Summary
Human CD1c is a protein that activates αβ T cells by presenting self antigens, synthetic mannosyl phosphodolichols and mycobacterial mannosyl phosphopolyketides. To determine which molecular structures of antigens mediate a T cell response, we measured activation by structurally divergent M. tuberculosis mannosyl-β1-phosphomycoketides as well as by synthetic analogs produced by two methods that yield either stereorandom or stereospecific methyl branching patterns. T cell responses required both a phosphate and a β-linked mannose unit, and showed preference for C30–34 lipid units with methyl branches in the S-configuration. Thus, in all cases T cell responses were strongest for synthetic compounds that mimicked the natural branched lipids produced by mycobacterial polyketide synthase 12. Incorporation of methylmalonate to form branched lipids is a common bacterial lipid synthesis pathway that is absent in vertebrates, so the preferential recognition of branched lipids may represent a new type of lipid-based pathogen associated molecular pattern (PAMP).
Keywords: Mycobacterium tuberculosis, CD1, polyketide, T cell, stereoselective synthesis
Introduction
Three related antigen presentation systems function to display structurally diverse antigens to αβ T cells, MHC class I, MHC class II and CD1. All three types of antigen presenting molecules fold in 3-dimensions to form hollow antigen binding grooves. Whereas MHC proteins capture peptides and glycopeptides for display to T cells, the grooves of CD1 proteins are larger and lined by hydrophobic amino acids [1], so that they are specialized to bind lipids (reviewed in [2]). The CD1 system consists of five homologous isoforms in humans (CD1a, CD1b, CD1c, CD1d, CD1e) [3]. CD1e is an intracellular protein, which functions to transfer lipids [4, 5]. The other four human CD1 proteins are expressed on the surface of antigen presenting cells (APCs) and bind lipids within their grooves in a way such that the more hydrophilic elements of the antigens (carbohydrates, peptides, phosphoesters) protrude from the groove to contact T cell receptors (TCRs) [6–9]. Crystal structures show that CD1a, CD1b and CD1d grooves differ in their overall size and shape [1, 10, 11]. CD1 isoforms are differentially expressed on B cells, Langerhans cells and myeloid dendritic cells, and in some cases multiple CD1 isoforms are expressed in the same APC [12]. This suggests that multiple CD1 proteins function together as a family, using their structurally divergent grooves to capture and present diverse classes of self and foreign lipids to T cells. The structures of known antigens are diverse and include molecules composed of mycolate [13, 14], diacylglycerol [15–17], sphingolipid [7, 18–20], polyisoprenol [21], polyketide [22], fatty acyl [23, 24] and other lipid anchors [25]. These antigens have been isolated from microbial pathogens, mammalian cells and synthetic sources, raising the question of how T cells might discriminate among different classes of self and foreign antigens.
In MHC systems, early observations that cells somehow convert full length proteins into recognizable forms through cellular “antigen processing” have been tested by genetic mapping of minimal peptide epitopes [26], elution of peptides from MHC proteins [27–29], synthesis of peptide analogs and crystallization of MHC-peptide complexes [30–32]. Collectively, these studies have provided chemically precise descriptions of the optimal size of antigens for MHC class I (nonamer peptides) and MHC class II (dodecamer peptides) [33, 34]. This basic information has supported innumerable studies of epitope mapping in autoimmune and infectious disease, subunit vaccine design and synthesis of partial agonists for T cells [35–37]. With growing numbers of antigens identified in the CD1 system, the search of chemical motifs that define antigens presented by each type of CD1 protein is now beginning. Unlike MHC encoded antigen presenting molecules, the genes encoding CD1 proteins show low levels of polymorphism, including sequences that encode residues located in the α1 and α2 domains of the CD1 heavy chain and form the structures that mediate antigen binding [38]. Therefore, the specificities of CD1 proteins for lipids likely does not vary among individuals in a population, but likely does vary among each of the distinct CD1 isoforms expressed on antigen presenting cells (APCs). The precise shapes and volumes of mouse CD1d (1650 Å3) and human CD1b (2200 Å3), CD1a (1300 Å3) and CD1d (1400 Å3) proteins have been determined from crystal structures [1, 10, 11, 39], This information, along with the discovery of many types of lipid antigens that differ in the number, size and molecular composition of their lipid anchors, provide two complementary approaches to basic questions about possible chemical motifs that govern lipid antigen presentation.
Chemical synthesis of antigenic lipids can address these basic questions and can also be used to produce analogs with desired immunological properties. The potent agonist of CD1d-restricted NK T cells, α-galactosyl ceramide, has been used to influence outcomes in a variety of animal models of autoimmune, infectious, allergic and neoplastic disease [40]. By producing α-galactosyl ceramides with slightly altered structures such as shorted or unsaturated alkyl chains, it has been possible to influence the half-life of action and the balance of Th1-Th2 cytokines produced by the responding cells [41–43]. Such altered lipid ligands can strongly influence the outcomes of in vivo animal models of multiple sclerosis, rheumatoid arthritis and inflammatory bowel disease [44–46] (and reviewed in [47]). Conversely, β-galactosyl ceramide, a non-activating ligand for CD1d, can suppress CD1d-mediated activation of NK T cells [7, 48, 49]. Although a variety of self and foreign lipids for CD1a, CD1b and CD1c proteins have been identified [65], most or all existing ligands show low potency (activating T cells in the micromolar range) or must be painstakingly purified in small amounts from bacteria. Therefore, there is a good rationale to synthesize antigenic lipids with higher yield in structurally varied forms that may activate, partially activate or deactivate T cells.
Among the human CD1 proteins, CD1c is the isoform for which there is currently the least information relating to its molecular mechanisms of presentation to T cells. CD1c is abundantly expressed on thymocytes, myeloid dendritic cells (DCs) and marginal zone B cells (reviewed in [12]) CD1c participates in regulating immunoglobulin class switching in vitro and human responses to M. tuberculosis infection in vivo [21, 50]. CD1c can directly activate both αβ and γδ T cells in the absence of exogenous antigens, suggesting that it likely presents self antigens to T cells in vitro [51–53] and ex vivo [54]. However, the molecular structures of these self antigens are unknown. CD1c is the only human antigen presenting molecule that has not been crystallized to date, so the only available information on the molecular basis of lipid presentation comes from studies of the foreign glycophospholipid antigens that it presents, synthetic mannosyl phosphodolichols (MPD) and mannosyl-β1-phosphomycoketides (MPM) from M. tuberculosis and M. avium [21]. The C30–34 alkyl chains in mycoketide antigens have methyl branches at C4 (δ-methyl) and every fourth carbon atom thereafter. This highly unusual branched lipid is made by the alternating incorporation of malonyl (C2) and methylmalonyl (C3) units by polyketide synthase 12 (Pks12), an enzyme for which there is no known homolog in non-mycobacterial cells [22].
Here we investigate the specificity of this T cell response using natural [21] and synthetic mycoketide-like compounds, including newly synthesized molecules made according to synthesis schemes whose methods have been previously described in detail [55, 56]. Aside from their use as tool to determine T cell specificity for antigens, the total synthesis of highly antigenic MPMs in high yield has been a longstanding priority in this field for several reasons. MPMs comprise approximately one part per million (ppm) of the M. tuberculosis cell wall. Its scarcity does not limit its ability to activate T cells or influence antibiotic resistance in M. avium species [21, 57]. However, the trace amounts of natural compound available even from very large mycobacterial cultures create a situation in which structural elucidation of natural antigens was accomplished solely through sensitive collision induced dissociation mass spectrometry (CID-MS) techniques. Thus, certain key elements of the structure of the natural antigens, including the anomeric linkage of the mannose unit and the position of the methyl branches were indirectly inferred from MS data. Therefore, direct comparison of natural compounds with bona fide synthetic standards more fully establishes the structure of natural antigens. Last, structure-function analysis found that that the alkyl chain length, number and stereochemistry of the methyl branches of the lipid moiety strongly influence antigenicity. In all cases synthetic MPMs that most closely mimic the structure of natural mycobacterial MPMs were found to be most potent. Because methyl branching is common in bacterial fatty acyl and polyketide systems and absent in mammalian fatty acids, identification of methyl branching as a determinant of antigenic potency now makes it a candidate lipid-based pathogen associated molecular pattern (PAMP) that may alert the immune system to infection.
Results
Synthetic mycoketides with a stereorandom branched alkyl chain activate CD1c-restricted T cells
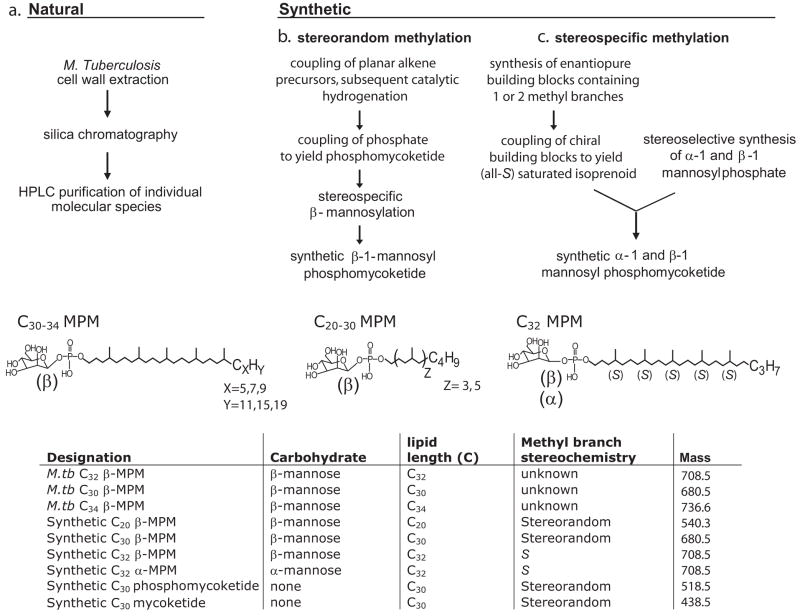
To determine the structural basis of antigen recognition, we prepared natural and synthetic MPM analogs. The biosynthesis of the 4, 8, 12, 16, 20 pentamethyl pentasyl mycoketide unit in mycobacteria involves the Pks12-mediated condensation of C2 and C3 in a strictly alternating fashion to build up the lipid in repeating C5 units [22]. The terminal portion of the lipid unit consists of an alkyl chain of somewhat varying length (C5–9) that is thought to initiate the elongation process (Fig. 1a). Therefore, we used HPLC based methods to isolate individual molecular species of natural M. tuberculosis β-mannosyl phosphomycoketides [22], which varied in overall lipid length (C30–34) by small increments involving the terminal alkyl unit (Fig. 1a). Second, we synthesized β-mannosyl phosphomycoketide analogs that varied in length by larger increments involving the number of repeating C5 units in the chain. This was accomplished using a method for stereospecifically coupling mannose in β-linkage with alkyl phosphate lipids made from polyisoprenols that had been saturated using Adams’ catalyst (platinum oxide) [55]. By altering the number of geranyl units used, it was possible to make stereorandom MPM analogs with C20 and C30 alkyl chains.
Figure 1.
Schematic of natural and synthetic mycoketide antigens
The low yields of mycobacterial antigens obtained to date from natural sources have not been sufficient to directly determine the stereochemistry of the 5 methyl branches on the alkyl chain. However, because the 12 catalytic subunits of Pks12 are proposed to elongate in 5 cycles of the same reaction, all methyl branches are likely to have the same stereochemistry. Further, the configuration of methylmalonyl-CoA has been determined to be S [58], and the carboxyl group of the methylmalonyl unit is replaced by a distal primer side group in a decarboxylative condensation reaction involving a mechanism that likely does not alter the stereochemistry [22]. Therefore, the biosynthetic mechanism strongly suggests that the mycoketide chain contains 5 S-stereocenters (all-S mycoketide). Thus, the first synthetic approach recapitulated the naturally recurring mycoketide unit insofar that the methyl branches were inserted at the equivalent positions (4, 8, 12, 16, 20) of the main alkyl chain, but resulting in a mixture of compounds that have either R or S stereochemistry at each of these chiral centers (synthetic β-MPM (stereorandom), Fig. 1b). Because the stereochemistry of the methyl branches of natural mycoketides had not been definitively determined, this was considered an expeditious route to the preparation of immunologically active synthetic antigens.
All compounds were tested on an equimolar basis by incubating with CD1c-expressing monocyte-derived dendritic cells or transformed B lymphoblastoid cells (C1R) and measuring the antigen-dependent proliferation or IL-2 release by CD8-1, a human αβ T cell line whose activation requires CD1c expression by antigen presenting cells (APC) ([53] and suppl Fig. 1). Although synthetic stereorandom C30 β-MPM stimulated an IL-2 response, the absolute potency (Dosehalf maximal stimulation) was 20 to 40-fold lower than M. tuberculosis C32 MPM, suggesting that either the small alterations in chain length (C30 versus C32) or the methyl branching stereochemistry strongly influenced recognition. Therefore, we sought to separately evaluate the role of lipid length and methyl branching on the T cell response.
Influence of mycoketide length on T cell activation
Purified mycobacterial MPMs with mycoketide units of C30, C32 or C34, which correspond to the natural range of lipid lengths naturally produced by mycobacteria, stimulated T cells with equivalent potency (Figs. 1a, 2b). Prior studies of mannosyl-β1-phosphodolichol (MPD) analogs suggested that lipids in the range of C55–95, have little or no ability to activate T cells compared to C35 analogs [21]. Further, we found that a synthetic stereorandom C20 MPM was more than thousand-fold less potent than the analogous compound C30 in length (Fig. 2c). Thus, CD8-1 was most potently activated by natural analogs whose chain length corresponds to those normally made by mycobacteria, as compared to synthetic analogs that are substantially longer or shorter. In addition, these results suggested that the 20 to 40 fold difference in potency observed in the comparison between synthetic stereorandom C30 MPM and natural M.tuberculosis C32 MPM (Fig. 2a) was not accounted for by the small difference in chain length. The difference in potency was, therefore, more likely due to the differences in the stereochemical orientation of the methyl branches.
Figure 2. Synthetic C30 β-MPM induces T cell activation.
Titrated amounts of synthetic stereorandom C30 β-MPM and M.tb C32 β-MPM were incubated with CD1c-positive antigen presenting cells and the CD8-1 T cell line for 24 hrs, after which the levels of IL-2 released in the culture supernatant were determined by measuring 3H-thymidine incorporation by IL-2 dependent HT-2 cell line (a). Similar T cell activation assays were performed with the naturally occurring M.tb mannosyl phosphomycoketides with chain lengths of C30, C32 or C34 (b), and the synthetic stereorandom C20 and C30 β-MPM (c). The results are reported as mean ± standard deviation of triplicate measurements. Similar results were obtained in two separate experiments.
Influence of methyl branching patterns and stereochemistry on T cell activation
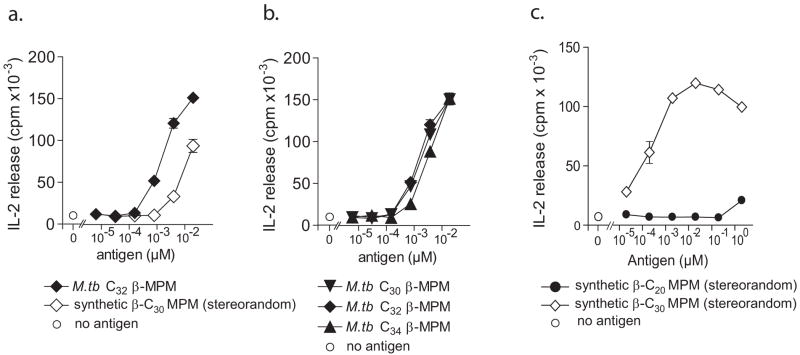
To directly address this question and to produce synthetic compounds that might have the greatest potency for T cell activation, we used a second approach involving the stereospecific synthesis of MPM and analogs with each methyl branch having S-configuration, corresponding to that predicted to be found in mycobacterial antigens (Fig. 1c). This was possible using a convergent strategy in which enantiopure syn-isoprenoid building blocks are assembled to yield lipid chains that have stereochemically defined methyl branches at all positions (all-S) [56]. Coupling of the C32 lipid chain alcohol to tetra-acetyl protected,α- or β-mannosyl phosphate afforded C32 α-MPM (all-S) and C32 β-MPM (all-S) after deprotection. In addition, three analogs of C32 β-MPM that contained the same overall length of the main chain, but differed in the number of methyl branches, were prepared. One analog lacked all methyl groups on the alkyl chain (analog C, Fig. 3a, and Suppl. data 2), and two analogs lacked all methyl groups except for the most proximal one at position 4, and were prepared with either S or R configuration (analogs A and B, respectively).
Figure 3. Synthesis and immunological evaluation of β-MPM analogs.
Three analogs (A, B, C) were prepared by coupling protected β-1-mannose phosphate to alkyl chains with or without a methyl group at position 4. The complete description of the syntheses and analyses are included in supplementary data (suppl. data 2). Titrated amounts of synthetic enantiopure all-S C32 β-MPM and M.tb C32 β-MPM (b) or synthetic C32 β-MPM analogs A, B, and C (c) were incubated with CD1c-positive antigen presenting cells and the CD8-1 T cell line for 24 hrs, after which the levels of IL-2 released in the culture supernatant were determined by measuring 3H-thymidine incorporation by IL-2 dependent HT-2 cell line. The results are reported as mean ± standard deviation of triplicate measurements, and are representative of results obtained in two (b) or three (c) separate experiments.
Consistent with prior high titer T cell responses to bacterial extracts and preliminary analysis of a synthetic compound [21, 56], quantitative analysis of natural and synthetic antigens showed that both were extremely potent in absolute terms with half-maximal T cell activation was seen at low nanomolar concentrations (Fig. 3b). These foreign or synthetic antigens are at least 1,000 fold more potent than self lipid antigens like gangliosides and sulfatides, which are recognized in the mid-micromolar range [18, 19, 59]. This is an absolute potency similar to that of the most potent lipid antigens known in the CD1 system, such as α-galactosyl ceramides. The equipotency of synthetic all-S C32 β-MPM and M. tuberculosis C32 MPM is consistent with the interpretation that they are identical compounds.
Comparison of the analogs differing in the presence and stereochemistry of the methyl branches showed that the compound lacking 4 out of 5 methyl branches (analog A) was 500 fold less potent (Fig. 3c) than the fully branched compound. The presence and stereochemistry of this single methyl group seems to influence the T cell response, because analog B, which is identical to A except for its stereochemistry at the methyl branch showed an even further reduction of potency, similar to an alkyl lipid lacking all methyl branches (analog C). It is also notable that the least potent analogs are predicted to be more water soluble than the most potent branched compound, suggesting that the differences are not primarily related to solubility in media. Overall, the reduced potency of all three analogs indicates that the presence of methyl branches on the lipid chain strongly influences the capacity of the compound to stimulate the T cell response.
Influence of the mannose glycosidic linkage on T cell activation
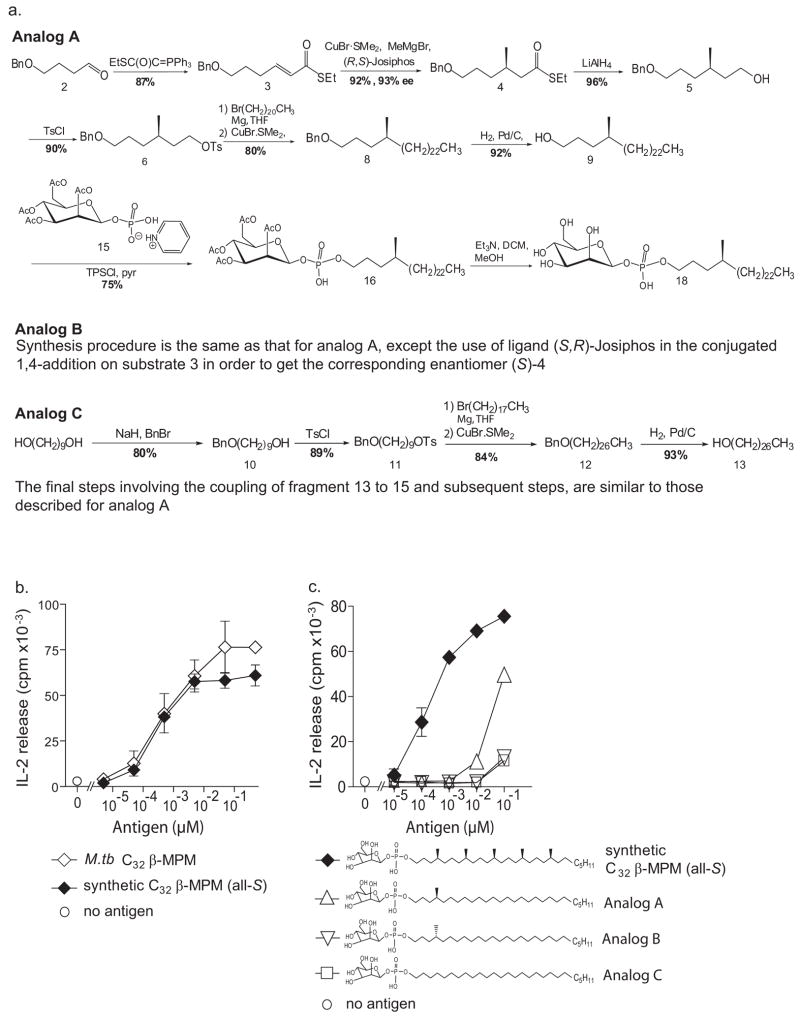
The synthesis of all-S C32 MPMs with both α- and β-anomerically linked mannose units provided authentic standards for definitively assessing the mannose linkage of natural MPM antigens. Electrospray ionization-MS in the negative mode showed that the M. tuberculosis compound and both synthetic MPMs generated the expected [M-H]− ion at m/z 707.6, corresponding to C32 mannosyl phosphomycoketide (Figure 4a, left). Low energy CID-MS of the 707.5 ion, however, revealed two fragmentation patterns. The α-anomer of the synthetic MPM showed the preferential loss of the mannose resulting in a predominant fragment ion of the alkyl phosphate (m/z 545.5), whereas collision of the β-anomer resulted in a main fragment ion of m/z 689.5 corresponding to a loss of H2O, and a cross-ring cleavage product of m/z 587.5. Previous analysis of model compounds has suggested that fragments equivalent to 587.5 are generated when the C2 hydroxyl is cis to the C1 phosphate, as in the case of a β-1 but not anα-1 mannosyl linkage [60]. The synthesis of authentic α- and β-anomeric compounds provided standards for comparison, which bear out this prediction. The identical collisional MS spectra of the M. tuberculosis-isolated C32 MPM and the synthetic β-anomer of MPM confirm the β-linkage of the mannose in the natural compound. Testing of these compounds, as well as phosphomycoketide and mycoketide intermediates generated in the synthesis of C30 MPM (stereorandom) (Fig. 4b–c), confirms that the presence of the phosphate and mannose units, as well as their linkage in the β-configuration is necessary for the CD1c-mediated T cell response.
Figure 4. T cell fine specificity for carbohydrate linkage.
(a) Electrospray ionization MS in the negative mode of M.tb C32 MPM and synthetic C32 α-MPM and C32β-MPM yielded an expected [M–H]-ion at m/z 707.5 in all three samples. Prior studies [60] have shown that the through-ring cleavage product of m/z 587.5 is favored when the substituents at C1 and C2 are cis, as in β-mannosyl phosphates. Low energy collision induced dissociation mass spectrometry showed identical fragmentation patterns of M.tb-isolated C32 MPM and the synthetic β-anomer, confirming the beta linkage of the mannose in the natural compound. Intermediates of the C30 β-MPM (stereorandom) synthesis, including unmannosylated phosphomycoketide and mycoketide (b) and C32 mannosyl phosphomycoketide with α-or β-anomerically linked mannose (c) were tested for their capacity to induce IL-2 release by CD1c-restricted T cells (CD8-1). The results are reported as mean ± standard deviation of triplicate measurements. Similar results were obtained in two (b) or three (c) separate experiments.
Discussion
Here we provide functional insight into how the fine structure of an antigen controls a CD1c-restricted T cell response. This has implications for the molecular mechanism of T cell activation as well as defining the types of organisms and biosynthetic pathways that normally produce such antigenic lipids. Other studies of glycolipids presented by CD1b and CD1d have found that T cell responses are not preserved after altering either the number or linkage of carbohydrates [7, 19, 61], pointing to carbohydrate linkage specificity as a general feature of glycolipid antigen recognition. The recently solved ternary CD1d-α-galactosylceramide-TCR crystal structure shows precisely how the carbohydrate moiety influences the T cell response based on its position at the interface of CD1d and the TCR [9]. In this case, the galactosyl unit is positioned at the opening of the groove so that the α-anomeric linkage causes the hexose ring to lie roughly in parallel to the surface of CD1d and fits in a small cavity at the CD1d-TCR interface. The β-linked anomer is predicted to cause the ring to protrude outwards from CD1d and impede the approach of the TCR. The CD1c-restricted T cell preference for the β-anomeric MPM, in contrast to the α-anomeric glycosphingolipid, points to a difference in the molecular mechanisms of carbohydrate positioning in these CD1c and CD1d antigen presentation events.
The carbohydrate linkage has also been used to infer the types of organisms that might produce antigenic glycolipids. In the case of monoglycosyl ceramides, mammalian cells typically produce β-linked species, whereas synthetic lipids, which recapitulate the structures of those found in Sphingomonas paucimobilis and related bacteria, produce α-linked ceramides [20, 62]. The correlation of α-linkage with bacterial biosynthesis pathways led to the more general speculation that the α-linked sugar is the key chemical element to allow certain subsets of sphingolipids to be recognized as foreign by CD1d-restricted NK T cells. This and prior studies identify 1-linked mannosyl phospholipids as CD1c-presented antigens [21, 56]. Here we provide further evidence that natural mycobacterial mannosyl phosphomycoketides contain β-anomerically linked mannose units. Because candidate self antigens, known as mannosyl-β1-phosphodolichols, have identical phosphomannose units, the search for a chemical basis for recognizing mycobacterial MPMs as foreign must consider aspects of the fine structure of the lipid moieties of related compounds.
It is notable that both known classes of antigens presented by CD1c, MPD and MPM, have repeating methyl branches [21, 22]. With one exception that contains both straight chain and branched lipids [23], all known antigens presented by all other CD1 isoforms lack such branches, raising the possibility that methyl branching represents an isoform-specific motif for CD1c presented antigens. Definitive proof of this hypothesis requires further identification of CD1c-presented antigens, study of the natural ligands eluting from CD1c proteins and crystal structures of CD1c-lipid complexes. However, direct evidence that methyl branches contribute to the T cell activation event supports this hypothesis (Fig. 3). A candidate mechanism for this effect has been proposed in which the repeating methyl branches might function like a ratchet to retain lipids within the groove [63], in contrast to the known interaction of straight chain lipids with the slender, unbranched A’ pocket of CD1a [10, 64].
The synthetic analogs studied here bridge among the natural structures found in three types of lipid anchors for CD1-presented antigens: alkyl units (fatty acids, sphingosines), polyisoprenol units (polyprenols, dolichols) and polyketide units (mycoketides, phthioceranic acids). All known self lipid antigens in the CD1 system and almost every non-terpenoid lipid in mammalian cells are composed of unbranched alkyl chains in the range of C12–24 [65]. Therefore, repeating methyl branches might represent a lipid motif that is a pathogen associated molecular pattern (PAMP) that allows for activation of CD1c-restricted T cells in the setting of infection [66] in much the same way the Vγ9Vδ2 T cells preferentially recognize bacterial hydroxymethylbutenyl pyrophosphate in preference to mammalian isopentenyl pyrophosphates [67–70]. The two known biosynthetic mechanisms for producing methyl branches involve polyketide synthases and polyisoprenol synthases. Polyketide synthases are only known in non-mammalian organisms, supporting the idea that these are intrinsically foreign structures [71]. However, polyisoprenoid lipids are made by all cellular organisms, and CD1c proteins can present synthetic mannosyl phosphodolichols, when their length is C35 [21]. However, the C35 chain length is a synthetic construct, and naturally occurring polyisoprenols in mammalian cells are made as C90–100 dolichols or as C10–20 prenyl modifications of proteins, which fall outside the optimal C30–34 length reported here.
Thus, T cells showed specificity for the carbohydrate linkage (β-anomer) as well as the structure of the lipid units with the overall size (C30), number of methyl groups (5), and their stereochemistry (S) corresponding to those made naturally by a foreign enzyme present in disease causing species of mycobacteria. While synthetic C32 β-mannosyl phosphomycoketide can potently activate a specific T cell line, studies in polyclonal T cells from healthy donors or M. tuberculosis infected patients will ultimately establish the capacity of this compound to activate populations of CD1c-restricted T cells ex vivo. Although the responses of any single T cell line may not be representative of CD1c-restricted T cells in general, the mycoketide moiety likely interacts with portions of groove structures formed by invariant regions of the α1-α2 superdomain and are therefore likely conserved in human CD1c-mediated antigen presentation events.
Significance
The first detailed structure-function study of CD1c-presented antigens serves to frame a general hypothesis about the structures of CD1 presented antigens in which CD1c may selectively bind or retain foreign lipids with unusual length (C30–34) and methyl branching, rather than the larger pool of unbranched, shorter self lipid antigens. Thus, branched alkyl lipids of intermediate length may be a lipidic pathogen-associated molecular pattern recognized by CD1c. Further, the direct comparison of natural MPMs with bona fide standards with defined stereochemistry strongly supports prior speculations that these antigens are composed of β-linked carbohydrates and methyl branches in the S-configuration, so that this study now provides a complete chemical identification of the natural immunomodulatory lipids. Last, whereas α-galactosyl ceramides have now been extensively studied in mice [40, 72], high potency ligands for the CD1 isoforms lacking in mice but present in humans have not yet been developed. CD1c is abundantly expressed on human marginal zone B cells, which play an important role in the early stages of immune response [73]. Therefore, the synthesis of a mannosyl-β1-mycoketide in good yield provides a highly potent reagent for testing the functions of human CD1c-restricted T cells ex vivo and for possible development as a component of a subunit vaccine [74] or immunomodulatory agent.
Experimental procedures
Natural mycobacterial β-mannosyl phosphomycoketides
Mannosyl phosphomycoketides (MPM) were purified from M. tuberculosis H37rv and M. avium using refinements of previously described methods [22]. Briefly, MPM was extracted from CHCl3/CH3OH extracts of whole mycobacteria using an open silica gel column (Alltech) eluted sequentially with chloroform, acetone and methanol. Methanol-eluting fractions were further purified using one-dimensional preparative thin layer chromatography (silica gel G plate, Analtech Inc.) with a solvent system of CHCl3:CH3OH:H2O:NH4OH (60:35:7.2:0.8 v/v/v/v). The lipid fraction with T cell activation activity was extracted with CHCl3/CH3OH (1:1) from the silica gel and subjected to further separation by HPLC with a monochrome Diol column coupled on-line to a LCQ Advantage ion-trap mass spectrometer. To separate MPM homologues with different alkyl backbones, reversed phase HPLC with a Vydac C8 reversed phase column was used, and the compounds were eluted with isopropanol: methanol:acetonitrile: hexane: water (37:30:18:3:12, v/v/v/v/v) containing 6 mM ammonium acetate. This method yielded peak to baseline separations of C30 MPM (m/z 679.6), C32 MPM (m/z 708.6) and C34 MPM (m/z 736.6). In order to quantify trace amounts of recoverable HPLC purified M.tuberculosis MPM by mass spectrometry, synthetic C20 MPM was used as internal standard. One μM synthetic C20 MPM (10μl) and HPLC purified M.tuberculosis MPM were mixed and detected by electrospray mass spectrometry in the negative mode. The concentration of M.tuberculosis MPM was determined by comparing the peak intensities of natural homologues of MPM (C32 MPM m/z 707.5, plus trace amounts of C30 MPM m/z 679.5 and C31 MPM m/z 693.5) to the intensity of synthetic C20 MPM (m/z 539.5). The same method was used to quantify the synthetic C32 α-MPM and C32 β-MPM that were used for the comparative assays.
Synthesis of β-mannosyl phosphomycoketide (β-MPM) analogs
The detailed description of the synthesis of the two lead compounds analyzed here, stereorandom C30 β-MPM and the all-S α- and β-C32 MPM, has previously been described [55, 56]. Full characterization of these compounds, including the confirmation of the anomeric linkage of the mannose by 1H-, 13C- and 31P-NMR and ESI-MS, is included in the supplementary data of these publications. In the β-anomer there is a Nuclear Overhauser Effect (NOE) between the anomeric H and H3 and H5, which is absent in the α-anomer. Additional support was given by comparison of the anomeric 1J CH coupling constants of both anomers (α: 169 Hz), β: 159 Hz), and by a distinct chemical shift in 1H-NMR between anomeric protons of both compounds (α: 5.38 ppm, β: 5.07 ppm). The synthesis of the analogs A and B was carried out using the same strategy as published for C32 β-MPM (all-S) [56], comprising a catalytic asymmetric conjugate addition of methylmagnesium bromide to 3 (Fig. 3a) using a CuBr/(R,S)-Josiphos catalyst for analog A (and its enantiomer, CuBr/(S,R)-Josiphos, for analog B). This afforded building block 4 in 93% enantiomeric excess and of known absolute configuration [57], which was subsequently converted into 6 by reduction and tosylation. Copper-mediated cross coupling with BrMg(CH2)20CH3 for analog A and B gave the corresponding protected alkyl alcohols (Fig. 3a, 8 for analog A). Deprotection, followed by coupling to tetra-acetyl protected β-1-mannose phosphate resulted in the desired protected compounds (Fig. 3a 16 for analog A) in good yield. These compounds were purified by column chromatography, fully characterized and shown to be pure by thin layer chromatography, 1H-, 13C- and 31P-NMR and ESI-MS. Final deprotection with Et3N in MeOH/CH2Cl2 gave the required MPM analogs with a single methyl branch in the R or S configuration at position 4.
To prepare analog C, containing a straight chain alkyl group, nonadiol was monobenzylated and monotosylated followed by cross coupling with octadecylmagnesium bromide leading to HO(CH2)26CH3 after debenzylation. Subsequent coupling with tetra-acetyl protected β-1-mannose phosphate and finally deprotection led to C. Structure and purity of the final compounds was determined by ESI-MS. A detailed description of the syntheses of the β-MPM analogs including characterisation with NMR and ESI-MS, is included in the supplementary data (Suppl. data 2).
Cellular assays
Monocyte-derived CD1c-expressing antigen presenting cells (APC) were prepared from human PBMC by centrifugation over Ficoll-Hypaque, adherence to plastic, and treatment with 300 IU/ml granulocyte/monocyte –colony stimulating factor and 200 IU/ml Interleukin-4 for 72 hrs, followed by γ-irradiation (5000 rad). Antigen presenting cell lines were generated from C1R lymphoblastoid cells and k562 cells by stable transfection with vectors containing cDNA encoding human CD1a, CD1b, CD1c or CD1d heavy chains (C1R transfected using vector pSR α-NEO, k562 transfected with vector pcDNA3.1). CD1c-restricted, mannosyl phosphomycoketide reactive T cell line (CD8-1) [6, 75] was tested for IL-2 release using the HT-2 bioassay. Briefly, 5 × 104 CD8-1 T cells and 5 × 104 γ-irradiated APC were incubated in 200 μl of T cell media containing serial dilutions of lipids antigen for 24 h after which 50 μl of supernatant was analyzed for IL-2 release [61]. Supernatant was added to wells containing 104 IL-2 dependent HT2 cells in 100 μl media, which were cultured for 24 h before adding 1 μCi 3H-thymidine for an additional 6 h of culture, followed by harvesting and counting β-emissions. Assays were performed in triplicate and reported as the mean ± standard deviation.
Supplementary Material
Acknowledgments
The authors acknowledge Michael Brenner for providing T cells, Steven Porcelli for providing CD1 transfectants as well as T. D. Tiemersma-Wegman (chromatography) and A. Kiewiet (MS) for technical support (Stratingh Institute, University of Groningen). This work was supported by grants from the NIAMS (048632), NIAID (049313), the Harvard University Center for AIDS Research (CFAR, P30 AI 060354), Pew Foundation Scholars in the Biomedical Sciences the Burroughs Wellcome Fund, Netherlands Organization for Scientific Research (NWO/CW) and a generous gift of Josiphos ligand from Solvias, Basel.
Footnotes
None of the authors have conflict or interest, financial or otherwise.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zeng Z, Casta¤o AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 2.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 3.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 4.Angenieux C, Fraisier V, Maitre B, Racine V, Van Der WN, Fricker D, Proamer F, Sachse M, Cazenave JP, Peters P, Goud B, Hanau D, Sibarita JB, Salamero J, de la SH. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6:286. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 5.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, Salamero J, Cazenave JP, Hanau D, Mori L, Puzo G, De Libero G. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 6.Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, Wilson IA, Porcelli S, Brenner MB. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 8.Gadola SD, Koch M, Marles-Wright J, Lissin NM, Shepherd D, Matulis G, Harlos K, Villiger PM, Stuart DI, Jakobsen BK, Cerundolo V, Jones EY. Structure and binding kinetics of three different human CD1d-α-galactosylceramide-specific T cell receptors. J Exp Med. 2006;203:699. doi: 10.1084/jem.20052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 10.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4:808. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 11.Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nature Immunol. 2002;3:721. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 12.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. CTMI. 2007;314:113–141. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]
- 13.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted αβ T cells. Nature. 1994;372:691. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 14.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 15.Rauch J, Gumperz J, Robinson C, Skold M, Roy C, Young DC, Lafleur M, Moody DB, Brenner MB, Costello CE, Behar SM. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem. 2003;278:47508. doi: 10.1074/jbc.M308089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 17.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 18.Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Shamshiev A, Donda A, Prigozy TI, Mori L, Chigorno V, Benedict CA, Kappos L, Sonnino S, Kronenberg M, De Libero G. The αβ T cell response to self-glycolipids shows a novel mechanism of CD1b loading and a requirement for complex oligosaccharides. Immunity. 2000;13:255. doi: 10.1016/s1074-7613(00)00025-x. [DOI] [PubMed] [Google Scholar]
- 20.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 21.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 22.Matsunaga I, Bhatt A, Young DC, Cheng TY, Eyles SJ, Besra GS, Briken V, Porcelli SA, Costello CE, Jacobs WR, Jr, Moody DB. Mycobacterium tuberculosis pks12 Produces a Novel Polyketide Presented by CD1c to T Cells. J Exp Med. 2004;200:1559. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated Sulfoglycolipids Are Novel Mycobacterial Antigens Stimulating CD1-restricted T Cells during Infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. T cell activation by lipopeptide antigens. Science. 2004;303:527. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 25.Van Rhijn I, Young DC, Im JS, Levery SB, Illarionov PA, Besra GS, Porcelli SA, Gumperz J, Cheng TY, Moody DB. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci USA. 2004;101:13578. doi: 10.1073/pnas.0402838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oldstone MB, Whitton JL, Lewicki H, Tishon A. Fine dissection of a nine amino acid glycoprotein epitope, a major determinant recognized by lymphocytic choriomeningitis virus-specific class I-restricted H-2Db cytotoxic T lymphocytes. J Exp Med. 1988;168:559. doi: 10.1084/jem.168.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotzschke O, Falk K, Deres K, Schild H, Norda M, Metzger J, Jung G, Rammensee HG. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990;348:252. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- 28.Rudensky AY, Preston-Hurlburt P, Hong SC, Barlow A, Janeway CA., Jr Sequence analysis of peptides bound to MHC class II molecules. Nature. 1991;353:622. doi: 10.1038/353622a0. [DOI] [PubMed] [Google Scholar]
- 29.Huczko EL, Bodnar WM, Benjamin D, Sakaguchi K, Zhu NZ, Shabanowitz J, Henderson RA, Appella E, Hunt DF, Engelhard VH. Characteristics of endogenous peptides eluted from the class I MHC molecule HLA-B7 determined by mass spectrometry and computer modeling. J Immunol. 1993;151:2572–2587. [PubMed] [Google Scholar]
- 30.Silver ML, Parker KC, Wiley DC. Reconstitution by MHC-restricted peptides of HLA-2 heavy chain with beta2-microglobulin, in vitro. Nature. 1991;350:619. doi: 10.1038/350619a0. [DOI] [PubMed] [Google Scholar]
- 31.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 32.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley Dc. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 33.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 34.Rudensky AY, Preston-Hurlburt P, al Ramadi BK, Rothbard J, Janeway CA., Jr Truncation variants of peptides isolated from MHC class II molecules suggest sequence motifs. Nature. 1992;359:429. doi: 10.1038/359429a0. [DOI] [PubMed] [Google Scholar]
- 35.Madrenas J, Wange RL, Wang JL, Isakov N, Samelson LE, Germain RN. Zeta phosphorylation without ZAP-70 activation induced by TCR antagonists or partial agonists. Science. 1995;267:515–518. doi: 10.1126/science.7824949. [DOI] [PubMed] [Google Scholar]
- 36.Sloan-Lancaster J, Evavold BD, Allen PM. Induction of T-cell anergy by altered T-cell-receptor ligand on live antigen-presenting cells. Nature. 1993;363:156–159. doi: 10.1038/363156a0. [DOI] [PubMed] [Google Scholar]
- 37.Zaremba S, Barzaga E, Zhu M, Soares N, Tsang KY, Schlom J. Identification of an enhancer agonist cytotoxic T lymphocyte peptide from human carcinoembryonic antigen. Cancer Res. 1997;57:4570–4577. [PubMed] [Google Scholar]
- 38.Han M, Hannick LI, DiBrino M, Robinson MA. Polymorphism of human CD1 genes. Tissue Antigens. 1999;54:122. doi: 10.1034/j.1399-0039.1999.540202.x. [DOI] [PubMed] [Google Scholar]
- 39.Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6:819. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 40.Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–1388. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu KO, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, Fujiwara N, Arias I, Miyake S, Yamamura T, Chang YT, Besra GS, Porcelli SA. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of alpha-galactosylceramides. Proc Natl Acad Sci USA. 2005;102:3383. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oki S, Chiba A, Yamamura T, Miyake S. The clinical implication and molecular mechanism of preferential IL-4 production by modified glycolipid-stimulated NKT cells. J Clin Invest. 2004;113:1631. doi: 10.1172/JCI20862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, Tarlton A, Reddy BG, Schmidt RR, Reiter Y, Griffiths GM, van der Merwe PA, Besra GS, Jones EY, Batista FD, Cerundolo V. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiba A, Oki S, Miyamoto K, Hashimoto H, Yamamura T, Miyake S. Suppression of collagen-induced arthritis by natural killer T cell activation with OCH, a sphingosine-truncated analog of alpha-galactosylceramide. Arthritis Rheumatism. 2004;50:305. doi: 10.1002/art.11489. [DOI] [PubMed] [Google Scholar]
- 45.Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- 46.Ueno Y, Tanaka S, Sumii M, Miyake S, Tazuma S, Taniguchi M, Yamamura T, Chayama K. Single dose of OCH improves mucosal T helper type 1/T helper type 2 cytokine balance and prevents experimental colitis in the presence of valpha14 natural killer T cells in mice. Inflamm Bowel Dis. 2005;11:35. doi: 10.1097/00054725-200501000-00005. [DOI] [PubMed] [Google Scholar]
- 47.Savage PB, Teyton L, Bendelac A. Glycolipids for natural killer T cells. Chem Soc Rev. 2006;35:771–779. doi: 10.1039/b510638a. [DOI] [PubMed] [Google Scholar]
- 48.Naidenko OV, Maher JK, Ernst WA, Sakai T, Modlin RL, Kronenberg M. Binding and antigen presentation of ceramide-containing glycolipids by soluble mouse and human CD1d molecules. J Exp Med. 1999;190:1069–1080. doi: 10.1084/jem.190.8.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parekh VV, Singh AK, Wilson MT, Olivares-Villagomez D, Bezbradica JS, Inazawa H, Ehara H, Sakai T, Serizawa I, Wu L, Wang CR, Joyce S, Van Kaer L. Quantitative and qualitative differences in the in vivo response of NKT cells to distinct alpha- and beta-anomeric glycolipids. J Immunol. 2004;173:3693–3706. doi: 10.4049/jimmunol.173.6.3693. [DOI] [PubMed] [Google Scholar]
- 50.Sieling PA, Porcelli SA, Duong BT, Spada F, Bloom BR, Diamond B, Hahn BH. Human double-negative T cells in systemic lupus erythematosus provide help for IgG and are restricted by CD1c. J Immunol. 2000;165:5338. doi: 10.4049/jimmunol.165.9.5338. [DOI] [PubMed] [Google Scholar]
- 51.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341:447. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 52.Spada FM, Grant EP, Peters PJ, Sugita M, Melian A, Leslie DS, Lee HK, van Donselaar E, Hanson DA, Krensky AM, Majdic O, Porcelli SA, Morita CT, Brenner MB. Self-recognition of CD1 by gamma/delta T cells: implications for innate immunity. J Exp Med. 2000;191:937–948. doi: 10.1084/jem.191.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beckman EM, Melian A, Behar SM, Sieling PA, Chatterjee D, Furlong ST, Matsumoto R, Rosat JP, Modlin RL, Porcelli SA. CD1c restricts responses of mycobacteria-specific T cells. Evidence for antigen presentation by a second member of the human CD1 family. J Immunol. 1996;157:2795. [PubMed] [Google Scholar]
- 54.Roura-Mir C, Catalfamo M, Cheng TY, Marqusee E, Besra GS, Jaraquemada D, Moody DB. CD1a and CD1c activate intrathyroidal T cells during Graves’ disease and Hashimoto’s thyroiditis. J Immunol. 2005;174:3773. doi: 10.4049/jimmunol.174.6.3773. [DOI] [PubMed] [Google Scholar]
- 55.Crich D, Dudkin V. Confirmation of the connectivity of 4,8,12,16,20-pentamethylpentacosylphoshoryl beta-D-mannopyranoside, an unusual beta-mannosyl phosphoisoprenoid from Mycobacterium avium, through synthesis. J Am Chem Soc. 2002;124:2263. doi: 10.1021/ja0123958. [DOI] [PubMed] [Google Scholar]
- 56.van Summeren RP, Moody DB, Feringa BL, Minnaard AJ. Total synthesis of enantiopure beta-D-mannosyl phosphomycoketides from Mycobacterium tuberculosis. J Am Chem Soc. 2006;128:4546–4547. doi: 10.1021/ja060499i. [DOI] [PubMed] [Google Scholar]
- 57.Philalay JS, Palermo CO, Hauge KA, Rustad TR, Cangelosi GA. Genes required for intrinsic multidrug resistance in Mycobacterium avium. Antimicrob Agents Chemother. 2004;48:3412–3418. doi: 10.1128/AAC.48.9.3412-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Retey J, Lynen F. The absolute configuration of methylmalonyl-CoA. Biochem Biophys Res Commun. 1964;16:358–361. doi: 10.1016/0006-291x(64)90040-3. [DOI] [PubMed] [Google Scholar]
- 59.Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolucka BA, Rush JS, Waechter CJ, Shibaev VN, De Hoffmann E. An electrospray-ionization tandem mass spectrometry method for determination of the anomeric configuration of glycosyl 1-phosphate derivatives. Analytical Biochemistry. 1998;255:244. doi: 10.1006/abio.1997.2421. [DOI] [PubMed] [Google Scholar]
- 61.Moody DB, Guy MR, Grant E, Cheng TY, Brenner MB, Besra GS, Porcelli SA. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med. 2000;192:965. doi: 10.1084/jem.192.7.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, III, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Walker D, Beutler B, Teyton L, Savage PB, Bendelac A. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 63.Zajonc DM, Wilson IA. Architecture of CD1 proteins. CTMI. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 64.Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J, Costello CE, Rudd PM, Dwek RA, Miller MJ, Brenner MB, Moody DB, Wilson IA. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 65.De Libero G, Mori L. Recognition of lipid antigens by T cells. Nat Rev Immunol. 2005;5:485–496. doi: 10.1038/nri1631. [DOI] [PubMed] [Google Scholar]
- 66.Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka Y, Morita CT, Nieves E, Brenner MB, Bloom BR. Natural and synthetic non-peptide antigens recognized by human gamma delta T cells. Nature. 1995;375:155. doi: 10.1038/375155a0. [DOI] [PubMed] [Google Scholar]
- 68.Belmant C, Espinosa E, Poupot R, Peyrat MA, Guiraud M, Poquet Y, Bonneville M, Fournie JJ. 3-Formyl-1-butyl pyrophosphate A novel mycobacterial metabolite-activating human gammadelta T cells. J Biol Chem. 1999;274:32079–32084. doi: 10.1074/jbc.274.45.32079. [DOI] [PubMed] [Google Scholar]
- 69.Altincicek B, Moll J, Campos N, Foerster G, Beck E, Hoeffler JF, Grosdemange-Billiard C, Rodriguez-Concepcion M, Rohmer M, Boronat A, Eberl M, Jomaa H. Cutting edge: human gamma delta T cells are activated by intermediates of the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis. J Immunol. 2001;166:3655–3658. doi: 10.4049/jimmunol.166.6.3655. [DOI] [PubMed] [Google Scholar]
- 70.Puan KJ, Jin C, Wang H, Sarikonda G, Raker AM, Lee HK, Samuelson MI, Marker-Hermann E, Pasa-Tolic L, Nieves E, Giner JL, Kuzuyama T, Morita CT. Preferential recognition of a microbial metabolite by human Vγ2Vδ2 T cells. Int Immunol. 2007;19:657–673. doi: 10.1093/intimm/dxm031. [DOI] [PubMed] [Google Scholar]
- 71.Hopwood DA. Cracking the polyketide code. PLoS Biol. 2004;2:E35. doi: 10.1371/journal.pbio.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 73.Lopes-Carvalho T, Foote J, Kearney JF. Marginal zone B cells in lymphocyte activation and regulation. Curr Opin Immunol. 2005;17:244–250. doi: 10.1016/j.coi.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 74.Hiromatsu K, Dascher CC, LeClair KP, Sugita M, Furlong ST, Brenner MB, Porcelli SA. Induction of CD1-Restricted Immune Responses in Guinea Pigs by Immunization with Mycobacterial Lipid Antigens. J Immunol. 2002;169:330. doi: 10.4049/jimmunol.169.1.330. [DOI] [PubMed] [Google Scholar]
- 75.Rosat JP, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, Modlin RL, Porcelli SA, Furlong ST, Brenner MB. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ αβ T cell pool. J Immunol. 1999;162:366. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.