Abstract
The aberrant expression of many genes is a common feature in the malignant transformation of cells. In mammalian cells, posttranscriptional gene regulatory processes are emerging as critical determinants controlling gene expression both in physiologic and pathologic conditions. These regulatory mechanisms are directed primarily by the interaction of mRNAs with specific RNA-binding proteins (RBP). There is an emerging body of data demonstrating that two RBPs, AUF1 and HuR, can antagonistically affect the posttranscriptional fate of target mRNAs, as well as concurrently bind to common target transcripts. Employing MCT-1 oncogene-mediated transformation of immortalized breast epithelial MCF10A cells, we characterized the largely reciprocal association of these two RBPs with target mRNAs and their influence on protein expression vis-a-vis cellular transformation. Using a ribonomics approach, we identified mRNAs from cancer-related pathways whose association with AUF1 and/or HuR were altered when comparing immortalized with transformed MCF10A cells. Significantly, we were able to show that knockdown of HuR expression using RNA interference reduced anchorage-independent growth capacity in transformed MCF10A cells and decreased protein expression of a number of validated target genes. Our data show that the global alterations in binding of HuR and AUF1 with target transcripts have a critical role in posttranscriptional regulation of genes encoding proteins involved in breast epithelial cell transformation. These findings further support the feasibility of using a ribonomics approach for the identification of cancer-related pathways.
Introduction
Analysis of gene expression profiles shows differential expression of many genes in normal compared with cancer cells. Modulation of gene expression pattern at the posttranscriptional level has emerged as an important regulatory paradigm primarily governed by RNA-binding proteins (RBP) that associate with mRNAs and regulate their processing (1). Given that RBPs influence the expression of genes that are essential to key cellular functions, including cell division cycle, carcinogenesis, senescence, and immune and stress responses, they have become the focus of intensive investigation (2). HuR and AUF1 RBPs overlap in their tissue distribution, preferentially localize in the nucleus, bind to many common AU-rich target mRNAs, and exert primarily opposing influences on target mRNA stability; therefore, a functional link between them has been postulated (3, 4). Through its association with target mRNAs, HuR was shown to enhance the expression of many proliferative and proto-oncogenic factors, like epidermal growth factor, cyclin A, cyclin B1, and c-myc (5, 6). AUF1 have also been shown to associate with stress response, immune response, mitogenic, cell cycle, and cancer-related mRNAs (3). HuR and AUF1 can compete with each other (7) or with other RBPs (8, 9) for binding to the same AU-rich elements on specific target mRNAs.
In this study, using a microarray approach, we show that the dynamic global changes in association of two RBPs, HuR and AUF1, with cancer-related mRNAs have important influence on cell transformation in an MCT-1–mediated breast epithelial transformation model. Gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG), and BioCarta pathway databases were used to identify functionally related groups of genes whose association with HuR and/or AUF1 were modulated significantly in MCT-1–transformed cells versus immortalized MCF10A cells. Genes whose association with RBPs were modulated were generally shown to be components of major pathways involved in cellular transformation. Knockdown of HuR expression reduced anchorage-independent growth capacity in transformed MCF10A cells. This study provides novel insights into the critical influence of RBPs on gene expression in a cellular transformation model and establishes, in principle, the utility of ribonomics toward the identification of cancer-related pathways.
Materials and Methods
Cell culture and transient transfections
Human mammary epithelial cells MCF10A stable transfected with the pLXSN empty vector (Vector) or pLXSN-MCT-1-V5-Histidine (MCT-1; ref. 10) were cultured in supplemented MEGM medium (Cambrex).
For transfections, Oligofectamine (Invitrogen) and small interfering RNA (siRNA) targeting the HuR coding region (AAGAGGCAATTACCAGTTTCA) and a control siRNA (Qiagen) were used at 20 nmol/L.
Immunoprecipitation of ribonucleoprotein complexes and micro-array analysis (RIP-Chip assay)
Immunoprecipitation (IP) of endogenous RNA-protein complexes were carried out as previously described (11).
RNA from IP material was biotin labeled with the Illumina TotalPrep RNA Amplification kit (Ambion). Illumina’s Sentrix Human Ref-6 Expression BeadChips containing 48,000 RefSeq transcripts (Illumina) were used for microarray analysis. Data were normalized by Z-score transformation (12) and tested for significant differences in signal intensity. Genes considered to be significantly changed (Z ratio, >1.5 or <−1.5; P < 0.05, fdr < 0.3) were analyzed for representation of GO terms to identify key biologically functional categories (13) that were significantly changed (P < 0.05) in MCT-1 versus Vector cells within the HuR IP’d mRNAs or AUF1 IP’d mRNAs populations. Analysis was performed using parametric analysis of gene set enrichment (PAGE) package (14). Differentially expressed genes were then analyzed according to predefined pathways annotated by KEGG (15) and Biocarta databases using the DAVID3 bioinformatics resource to identify the critical pathways and visualize genes on BioCarta and KEGG pathway maps. See GEO database4 for complete microarray data.
Real-time RT-qPCR analysis of target mRNAs
RNA from IP material was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) and quantified by real-time qPCR analysis using gene-specific primer pairs (Supplementary Table S1) with iQ SYBR Green Supermix (Bio-Rad) and Bio-Rad iCycler instrument.
Western blotting
Proteins were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Blots were probed with monoclonal antibodies recognizing HuR (Santa Cruz Biotechnology, Inc.), β-actin (Abcam), V5 (MCT-1-V5; Invitrogen), BAX (BD PharMingen) or polyclonal EIF4E-BP2 (Cell Signaling Technology), AUF1, and caspase-2 antibodies from Upstate Biotech. Signals were detected by enhanced chemiluminescence (Pierce).
Anchorage-independent growth assay
At 24 h after transfection, cells were plated at 7,500 cells/mL in a 1:3 agarose to MEGM mixture on the top layer (1:1 agarose/MEGM) and cultured after covering with MEGM medium. Two weeks later, colonies were counted and images were taken using a Nikon Eclipse TE-2000S microscope and a Nikon Plan Flour 10× Ph/DLL ∞ /0.17WD 16.0 optic.
Results
Functional annotation of the mRNAs whose association with HuR and/or AUF1 were altered in MCT-1–transformed cells
We have shown previously that overexpressed MCT-1 protein can transform the nontumorigenic MCF10A cells with induction of anchorage-independent growth (10). Here, we corroborate and extend these data by showing that MCF10A cells overexpressing MCT-1 resulted in formation of tumors in a human xenograft mice model (Supplementary Fig. S1).
Given that HuR and AUF1 usually exert opposing influences on the posttranscriptional fate of target mRNAs and has been shown to associate with many cancer-related genes, we set out to investigate whether the association of these two RBPs with their target transcripts changed in breast epithelial MCF10A cells when transformed through enhanced expression of the MCT-1 oncogene. To this end, HuR-associated and AUF1-associated mRNAs were detected by IP of endogenous HuR and AUF1 from nontransformed and MCT-1–transformed MCF10A cells using anti-HuR or anti-AUF1 antibodies and then followed by global microarray hybridization of the mRNAs obtained in each IP material. Microarray data revealed 2,072 transcripts with significantly (P < 0.05) changed association with AUF1 and 1,676 transcripts with altered binding to HuR in MCT-1–transformed compared with nontransformed Vector cells. The association of 712 targets was significantly altered in tandem with both AUF1 and HuR binding proteins. GO analysis revealed main categories that had significant representation of differentially expressed mRNAs associated with HuR and/or AUF1 in transformed cells (Fig. 1; Supplementary Table S4). GO categories related to cellular transformation, such as cell cycle regulation, signal transduction, DNA damage response, translation regulation, and angiogenesis, were particularly prominent (Fig. 1). Target mRNAs within a majority of the above pathways showed reciprocal associations with HuR and AUF1 in the transformed phenotype (Fig. 1) that confirmed previously shown opposing influences of these two RBPs on the posttranscriptional fate of common target mRNAs.
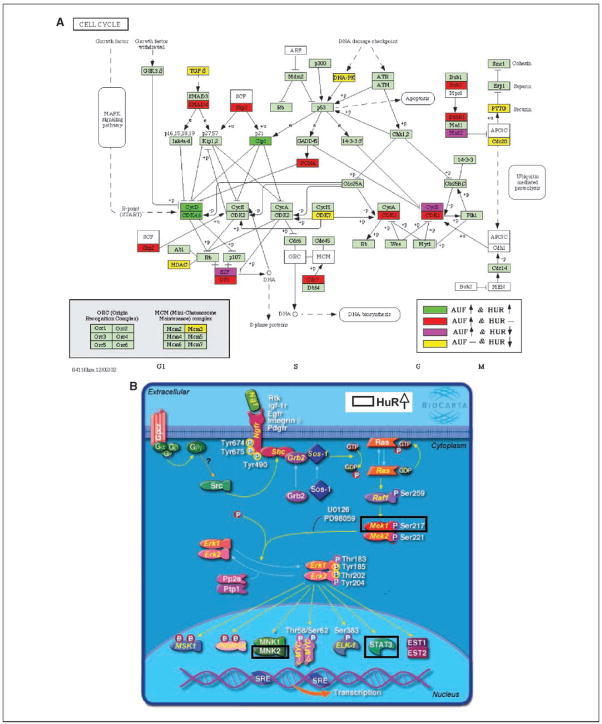
Figure 1.
Functional annotation of AUF and/or HuR association with mRNAs in transformed compared with nontransformed MCF10A cells. Genes significantly changed (Z ratio, >1.5 or <−1.5; P < 0.05) were analyzed for representation of GO terms to identify functional categories. Values increase from green to red via black. Insert represents GO categories with the highest Z-score. All illustrated in cluster categories are listed in Supplementary Table S4.
Modulation of HuR and/or AUF1 association with mRNAs results in differential expression of genes involved in cell cycle and cell signaling pathways in MCT-1–transformed cells
Using GO analysis, we identified important functional pathways involved in cellular transformation; however, many GO classes are overlapping or redundant. Therefore, KEGG and BioCarta pathway databases integrating individual components into unified pathways (15) were used to characterize the enrichment of specific pathway components into functionally regulated gene groups. Nine KEGG pathways were significantly enriched (P < 0.05) in genes showing altered association with HuR and/or AUF1 in transformed cells: cell cycle, cell communication, p53 signaling pathway, ribosome, oxidative phosphorylation, purine metabolism, focal adhesion, ubiquitin-mediated proteolysis, regulation of actin cytoskeleton. Prominent among these KEGG pathways was the cell cycle pathway which was characterized by a significant number of transcripts differentially associated with HuR and/or AUF1 in transformed phenotype as illustrated on pathway map (Fig. 2A; Supplementary Table S2). Using BioCarta pathway database, we found seven pathways significantly (P < 0.05) enriched in genes whose association with HuR and/or AUF1 were altered in transformed compared with nontransformed cells; caspase cascade in apoptosis, cyclins, and cell cycle regulation, Erk1/Erk2 Mapk signaling, Erk, and phosphatidylinositol 3-kinase are necessary for collagen binding in corneal epithelia, role of Ran in mitotic spindle regulation, phosphoinositides and their downstream targets, and HIV type I Nef (negative effector of Fas and TNF). Importantly, increased association of HuR with multiple target genes involved in Erk1/Erk2 Mapk signaling pathway in transformed compared with nontransformed MCF10A cells was identified using the BioCarta pathway database (Fig. 2B; Supplementary Table S3).
Figure 2.
HuR and/or AUF1 differentially regulate mRNAs encoding cell cycle and Erk1/Erk2 Mapk signaling-related genes in MCT-1–transformed cells. mRNAs from HuR and/or AUF1 IP materials differentially expressed in transformed cells were visualized on KEGG cell cycle (A) and BioCarta (B) Erk1/Erk2 Mapk signaling pathway maps. Known pathway-related genes and their relations are shown. Genes showing altered association with HuR and/or AUF1 in transformed phenotype are colored (A) or framed (B) as indicated. Arrows indicate an increased or decreased association with HuR/AUF1 in malignant cells. All genes found to be altered in each pathway are listed in Supplementary Tables S2 and S3.
Protein levels of selected cancer-related genes altered in accordance with differential HuR and/or AUF1 association with their mRNAs in transformed cells
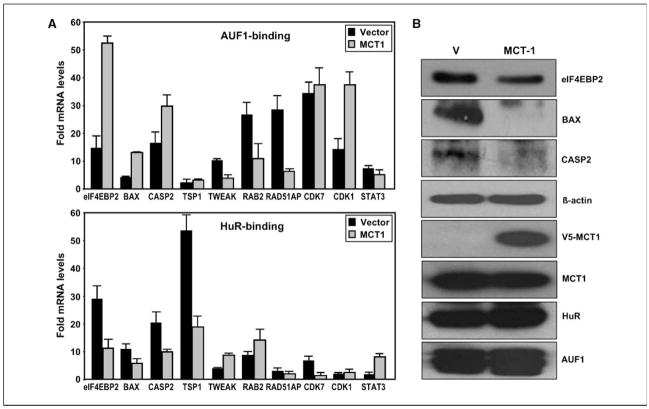
We selected 10 candidates from the significantly changed genes showing differential association with HuR and/or AUF1 in transformed MCF10A cells for further detailed analysis (Fig. 3A; Supplementary Table S1). These candidates were chosen because they were representative of a wide range of pathways involved in tumor development and metastasis, such as translation regulation (eIF4EBP2), angiogenesis (TWEAK, TSP1), apoptosis (BAX, CASP2), cell cycle (CDK7, CDK1), signal transduction (RAB2, STAT3), or DNA damage response (RAD51AP1). We initially validated the association of the 10 genes with HuR and AUF1 in nontransformed and MCT-1–transformed MCF10A cells by IP of HuR-mRNA and AUF1-mRNA complexes followed by reverse transcription of IP’d RNA (RT) and real-time qPCR (Fig. 3A). Importantly, these transcripts predicted to change in concordance with the modulated binding state of HuR and AUF1 in MCT-1–transformed cells, manifested concurrently altered protein levels of these genes (Fig. 3B). In accord with the opposed influence of these two RBPs on mRNAs (AUF1, destabilization; HuR, stabilization), we observed that the protein expression of examined genes became significantly down-regulated with decreased HuR and increased AUF binding to their mRNAs in transformed cells.
Figure 3.
Validation of association of HuR and AUF with target mRNAs in nontransformed and MCT-1–transformed MCF10A cells. A, endogenous HuR and AUF target transcripts were detected by RT-qPCR analysis of the RNA obtained from IP material. Each reaction was carried out in triplicate and three independent experiments were run. Glyceraldehyde-3-phosphate dehydrogenase mRNA background amplification in the IP material served as the internal control. B, several of the transcripts from A were further validated by Western blotting to assess the protein expression of examined genes in vector compared with MCT-1–transformed cells. Total proteins (40 μg) were resolved and the abundance of BAX, eIF4EBP2, caspase-2, HuR, AUF1, MCT-1-V5, and β-actin were measured.
HuR knockdown affected the phenotype of MCT-1–transformed human mammary epithelial cells
Once the alteration in association of the selected mRNAs with HuR and AUF1 in transformed phenotype had been confirmed, we sought to determine the functional consequence of modulating levels of RBPs. We selected HuR to address this question and transfected the MCF10A (Vector and MCT-1 overexpressing) cell lines with either siRNA targeting HuR or control siRNA. As shown in Fig. 4A, reduction of HuR was found to decrease protein levels of the above-validated target genes. Conversely, siRNA knockdown of AUF1 levels resulted in increased expression of validated genes (Supplementary Fig. S2). Finally, we wished to formally show that altered HuR binding to target genes affected the phenotype of MCT-1–transformed human mammary epithelial cells. To this end, we carried out soft agar colony formation assays after siRNA-mediated knockdown of HuR. Reduced HuR levels decreased several fold the ability of MCF10A MCT-1–overexpressing cells to form anchorage-independent colonies compared with control siRNA-transfected cells (Fig. 4B).
Figure 4.
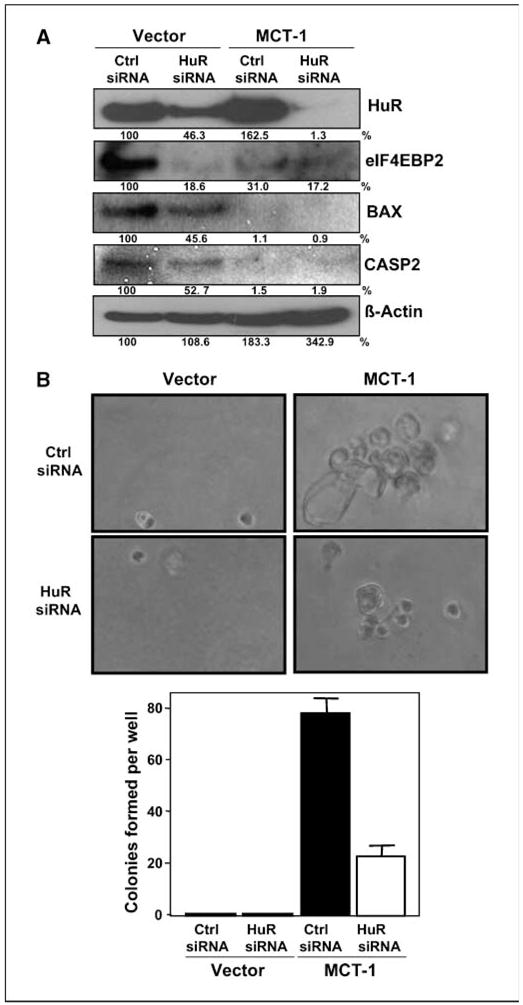
Suppression of HuR functionally alters MCT-1–transformed MCF10A cells. A, at 72 h after transfection of MCF10A (Vector and MCT-1) cells with control siRNA (Ctrl siRNA) or siRNA targeting HuR (HuR siRNA), the expression levels of HuR, eIF4EBP2, BAX, caspase-2, and β-actin for loading control were measured by Western blotting. B, cells were transfected with HuR siRNA as in Fig. 3A and cultured in agarose/medium. Colony formation on soft agar was counted, and representative pictures were taken (10×) 2 wk later. Graphs represent the mean and SE of three independent experiments; P < 0.01.
Discussion
The data presented here provided systematic evidence that dynamic association of two RBPs, HuR and AUF1, with target mRNAs have a critical role in posttranscriptional regulation of genes encoding proteins involved in breast epithelial cell transformation. Functional analysis of target mRNAs associated with HuR and/or AUF1 altered in the transformed phenotype led us to some major cellular transformation pathways, such as cell cycle regulation, signal transduction, DNA damage response, translation regulation, and angiogenesis (Fig. 1; Supplementary Table S4). Two pathways important in cancer development, cell cycle and Erk1/ Erk2 Mapk signaling, were found to be notably represented by transcripts differentially associated with HuR and/or AUF1 in transformed compared with nontransformed cells underscoring the role of RBPs in cellular transformation. These findings are consistent with the ‘‘posttranscriptional operon’’ concept, in which RBPs can jointly regulate subsets of mRNAs encoding functionally related proteins (16) and emphasizes the importance of this emerging paradigm in cancer research.
Target mRNAs within a majority of the above pathways showed reciprocal, alternating association with HuR and AUF1 in the transformed phenotype (Fig. 1) that support prior work showing frequent opposing influences of these two RBPs on the posttran-scriptional fate of their common target mRNAs. HuR has been shown to stabilize target mRNAs and/or induce their translation, as well as rarely repress mRNA translation, it has also been postulated to be involved in splicing and export (2, 17, 18). AUF1 has been widely described to participate in destabilization of target mRNAs (19). In contrast, some data indicate that AUF1 may act either as a stabilizing or a destabilizing factor (20) and, recently, to promote translation by competitively binding with other RBPs (9). Our data support the notion that determinants of whether HuR and AUF1 bind concurrently or competitively to a given mRNA depends on the target mRNA sequence and the cellular context in which the association of ribonucleoprotein complex is investigated (7). The effect on validated target gene protein levels after HuR and AUF knockdown underscore the translational relevance of differential HuR and/or AUF1 association with these mRNAs. While we cannot completely discount the contribution of other mechanism(s) and possible involvement of other posttranscriptional regulatory factors, our results support the biologically relevant influence of these two RBPs on cell transformation.
Together, these data illustrate that the dynamic global alterations in binding of both HuR and AUF1 (and likely other RBPs) to cancer-related mRNAs have important biological implications as shown in our breast epithelial transformation model. The robust predictive value of our approach allowed the identification and successful validation of critical transcripts whose association with HuR/AUF1 was altered as the cellular phenotype changed. Significantly, the data presented herein reinforce the usefulness of exploiting ribonomics to identify pathways important for cancer development.
Acknowledgments
Grant support: Merit Review Award from the Department of Veterans Affairs (R.B. Gartenhaus) and Clinical Innovator Award from the Flight Attendant Research Institute (R.B. Gartenhaus).
We thank Sharon Corl for technical support.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Benjamin D, Moroni C. mRNA stability and cancer: an emerging link? Expert Opin Biol Ther. 2007;7:1515–29. doi: 10.1517/14712598.7.10.1515. [DOI] [PubMed] [Google Scholar]
- 2.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya S, Giordano T, Brewer G, Malter J. Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res. 1999;27:1464–72. doi: 10.1093/nar/27.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol. 2002;22:7268–78. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheflin LG, Zou AP, Spaulding SW. Androgens regulate the binding of endogenous HuR to the AU-rich 3′UTRs of HIF-1α and EGF mRNA. Biochem Biophys Res Commun. 2004;322:644–51. doi: 10.1016/j.bbrc.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:1–12. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ming XF, Stoecklin G, Lu M, Looser R, Moroni C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol. 2001;21:5778–89. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–8. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 10.Hsu H, Shi B, Gartenhaus R. The MCT-1 oncogene product impairs cell cycle checkpoint control and transforms human mammary epithelial cells. Oncogene. 2005;24:4956–64. doi: 10.1038/sj.onc.1208680. [DOI] [PubMed] [Google Scholar]
- 11.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–27. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S-Y, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 17.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–9. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leandersson K, Riesbeck K, Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34:3988–99. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–6. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–95. [PubMed] [Google Scholar]