125. doi: 10.3322/canjclin.46.2.113. [DOI] [PubMed] [Google Scholar]
- 4.Walker B, Figgs LH, Zahm S. Differences in cancer incidence, mortality, and survival between African Americans and whites. Environ Health Perspect. 1995;103 doi: 10.1289/ehp.95103s8275. 275-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC. Mendelian inheritance of familial prostate cancer. Proc Natl Acad Sci USA. 1992;89:3367–3371. doi: 10.1073/pnas.89.8.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaid DJ, McDonnell SK, Blute ML, Thibodeau SN. Evidence for autosomal dominant inheritance of prostate cancer. Am J Hum Genet. 1998;62:1425–1438. doi: 10.1086/301862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grönberg H, Damber L, Damber J-E, Iselius L. Segregation analysis of prostate cancer in Sweden: Support for dominant inheritance. Am J of Epid. 1997;146:552–557. doi: 10.1093/oxfordjournals.aje.a009313. [DOI] [PubMed] [Google Scholar]
- 8.Cui J, Staples MP, Hopper JL, English DR, McCredie MR, Giles GG. Segregation analyses of 1,476 population-based australian families affected by prostate cancer. Am J Hum Genet. 2001;68:1207–1218. doi: 10.1086/320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanford JL, Ostrander EA. Familial prostate cancer. Epidemiol Rev. 2001;23:19–23. doi: 10.1093/oxfordjournals.epirev.a000789. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg GD, Carter BS, Beaty TH, Childs B, Walsh PC. Family history and the risk of prostate cancer. Prostate. 1990;17:337–347. doi: 10.1002/pros.2990170409. [DOI] [PubMed] [Google Scholar]
- 11.Cannon L, Bishop DT, Skolnick M, Hunt S, Lyon JL, Smart CR. Genetic epidemiology of prostate cancer in the Utah Mormon genealogy. Cancer Surv. 1982;1:47–69. [Google Scholar]
- 12.Page WF, Braun MM, Partin AW, Caporaso N, Walsh P. Heredity and prostate cancer: a study of World War II veteran twins. Prostate. 1997;33:240–245. doi: 10.1002/(sici)1097-0045(19971201)33:4<240::aid-pros3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 13.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland [see comments] N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 14.Smith JR, Freije D, Carpten JD, Grönberg H, Xu J, Isaacs S, Brownstein MJ, Bova GS, Guo H, Bujnovszky P, Nusskern DR, Damber J-E, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274:1371–1374. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 15.Suarez BK, Lin J, Burmester JK, Broman KW, Weber JL, Banerjee TK, Goddard KA, Witte JS, Elston RC, Catalona WJ. A genome screen of multiplex sibships with prostate cancer. Am J Hum Genet. 2000;66:933–944. doi: 10.1086/302818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbs M, Stanford JL, Jarvik GP, Janer M, Badzioch M, Peters MA, Goode EL, Kolb S, Chakrabarti L, Shook M, Basom R, Ostrander EA, Hood L. A genomic scan of families with prostate cancer identifies multiple regions of interest. Am J Hum Genet. 2000;67:100–109. doi: 10.1086/302969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suarez BK, Lin J, Witte JS, Conti DV, Resnick MI, Klein EA, Burmester JK, Vaske DA, Banerjee TK, Catalona WJ. Replication linkage study for prostate cancer susceptibility genes. Prostate. 2000;45:106–114. doi: 10.1002/1097-0045(20001001)45:2<106::aid-pros4>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 18.Goddard KA, Witte JS, Suarez BK, Catalona WJ, Olson JM. Model-free linkage analysis with covariates confirms linkage of prostate cancer to chromosomes 1 and 4. Am J Hum Genet. 2001;68:1197–1206. doi: 10.1086/320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham JM, McDonnell SK, Marks A, Hebbring S, Anderson SA, Peterson BJ, Slager S, French A, Blute ML, Schaid DJ, Thibodeau SN. Genome linkage screen for prostate cancer susceptiblity loci: Results from the Mayo Clinic Familial Prostate Cancer Study. Prostate. 2003;57:335–346. doi: 10.1002/pros.10308. [DOI] [PubMed] [Google Scholar]
- 20.Edwards S, Meitz J, Eles R, Evans C, Easton D, Hopper J, Giles G, Foulkes WD, Narod S, Simard J, Badzioch M, Mahle L. Results of a genome-wide linkage analysis in prostate cancer families ascertained through the ACTANE consortium. Prostate. 2003;57:270–279. doi: 10.1002/pros.10301. [DOI] [PubMed] [Google Scholar]
- 21.Janer M, Friedrichsen DM, Stanford JL, Badzioch MD, Kolb S, Deutsch K, Peters MA, Goode EL, Welti R, DeFrance HB, Iwasaki L, Li S, Hood L, Ostrander EA, Jarvik GP. Genomic scan of 254 hereditary prostate cancer families. Prostate. 2003;57:309–319. doi: 10.1002/pros.10305. [DOI] [PubMed] [Google Scholar]
- 22.Lange EM, Gillanders EM, Davis CC, Brown WM, Campbell JK, Jones M, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Giri V, Dimmer JB, Montie JE, Trent JM, Cooney KA. Genome-wide scan for prostate cancer susceptibility genes using families from the University of Michigan prostate cancer genetics project finds evidence for linkage on chromosome 17 near BRCA1. Prostate. 2003;57:326–334. doi: 10.1002/pros.10307. [DOI] [PubMed] [Google Scholar]
- 23.Schleutker J, Baffoe-Bonnie AB, Gillanders E, Kainu T, Jones MP, Freas-Lutz D, Markey C, Gildea D, Riedesel E, Albertus J, Gibbs KD, Jr, Matikainen M, Koivisto PA, Tammela T, Bailey-Wilson JE, Trent JM, Kallioniemi OP. Genome-wide scan for linkage in finnish hereditary prostate cancer (HPC) families identifies novel susceptibility loci at 11q14 and 3p25-26. Prostate. 2003;57:280–289. doi: 10.1002/pros.10302. [DOI] [PubMed] [Google Scholar]
- 24.Wiklund F, Jonsson BA, Goransson I, Bergh A, Gronberg H. Linkage analysis of prostate cancer susceptibility: confirmation of linkage at 8p22-23. Hum Genet. 2003;112:414–418. doi: 10.1007/s00439-003-0916-6. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Gillanders EM, Wiley KE, Chang B-I, Isaacs SD, Zheng SL, Jones M, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Meyers DA, Walsh PC, Trent JM, Isaacs WB. Genome-wide scan for prostate cancer susceptilbity genes in the Johns Hopkins Hereditary prostate cancer families. Prostate. 2003;57:320–325. doi: 10.1002/pros.10306. [DOI] [PubMed] [Google Scholar]
- 26.Matsui H, Suzuki K, Ohtake N, Nakata S, Takeuchi T, Yamanaka H, Inoue I. Genomewide linkage analysis of familial prostate cancer in the Japanese population. J Hum Genet. 2004;49:9–15. doi: 10.1007/s10038-003-0099-y. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh CL, Oakley-Girvan I, Balise RR, Halpern J, Gallagher RP, Wu AH, Kolonel LN, O'Brien LE, Lin IG, Van Den Berg DJ, Teh CZ, West DW, Whittemore AS. A genome screen of families with multiple cases of prostate cancer: evidence of genetic heterogeneity. Am J Hum Genet. 2001;69:148–158. doi: 10.1086/321281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillanders EM, Xu J, Chang BL, Lange EM, Wiklund F, Bailey-Wilson JE, Baffoe-Bonnie A, Jones M, Gildea D, Riedesel E, Albertus J, Isaacs SD, Wiley KE, Mohai CE, Matikainen MP, Tammela TL, Zheng SL, Brown WM, Rokman A, Carpten JD, Meyers DA, Walsh PC, Schleutker J, Gronberg H, Cooney KA, Isaacs WB, Trent JM. Combined genome-wide scan for prostate cancer susceptibility genes. J Natl Cancer Inst. 2004;96:1240–1247. doi: 10.1093/jnci/djh228. [DOI] [PubMed] [Google Scholar]
- 29.Camp NJ, Farnham JM, Cannon Albright LA. Genomic search for prostate cancer predisposition loci in Utah pedigrees. Prostate. 2005;65:365–374. doi: 10.1002/pros.20287. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Dimitrov L, Chang BL, Adams TS, Turner AR, Meyers DA, Eeles RA, Easton DF, Foulkes WD, Simard J, Giles GG, Hopper JL, Mahle L, Moller P, Bishop T, Evans C, Edwards S, Meitz J, Bullock S, Hope Q, Hsieh CL, Halpern J, Balise RN, Oakley-Girvan I, Whittemore AS, Ewing CM, Gielzak M, Isaacs SD, Walsh PC, Wiley KE, Isaacs WB, Thibodeau SN, McDonnell SK, Cunningham JM, Zarfas KE, Hebbring S, Schaid DJ, Friedrichsen DM, Deutsch K, Kolb S, Badzioch M, Jarvik GP, Janer M, Hood L, Ostrander EA, Stanford JL, Lange EM, Beebe-Dimmer JL, Mohai CE, Cooney KA, Ikonen T, Baffoe-Bonnie A, Fredriksson H, Matikainen MP, Tammela T, Bailey-Wilson J, Schleutker J, Maier C, Herkommer K, Hoegel JJ, Vogel W, Paiss T, Wiklund F, Emanuelsson M, Stenman E, Jonsson BA, Gronberg H, Camp NJ, Farnham J, Cannon-Albright LA, Seminara D. A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics. Am J Hum Genet. 2005;77:219–229. doi: 10.1086/432377. Epub 2005 Jun 2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ostrander EA, Stanford JL. Genetics of prostate cancer: too many loci, too few genes. Am J Hum Genet. 2000;67:1367–1375. doi: 10.1086/316916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrander EA, Markianos K, Stanford JL. Finding prostate cancer susceptibility genes. Annu Rev Genomics Hum Genet. 2004;5:151–175. doi: 10.1146/annurev.genom.5.061903.180044. [DOI] [PubMed] [Google Scholar]
- 33.Easton DF, Schaid DJ, Whittemore AS, Isaacs WJ. Where are the prostate cancer genes? A summary of eight genome wide searches. Prostate. 2003;57:261–269. doi: 10.1002/pros.10300. [DOI] [PubMed] [Google Scholar]
- 34.Verhage BA, Kiemeney LA. Inherited predisposition to prostate cancer. Eur J Epidemiol. 2003;18:1027–1036. doi: 10.1023/a:1026101914592. [DOI] [PubMed] [Google Scholar]
- 35.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13:R103–R121. doi: 10.1093/hmg/ddh072. Epub 2004 Jan 2028. [DOI] [PubMed] [Google Scholar]
- 36.Carter BS, Bova GS, Beaty TH, Steinberg GD, Childs B, Isaacs WB, Walsh PC. Hereditary prostate cancer: epidemiologic and clinical features. J Urol. 1993;150:797–802. doi: 10.1016/s0022-5347(17)35617-3. [DOI] [PubMed] [Google Scholar]
- 37.Singh R, Eeles RA, Durocher F, Simard J, Edwards S, Badzioch M, Kote-Jarai Z, Teare D, Ford D, Dearnaley D, Ardern-Jones A, Murkin A, Dowe A, Shearer R, Kelly J, Labrie F, Easton D, Narod SA, Tonin PN, Foulkes WD. High risk genes predisposing to prostate cancer development-do they exist? Prostate Cancer Prostatic Dis. 2000;3:241–247. doi: 10.1038/sj.pcan.4500478. [DOI] [PubMed] [Google Scholar]
- 38.Easton D, Peto J. The contribution of inherited predisposition to cancer incidence. Cancer Surv. 1990;9:395–416. [PubMed] [Google Scholar]
- 39.Nupponen NN, Carpten JD. Prostate cancer susceptibility genes: many studies, many results, no answers. Cancer Metastasis Rev. 2001;20:155–164. doi: 10.1023/a:1015557308033. [DOI] [PubMed] [Google Scholar]
- 40.Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, Desrochers M, Dumont M, Farnham JM, Frank D, Frye C, Ghaffari S, Gupte JS, Hu R, Iliev D, Janecki T, Kort EN, Laity KE, Leavitt A, Leblanc G, McArthur-Morrison J, Pederson A, Penn B, Peterson KT, Reid JE, Richards S, Schroeder M, Smith R, Snyder SC, Swedlund B, Swensen J, Thomas A, Tranchant M, Woodland AM, Labrie F, Skolnick MH, Neuhausen S, Rommens J, Cannon-Albright LA. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 41.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, Moses T, Ewing C, Gillanders E, Hu P, Bujnovszky P, Makalowska I, Baffoe-Bonnie A, Faith D, Smith J, Stephan D, Wiley K, Brownstein M, Gildea D, Kelly B, Jenkins R, Hostetter G, Matikainen M, Schleutker J, Klinger K, Connors T, Xiang Y, Wang Z, De Marzo A, Papadopoulos N, Kallioniemi OP, Burk R, Meyers D, Gronberg H, Meltzer P, Silverman R, Bailey-Wilson J, Walsh P, Isaacs W, Trent J. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Zheng SL, Carpten JD, Nupponen NN, Robbins CM, Mestre J, Moses TY, Faith DA, Kelly BD, Isaacs SD, Wiley KE, Ewing CM, Bujnovszky P, Chang B, Bailey-Wilson J, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB. Evaluation of linkage and association of HPC2/ELAC2 in patients with familial or sporadic prostate cancer. Am J Hum Genet. 2001;68:901–911. doi: 10.1086/319513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards SM, Kote-Jarai Z, Meitz J, Hamoudi R, Hope Q, Osin P, Jackson R, Southgate C, Singh R, Falconer A, Dearnaley DP, Ardern-Jones A, Murkin A, Dowe A, Kelly J, Williams S, Oram R, Stevens M, Teare DM, Ponder BA, Gayther SA, Easton DF, Eeles RA. Two percent of men with early-onset prostate cancer harbor germline mutations in the BRCA2 gene. Am J Hum Genet. 2003;72:1–12. doi: 10.1086/345310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rokman A, Baffoe-Bonnie AB, Gillanders E, Fredriksson H, Autio V, Ikonen T, Gibbs KD, Jr, Jones M, Gildea D, Freas-Lutz D, Markey C, Matikainen MP, Koivisto PA, Tammela TL, Kallioniemi OP, Trent J, Bailey-Wilson JE, Schleutker J. Hereditary prostate cancer in Finland: fine-mapping validates 3p26 as a major predisposition locus. Hum Genet. 2005;116:43–50. doi: 10.1007/s00439-004-1214-7. [DOI] [PubMed] [Google Scholar]
- 45.Rennert H, Bercovich D, Hubert A, Abeliovich D, Rozovsky U, Bar-Shira A, Soloviov S, Schreiber L, Matzkin H, Rennert G, Kadouri L, Peretz T, Yaron Y, Orr-Urtreger A. A novel founder mutation in the RNASEL gene, 471delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am J Hum Genet. 2002;71:981–984. doi: 10.1086/342775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rokman A, Ikonen T, Seppala EH, Nupponen N, Autio V, Mononen N, Bailey-Wilson J, Trent J, Carpten J, Matikainen MP, Koivisto PA, Tammela TL, Kallioniemi OP, Schleutker J. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am J Hum Genet. 2002;70:1299–1304. doi: 10.1086/340450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witte JS, Suarez BK, Thiel B, Lin J, Yu A, Banerjee TK, Burmester JK, Casey G, Catalona WJ. Genome-wide scan of brothers: replication and fine mapping of prostate cancer susceptibility and aggressiveness loci. Prostate. 2003;57:298–308. doi: 10.1002/pros.10304. [DOI] [PubMed] [Google Scholar]
- 48.Xu J. ICPCG Combined analysis of hereditary prostate cancer linkage to 1q24-25: Results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet. 2000;66:945–957. doi: 10.1086/302807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaid DJ, Chang BL. Description of the International Consortium For Prostate Cancer Genetics, and failure to replicate linkage of hereditary prostate cancer to 20q13. Prostate. 2005;63:276–290. doi: 10.1002/pros.20198. [DOI] [PubMed] [Google Scholar]
- 50.Schaid DJ, McDonnell SK, Zarfas KE, Cunningham JM, Hebbring S, S.N. T, Eeles RA, Easton DF, Foulkes WD, Simard J, Giles GG, J.L. H, Mahle L, Moller P, Bishop DT, Evans C, Edwards S, Meitz J, Bullock S, Hope Q, The ACTANE Consortium. Hsieh C-l, Halpern J, Balise RR, Oakley-Girvan I, Whittemore AS, Xu J, Dimitrov L, Chang B-L, Adams TS, Turner AR, Meyers DA, Friedrichsen DM, Deutsch K, Kolb S, Janer M, Hood L, Ostrander EA, Stanford JL, Ewing CM, Gielzak M, Isaacs SD, Walsh PC, Wiley KE, Isaacs WB, Lange EM, Ho LA, Beebe-Dimmer JL, Wood DP, Cooney KA, Seminara D, I, Konen T, Baffoe-Bonnie A, Fredriksson H, Matikainen MP, Tammela TLK, Bailey-Wilson J, Schleutker J, Maier C, Herkommer K, Hoegel JJ, Vogel W, Paiss T, Wiklund F, Emanuelsson M, Stenman E, Jonsson B-A, Gronberg H, Camp NJ, Farnham J, Cannon-Albright LA, Catalona WJ, Suarez BK, Roehl KA. Pooled genome linkage scan of aggressive prostate cancer: Results from the International Consortium for Prostate Cancer Genetics. Human Genetics. 2006 doi: 10.1007/s00439-006-0219-9. In Press. [DOI] [PubMed] [Google Scholar]
- 51.Ostrander EA, Kwon EM, Stanford JL. Genetic susceptibility to aggressive prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1761–1764. doi: 10.1158/1055-9965.EPI-06-0730. [DOI] [PubMed] [Google Scholar]
- 52.Gleason D. Histologic grading and clinical staging of prostatic carcinoma. Philadelphia: Lea and Febiger; 1977. [Google Scholar]
- 53.Gleason DF. Histologic grading of prostate cancer: a perspective. Hum Pathology. 1992;23:273–279. doi: 10.1016/0046-8177(92)90108-f. [DOI] [PubMed] [Google Scholar]
- 54.Witte JS, Goddard KA, Conti DV, Elston RC, Lin J, Suarez BK, Broman KW, Burmester JK, Weber JL, Catalona WJ. Genomewide scan for prostate cancer-aggressiveness loci. Am J Hum Genet. 2000;67:92–99. doi: 10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neville PJ, Conti DV, Paris PL, Levin H, Catalona WJ, Suarez BK, Witte JS, Casey G. Prostate cancer aggressiveness locus on chromosome 7q32-q33 identified by linkage and allelic imbalance studies. Neoplasia. 2002;4:424–431. doi: 10.1038/sj.neo.7900254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neville PJ, Conti DV, Krumroy LM, Catalona WJ, Suarez BK, Witte JS, Casey G. Prostate cancer aggressiveness locus on chromosome segment 19q12-q13.1 identified by linkage and allelic imbalance studies. Genes Chromosomes Cancer. 2003;36:332–339. doi: 10.1002/gcc.10165. [DOI] [PubMed] [Google Scholar]
- 57.Paiss T, Worner S, Kurtz F, Haeussler J, Hautmann RE, Gschwend JE, Herkommer K, Vogel W. Linkage of aggressive prostate cancer to chromosome 7q31-33 in German prostate cancer families. Eur J Hum Genet. 2003;11:17–22. doi: 10.1038/sj.ejhg.5200898. [DOI] [PubMed] [Google Scholar]
- 58.Slager SL, Schaid DJ, Cunningham JM, McDonnell SK, Marks AF, Peterson BJ, Hebbring SJ, Anderson S, French AJ, Thibodeau SN. Confirmation of linkage of prostate cancer aggressiveness with chromosome 19q. Am J Hum Genet. 2003;72:759–762. doi: 10.1086/368230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slager SL, Zarfas KE, Brown WM, Lange EM, McDonnell SK, Wojno KJ, Cooney KA. Genome-wide linkage scan for prostate cancer aggressiveness loci using families from the University of Michigan Prostate Cancer Genetics Project. Prostate. 2006;66:173–179. doi: 10.1002/pros.20332. [DOI] [PubMed] [Google Scholar]
- 60.Schaid DJ, McDonnell SK, Zarfas KE, Cunningham JM, Hebbring S, Thibodeau SN, Eeles RA, Easton DF, Foulkes WD, Simard J, Giles GG, Hopper JL, Mahle L, Moller P, Bishop DT, Evans C, Edwards S, Meitz J, Bullock S, Hope Q, Hsieh C-l, Halpern J, Balise RR, Oakley-Girvan I, Whittemore AS, Xu J, Dimitrov LC, Hang B-L, Adams TS, Turner AR, Meyers DA, Friedrichsen DM, Deutsch K, Kolb S, Janer M, Hood L, Ostrander EA, Stanford JL, Ewing CM, Gielzak M, Isaacs SD, Walsh PC, Wiley KE, Isaacs WB, Lange EM, Ho LA, Beebe-Dimmer JL, Wood DP, Cooney KA, Seminara D, Ikonen T, Baffoe-Bonnie A, Fredriksson H, Matikainen MP, Tammela TLK, Bailey-Wilson J, Schleutker J, Maier C, Herkommer K, Hoegel JJ, Vogel W, Paiss T, Wiklund F, Emanuelsson M, Stenman E, Jonsson B-A, Gronberg H, Camp NJ, Farnham J, Cannon-Albright LA, Catalona WJ, Suarez BK, K.A. R The ACTANE Consortium. Pooled genome linkage scan of aggressive prostate cancer: Results from the International Consortium for Prostate Cancer Genetics Human Genetics. 2006 doi: 10.1007/s00439-006-0219-9. In Press. [DOI] [PubMed] [Google Scholar]
- 61.Chang B, Isaacs S, Wiley K, Gillanders E, Zheng S, Meyers D, Walsh P, Tren J, Xu J, Isaacs W. Genome-wide screen for prostate cancer susceptibility genes in men with clinically significant disease. Prostate. 2005;64:356–361. doi: 10.1002/pros.20249. [DOI] [PubMed] [Google Scholar]
- 62.Stanford JL, McDonnell SK, Friedrichsen DM, Carlson EE, Kolb S, Deutsch K, Janer M, Hood L, Ostrander EA, Schaid DJ. Prostate cancer and genetic susceptibility: a genome scan incorporating disease aggressiveness. Prostate. 2006;66:317–325. doi: 10.1002/pros.20349. [DOI] [PubMed] [Google Scholar]
- 63.Johanneson B, Deutsch K, McIntosh L, Friedrichsen-Karyadi DM, Janer M, Kwon EM, Iwasaki L, Hood L, Ostrander EA, J.L. S. Suggestive genetic linkage to chromosome 11q12.1 in hereditary prostate cancer families with primary kidney cancer. Prostate. 2006 doi: 10.1002/pros.20528. In Press. [DOI] [PubMed] [Google Scholar]
- 64.Gronberg H, Bergh A, Damber JE, Emanuelsson M. Cancer risk in families with hereditary prostate carcinoma. Cancer. 2000;89:1315–1321. doi: 10.1002/1097-0142(20000915)89:6<1315::aid-cncr17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 65.Vaittinen P, Hemminki K. Familial cancer risks in offspring from discordant parental cancers. Int J Cancer. 1999;81:12–19. doi: 10.1002/(sici)1097-0215(19990331)81:1<12::aid-ijc3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 66.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. Epub 2006 May 2007. [DOI] [PubMed] [Google Scholar]
- 67.Eldon BJ, Jonsson E, Tomasson J, Tryggvadottir L, Tulinius H. Familial risk of prostate cancer in Iceland. BJU Int. 2003;92:915–919. doi: 10.1111/j.1464-410x.2003.04536.x. [DOI] [PubMed] [Google Scholar]
- 68.Jonsson E, Sigbjarnarson HP, Tomasson J, Benediktsdottir KR, Tryggvadottir L, Hrafnkelsson J, Olafsdottir EJ, Tulinius H, Jonasson JG. Adenocarcinoma of the prostate in Iceland: a population-based study of stage, Gleason grade, treatment and long-term survival in males diagnosed between 1983 and 1987. Scand J Urol Nephrol. 2006;40:265–271. doi: 10.1080/00365590600750110. [DOI] [PubMed] [Google Scholar]
- 69.Rokman A, Baffoe-Bonnie AB, Gillanders E, Fredriksson H, Autio V, Ikonen T, Gibbs KD, Jr, Jones M, Gildea D, Freas-Lutz D, Markey C, Matikainen MP, Koivisto PA, Tammela TL, Kallioniemi OP, Trent J, Bailey-Wilson JE, Schleutker J. Hereditary prostate cancer in Finland: fine-mapping validates 3p26 as a major predisposition locus. Hum Genet. 2005;116:43–50. doi: 10.1007/s00439-004-1214-7. Epub 2004 Nov 2011. [DOI] [PubMed] [Google Scholar]
- 70.Hamel N, Kotar K, Foulkes WD. Founder mutations in BRCA1/2 are not frequent in Canadian Ashkenazi Jewish men with prostate cancer. BMC Med Genet. 2003;4:7. doi: 10.1186/1471-2350-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orr-Urtreger A, Bar-Shira A, Bercovich D, Matarasso N, Rozovsky U, Rosner S, Soloviov S, Rennert G, Kadouri L, Hubert A, Rennert H, Matzkin H. RNASEL mutation screening and association study in Ashkenazi and non-Ashkenazi prostate cancer patients. Cancer Epidemiol Biomarkers Prev. 2006;15:474–479. doi: 10.1158/1055-9965.EPI-05-0606. [DOI] [PubMed] [Google Scholar]
- 72.Poynter JN, Cooney KA, Bonner JD, White KA, Tomsho LP, Rennert G, Gruber SB. APC I1307K and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:468–473. doi: 10.1158/1055-9965.EPI-05-0584. [DOI] [PubMed] [Google Scholar]
- 73.Bar-Shira A, Matarasso N, Rosner S, Bercovich D, Matzkin H, Orr-Urtreger A. Mutation screening and association study of the candidate prostate cancer susceptibility genes MSR1, PTEN, and KLF6. Prostate. 2006;66:1052–1060. doi: 10.1002/pros.20425. [DOI] [PubMed] [Google Scholar]
- 74.Dagan E, Laitman Y, Levanon N, Feuer A, Sidi AA, Baniel J, Korach Y, Baruch GB, Friedman E, Gershoni-Baruch R. The 471delAAAG Mutation and C353T Polymorphism in the RNASEL Gene in Sporadic and Inherited Cancer in Israel. Fam Cancer. 2006;31:31. doi: 10.1007/s10689-006-0010-z. [DOI] [PubMed] [Google Scholar]
- 75.Friedrichsen DM, Stanford JL, Isaacs SD, Janer M, Chang BL, Deutsch K, Gillanders E, Kolb S, Wiley KE, Badzioch MD, Zheng SL, Walsh PC, Jarvik GP, Hood L, Trent JM, Isaacs WB, Ostrander EA, Xu J. Identification of a prostate cancer susceptibility locus on chromosome 7q11-21 in Jewish families. Proc Natl Acad Sci U S A. 2004;101:1939–1944. doi: 10.1073/pnas.0308336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cook EH., Jr Merlin: faster linkage analysis with improved genotyping error detection. Pharmacogenomics J. 2002;2:139–140. doi: 10.1038/sj.tpj.6500102. [DOI] [PubMed] [Google Scholar]