Abstract
Hydrolytic metabolism of pyrethroid insecticides in humans is one of the major catabolic pathways that clear these compounds from the body. Rodent models are often used to determine the disposition and clearance rates of these esterified compounds. In this study the distribution and activities of esterases that catalyze pyrethroid metabolism have been investigated in vitro using several human and rat tissues, including small intestine, liver, and serum. The major esterase in human intestine is carboxylesterase 2 (hCE2). We found that the pyrethroid trans-permethrin is effectively hydrolyzed by a sample of pooled human intestinal microsomes (5 individuals), while deltamethrin and bioresmethrin are not. This result correlates well with the substrate specificity of recombinant hCE2 enzyme. In contrast, a sample of pooled rat intestinal microsomes (5 animals) hydrolyze trans-permethrin 4.5-fold slower than the sample of human intestinal microsomes. Furthermore, it is demonstrated that pooled samples of cytosol from human or rat liver are ~2-fold less hydrolytically active (normalized per mg protein) than the corresponding microsomal fraction toward pyrethroid substrates; however, the cytosolic fractions do have significant amounts (~40%) of the total esteratic activity. Moreover, a 6-fold interindividual variation in carboxylesterase 1 protein expression in human hepatic cytosols was observed. Human serum was shown to lack pyrethroid hydrolytic activity, but rat serum has hydrolytic activity that is attributed to a single CE isozyme. We purified the serum CE enzyme to homogeneity to determine its contribution to pyrethroid metabolism in the rat. Both trans-permethrin and bioresmethrin were effectively cleaved by this serum CE, but deltamethrin, esfenvalerate, alpha-cypermethrin, and cis-permethrin were slowly hydrolyzed. Lastly, two model lipase enzymes were examined for their ability to hydrolyze pyrethroids. However, no hydrolysis products could be detected. Together, these results demonstrate that extrahepatic esterolytic metabolism of specific pyrethroids may be significant. Moreover, hepatic cytosolic and microsomal hydrolytic metabolism should each be considered during the development of pharmacokinetic models that predict the disposition of pyrethroids and other esterified compounds.
Keywords: carboxylesterases, pyrethroids, xenobiotic metabolism
1. Introduction
Carboxylesterases (CEs) are members of the α,β-serine hydrolase multigene family (Cygler et al., 1993). These enzymes catalyze the hydrolysis of esters, amides, and thioesters and play an important role in endobiotic and xenobiotic metabolism (Satoh and Hosokawa, 1998). The two predominant CE isozymes in humans, hCE1 and hCE2, are expressed at high levels in the liver. hCE1 and hCE2 share 48% amino acid sequence homology and thus are placed in two separate CE classes, CES1 and CES2 respectively. The large quantities of CEs found in liver likely compensate for the fact that they are rather inefficient enzymes (Testa and Mayer, 2003). In addition to their abundant expression in liver, tissue-specific expression of human CE isozymes has been observed (Satoh et al., 2002). For example, hCE2 is expressed at relatively high levels in the small intestine, while hCE1 is not expressed in this tissue (Schwer et al., 1997). The selective expression of hCE2 in intestinal enterocytes may represent a defensive barrier that esterified xenobiotics need to breech before the organism is exposed following ingestion. Thus, xenobiotics that are metabolized by hCE2 in the small intestine may exhibit reduced bioavailability compared to compounds that are not metabolized by hCE2 (Imai, 2006). This may significantly affect the disposition of certain esterified xenobiotics following oral doses.
The physiological role(s) of CEs is an area of active research. Recent data strongly suggests a role for CEs in lipid metabolism utilizing their esterolytic activities (Dolinsky et al., 2004; Zhao et al., 2005). Many environmental toxicants, pharmaceuticals, and illicit drugs are esterified compounds (reviewed in Potter and Watkins, 2006). Examples include pyrethroid insecticides, phthalate esters, chemotherapeutic prodrugs (e.g., irinotecan), and narcotics such as cocaine and heroin. These compounds are cleared (or bioactivated in the case of irinotecan) from the body in part by the action of the CEs. Pyrethroid insecticides are esterified toxicants of particular importance because of their extensive use in agriculture (Hodgson and Levi, 1996) and public health practices (Takken, 2002). Pyrethroids will increasingly be used in agriculture as a result of the curtailed usage of organophosphate (OP) insecticides. As a result, human exposure to these compounds will likely increase in the future. The importance of esterases in the detoxification of pyrethroids has previously been demonstrated (Gaughan et al., 1980). For instance, when the OP compounds profenofos, sulprofos, O-ethyl O-(4-nitrophenyl) phenylphosphorothioate, or S,S,S-tributyl phosphorothioate were administered to mice in vivo before dosing with the pyrethroid trans-permethrin, all four OP compounds strongly inhibited liver microsomal esterase activity and in turn increased the in vivo toxicity of the pyrethroid. This provides direct evidence for the importance of hydrolytic metabolism in pyrethroid detoxification.
Species differences exist in the expression of CEs. These differences can impact the extrapolation of animal studies to humans. Rats and mice, the two most widely used animal models in pharmacokinetic studies, express high levels of CEs in their plasma. In contrast, human plasma has no detectable CE enzyme (Li et al., 2005). This difference may significantly affect the disposition of esterified xenobiotics and thus prevent the accurate prediction of the pharmacokinetic behavior of such compounds in humans. Another difference is the large number and complex distribution of CE isozymes present in the rodent liver, which contrasts to the less complex situation in human liver, where hCE1 and hCE2 are the two major isoforms expressed. The metabolism of pyrethroids by rodent and human hepatic microsomes, which possess high concentrations of CEs, has been investigated in depth (Soderlund and Casida, 1977; Choi et al., 2002; Anand et al., 2006; Godin et al., 2006; Ross et al., 2006). However, other tissues in humans and rats, such as adipose tissue, contain lipases that may contribute to pyrethroid metabolism but have not been adequately studied. In addition, pancreatic lipases (also called carboxyl ester lipases, EC 3.1.1.3), secreted into the lumen of the small intestine to facilitate the hydrolysis of triacylglycerols and cholesteryl esters (Hui and Howles, 2002), may also contribute to pyrethroid metabolism following oral exposures. The enzymatic activities of these digestive lipases are stimulated by bile salts, which are present in the intestinal lumen at high concentrations.
The first objective of this study was to investigate further the expression and hydrolytic activity of CEs present in the soluble hepatic fraction (i.e., cytosol) and in various other human and rat tissues that are likely important in the disposition and clearance of pyrethroid insecticides. We have focused on trans-permethrin and bioresmethrin because they are prototypical type 1 pyrethroids and widely used in agriculture. We have made comparison to some type 2 pyrethroids when appropriate; however, a more in depth study of deltamethrin and esfenvalerate will be forthcoming (Godin et al., personal communication). The tissues investigated include human intestinal microsomes, rat intestinal microsomes and cytosol, human and rat hepatic microsomes and cytosol, and human and rat serum. An additional objective of this study was to determine the substrate specificity and kinetic properties of purified rat serum carboxylesterase. The final objective was to investigate the hypothesis that lipases, which are expressed at high levels in adipose tissue, can hydrolyze pyrethroids since these compounds are hydrophobic and likely to partition into adipose reservoirs. We also examined a model pancreatic lipase, which is secreted into the lumen of the gastrointestinal tract, because it will likely encounter pyrethroid esters following oral exposures.
2. Materials and Methods
2.1 Chemicals
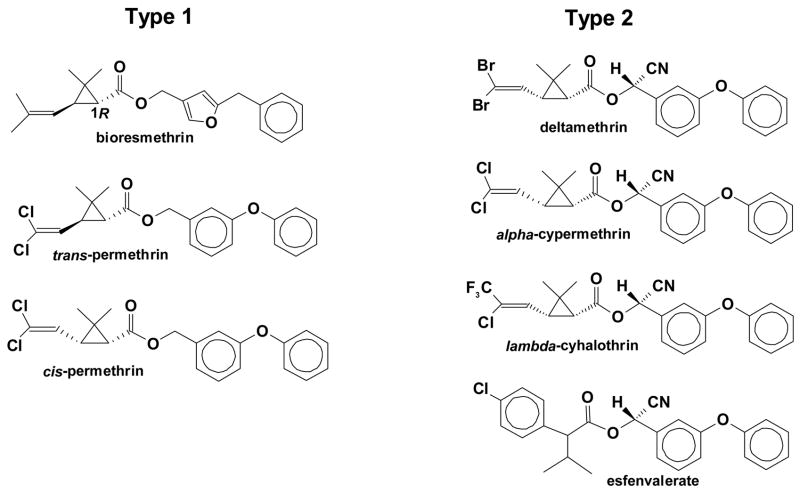
The pyrethroids shown in Fig. 1 were obtained from Chem Service (West Chester, PA). Type 1 pyrethroids included 1RS trans-permethrin (98% pure, 93% trans and 5% cis), 1RS cis-permethrin (purity 99%), and 1R trans-resmethrin (bioresmethrin, 99% pure, 97% trans and 2% cis); type 2 pyrethroids, which contain a cyano functionality on the α carbon, included alpha-cypermethrin (99% pure, mixture of isomers), lambda-cyhalothrin (99% pure, mixture of isomers), and deltamethrin (99% pure). 1RS/1R and cis/trans indicate the absolute stereochemistry at the C1 and C3 atoms of the cyclopropane ring, respectively; however, the designation 1RS is omitted for clarity in the text. The following authentic standards of pyrethroid metabolites were purchased from Sigma-Aldrich (St. Louis, MO): 3-phenoxybenzyl alcohol (3PBAlc), cis/trans-3-(2′,2′-dichlorovinyl)-2,2-dimethylcyclopropane carboxylic acid (a 1:1 mixture of cis and trans isomers) [also called cis/trans-dichlorochrysanthemic acid (Cl2CA)], 1R trans-chrysanthemic acid (CA), and 3-phenoxybenzaldehyde (3PBAld, purity). 3-(4-Methoxy)-phenoxybenzaldehyde, para-nitrophenyl acetate (pNPA), 4-methylumbelliferyl acetate (4-MUBA), 4-methylumbelliferyl stearate (4-MUBS), and 2-chloro-3,4-dimethoxybenzil (CDMB) were obtained and used without further purification from Sigma-Aldrich (St. Louis, MO). Para-nitrophenyl valerate (pNPV) was synthesized and kindly provided by Dr. Howard Chambers (Department of Entomology, Mississippi State University).
Fig. 1.
Structures of the type 1 pyrethroids and type 2 pyrethroids. The structural features of the pyrethroids that determine their rates of hydrolysis have been previously reviewed (Soderlund, 1992).
2.2. CE enzymes
The recombinant human CE proteins (hCE1 and hCE2) were expressed in baculovirus-infected Spodoptera frugiperda cells and purified (Morton and Potter, 2000). Porcine pancreatic lipase and Pseudomonas lipase were purchased from Sigma-Aldrich (St. Louis, MO). Rat serum CE was purified to homogeneity using a modification of the method of Hashinotsume et al. (1978) and Sanghani et al. (2002). Specifically, ten ml of pooled rat serum (from 5 adult male Sprague-Dawley) rats was subjected to ammonium sulfate fractionation. The protein pellet from the 50–70% ammonium sulfate fraction was dissolved in 10 mM Tris-HCl (pH 7.4) (buffer A), applied to a DEAE-cellulose column equilibrated in buffer A and eluted with a linear gradient of 0 to 1 M NaCl in buffer A. Fractions were collected and analyzed for CE activity by subjecting them to native polyacrylamide gel electrophoresis (PAGE) using the Ornstein-Davis buffer system as previously described (Dean et al., 1995). The gels were stained for esterase activity by incubating them in 100 mM potassium phosphate buffer (pH 6.5) containing 100-μM 4-MUBA. All subsequent fractions during purification were analyzed for esterase activity in this manner. Fractions containing the slowest migrating esterase activity were pooled, dialyzed into buffer A, applied to a column of QAE-cellulose equilibrated in buffer A and eluted with a linear gradient of NaCl from 0 to 1 M in buffer A. Fractions containing the esterase activity were pooled, dialyzed into buffer A containing 1 mM MgCl2, 1 mM MnCl2, and 1 mM CaCl2 and applied to a column of concanavalin A equilibrated in the same buffer. Glycosylated proteins were eluted from the column with buffer A containing 0.2 M NaCl and 0.5 M α-D-methyl-mannopyranoside. Fractions containing esterase activity were pooled, dialyzed into buffer A, and concentrated. The resulting solution was subjected to preparative nondenaturing PAGE using a 4% stacking gel and a 10% resolving gel. The resulting fractions containing the slowest migrating esterase activity were pooled, dialyzed into buffer A, and concentrated. The resulting solution was subjected to native gel electrophoresis. Silver staining demonstrated a single discrete band, which co-migrated with the esterolytic activity toward 4-methylumbelliferyl acetate. Electrophoresis of the isolated rat serum CE protein on SDS-PAGE gels and subsequent silver staining also demonstrate a discrete single band of approximately 70 kDa. Antiserum, previously shown to recognize only rat serum CE in whole serum, showed the identity of the protein as rat serum CE. Moreover, MADLI-TOF analysis of a trypsin-digested sample of the purified protein confirmed its identity as rat serum CE. The purified rat serum CE was used in the subsequent enzymatic assays.
2.3. Antibodies
Polyclonal antibodies, which were raised against human CE1, rat hydrolase A, and rat hydrolase B, were kindly provided by Dr. M. Hosokawa (Chiba University, Japan). The antihydrolase A antibody cross reacts with rat serum CE.
2.4. Tissue subcellular fractions and serum
Pooled human small intestinal microsomes (cat no. 452210, from 5 individuals) and pooled human liver microsomes (cat no. 452161, from 18 individuals) were purchased from BD Biosciences (Woburn, MA). Individual human intestinal microsomes and cytosol is currently not commercially available. Eleven individual human liver cytosolic samples and a pooled human liver cytosolic sample (cat no. 452861, from 20 individuals) were also obtained from BD Biosciences. Blood was collected from five adult male Sprague-Dawley rats (70–110-day old). The blood was allowed to stand for one hour to clot and subsequently centrifuged at 2,000 × g for 20 min to enable serum collection. Human serum obtained from a pool of adult male donors was purchased from Sigma-Aldrich (St. Louis, MO). Rat liver and small intestinal microsomes and cytosols were prepared as described previously (Ross et al., 2006; Ross and Pegram, 2004). Animals used as tissue donors were housed in an AALAC-approved facility prior to tissue and blood collection. All animal procedures were performed in accordance with the Mississippi State University guidelines for the ethical treatment of animals.
2.5. In-gel hydrolysis assays
Samples of human intestinal microsomes and human liver cytosols and microsomes were examined by native PAGE using 10% polyacrylamide mini-gels followed by an in-gel hydrolysis assay with the substrate 4-MUBA as described above (Dean et al., 1995).
2.6. Pyrethroid hydrolysis reactions
2.6.1. Human and rat tissue microsomes and cytosols
Hydrolytic reactions catalyzed by the various tissue fractions were performed in a total volume of 250 μL (Ross et al., 2006). Variable amounts of pyrethroid substrate (5–100 μM) were pre-incubated in 50 mM Tris-HCl buffer (pH 7.4) for 5 min (37°C). Reactions were started by the addition of the subcellular fraction (final protein concentration, 0.5 mg/ml). After 15 or 30 min incubations, the reactions were quenched by the addition of an equal volume of ice-cold acetonitrile containing 10 μM of internal standard [3-(4-methoxy)-phenoxybenzaldehyde]. After centrifugation (5 min, 16,100 × g), the hydrolysis products were quantified by HPLC (see below). Previous reports from our lab (Ross et al., 2006; Godin et al., 2006) have examined the time-course of pyrethroid metabolism by tissue subcellular fractions and pure CE enzymes. The incubation times that we have used in the current manuscript (15 min and 30 min) are within the linear range of the enzymatic reaction rate for the pyrethroids. Moreover, the substrate concentration of pyrethroid (50 μM) used in several enzymatic assays is high enough to saturate the CE enzyme but is below the solubility limit of pyrethroids in aqueous buffer. The non-enzymatic rates of the pyrethroid hydrolysis reactions are negligible. There is no detectable hydrolysis of these compounds at 37°C in buffer solutions during the 30 min incubation period.
2.6.2. Human and rat serum
Pyrethroid hydrolysis reactions with human or rat serum were conducted as follows. The pyrethroids (see Figure 2) were pre-incubated in 200–225 μL of 50 mM Tris-HCl buffer (pH 7.4) for 5 min before adding 25 μL of pooled rat serum or 50 μL of pooled human serum to each sample. The final concentration of pyrethroid was 50 μM in each sample. The samples were incubated at 37°C for 30 min before quenching with an equal volume of cold acetonitrile. Following centrifugation, hydrolysis products were analyzed by HPLC. When variable pyrethroid concentrations were utilized in serum incubations, the range of concentrations was from 5–100 μM.
Fig. 2.
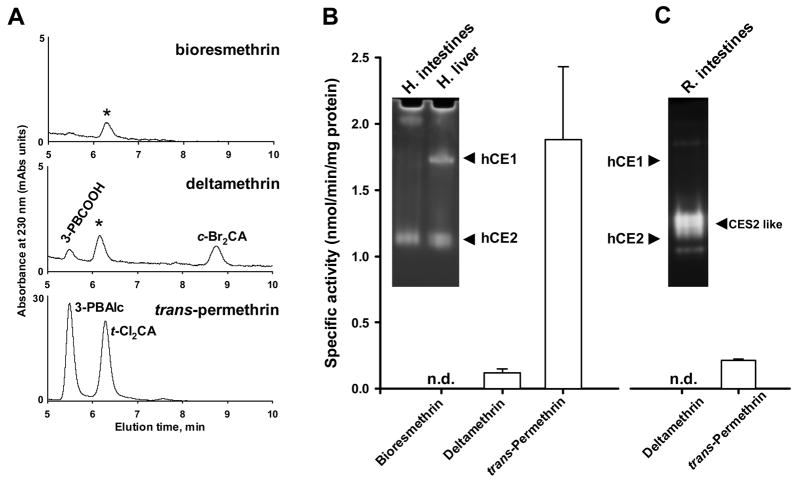
Hydrolysis of pyrethroids by a pool (n=1) of human intestinal microsomes from 5 individuals. A, Representative HPLC chromatograms of hydrolysis products of bioresmethrin (top), deltamethrin (middle), and trans-permethrin (bottom). Peaks that are labeled with the asterisk are derived from the human intestinal microsomes and are not from the pyrethroids. Abbreviations: 3-PBCOOH, 3-phenoxybenzoic acid; c-Br2CA, cis-dibromochrysanthemic acid; 3-PBAlc, 3-phenoxybenzyl alcohol; t-Cl2CA, trans-dichlorochrysanthemic acid. B, Specific hydrolysis rates for each pyrethroid catalyzed by human intestinal microsomes. n.d., hydrolysis products not detectable. Values are mean ± S.D. (n = 3–4 replicates of the single pool). Inset, native PAGE gel stained with 4-MUBA following separation of pooled human intestinal and liver microsomes. The migration locations of hCE1 and hCE2 are indicated. C, Specific hydrolysis rates for deltamethrin and trans-permethrin catalyzed by a pool (n=1) of rat intestinal microsomes from 5 individual animals. Inset, native PAGE gel stained with 4-MUBA following separation of rat intestinal cytosol proteins. The locations of hCE1 and hCE2 proteins are indicated. Cytosol exhibited the same distribution of CEs as found in intestinal microsomes, but esterases appeared as more discrete bands possibly because cytosolic fractions contain much less lipid than microsomal fractions.
2.6.3. Pure rat serum carboxylesterase or lipases
Hydrolysis of pyrethroids by purified rat serum CE was performed in 100-μL reaction volumes. Varying amounts of pyrethroid (5–100 μM) were pre-incubated for 5 min in 50 mM Tris-HCl buffer (pH 7.4) at 37°C. The hydrolytic reactions were initiated by addition of the pure CE (2.5 μg protein per reaction) and allowed to proceed for 30 min at 37°C. The reactions were quenched by the addition of an equal volume of ice-cold acetonitrile containing the internal standard. The samples were centrifuged and an aliquot was analyzed by HPLC to quantify the hydrolysis products. Non-enzymatic controls were also performed and found to have negligible rates. CE reactions at each substrate concentration were performed in duplicate.
Lipase-catalyzed pyrethroid hydrolysis reactions were also performed in the same manner as the pure rat serum CE, except in certain cases deoxycholic or cholic acid was added to the incubation medium at a final concentration of 3 mM. The two lipases examined were porcine pancrease lipase and Pseudomonas lipase both purchased from Sigma-Aldrich (St. Louis, MO).
2.6.4. HPLC analysis
HPLC-UV analysis of pyrethroid hydrolytic products was performed on a Surveyor LC system (Thermo Electron, San Jose, CA) using a reversed-phase HPLC column (2.1 mm × 100 mm, C18, Thermo Electron, San Jose, CA) as previously described (Ross et al., 2006).
2.7. Spectrophotometric assay for esterase activity
The esterase activities of pure CEs and mammalian liver subcellular fractions were routinely assayed by measuring the production of p-nitrophenol liberated from pNPV at 405 nm on a spectrophotometer (Wheelock et al., 2003). An extinction coefficient of 13 cm−1mM−1 (Morgan et al., 1994) was used to convert the slopes of each activity curve to specific enzyme activities. All enzymatic reaction rates were corrected for non-enzymatic hydrolysis rates.
2.8. Immunoblotting of human liver microsomes, liver cytosols, and intestinal microsomes
Human liver microsomes, liver cytosols, and intestinal microsomes were subjected to SDS-PAGE using standard protocols. After electrophoresis, the proteins were transferred to polyvinyldifluoride membranes and probed with rabbit anti-human CE1 polyclonal antibody diluted 1:4000 (v/v) in Tris-buffered saline/5% milk. Immuno-complexes were localized with a horse radish peroxidase-conjugated goat anti-rabbit secondary antibody and the SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL). The chemiluminescent signal was captured using a digital camera (Alpha Innotech gel documentation system). Bands on the digital images were quantified using NIH Image J software (v.1.33u).
2.9. Human liver cytosolic esterases and sensitivity to OP inhibition
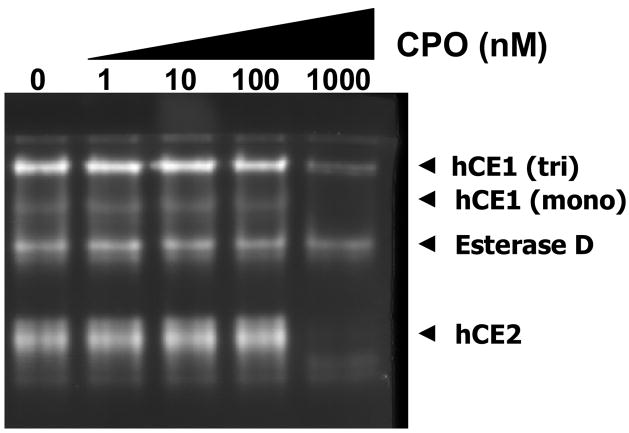
A representative individual human liver cytosolic sample (HG42) was diluted to 0.5 mg/ml protein in 50 mM Tris-HCl buffer (pH 7.4) and treated with chlorpyrifos oxon (CPO) at increasing concentrations (0–1000 nM) for 10 min at room temperature. Following CPO-treatment, the cytosolic proteins were mixed with native gel loading buffer (0.05% bromophenol blue, 40% glycerol) and immediately loaded on a native PAGE gel. Following electrophoresis, the gel was stained with 4-MUBA. Similar CPO treatments and native PAGE analysis were done on rat serum proteins.
2.10. Kinetic analysis and statistics
Non-linear regression of substrate concentration versus reaction velocity curves were analyzed using SigmaPlot v. 8.02 software by fitting experimental data to the Michaelis-Menten equation. Each substrate concentration in the kinetic experiments was evaluated in duplicate. The specific activity data obtained for intestinal microsomes, hepatic microsomes and cytosols, and serum using standard ester substrates and pyrethroids are reported as the mean (± S.D.) of three replicates.
2.11. Determination of protein concentration
Protein concentrations were determined using the BCA reagent according to the manufacturer’s instructions (Pierce, Rockford, IL).
3. Results
3.1. Pyrethroid hydrolysis catalyzed by human and rat intestinal microsomes
A pool of human intestinal microsomes (n=1) was found to effectively hydrolyze trans-permethrin (specific activity of 1.88 ± 0.55 nmol/min/mg protein); however, bioresmethrin and deltamethrin were not metabolized to any appreciable extent (see Fig. 2A and B). trans-Permethrin was hydrolyzed nearly 20-fold faster than deltamethrin. These results are consistent with the substrate specificity of the hCE2 enzyme, which is the major CE isoform found in human small intestine. Pure hCE2 was previously shown to hydrolyze trans-permethrin efficiently, but it does not cleave bioresmethrin or deltamethrin to any appreciable extent (Ross et al., 2006; Godin et al., 2006). Furthermore, addition of the specific hCE2 inhibitor 2-chloro-3,4-dimethoxybenzil (Wadkins et al., 2005) significantly reduced the trans-permethrin-hydrolytic activity of human intestinal microsomes further demonstrating that hCE2 is the primary esterase in human intestine responsible for pyrethroid hydrolysis (data not shown). This result was also supported by in-gel hydrolysis data that demonstrated negligible hCE1 protein staining by 4-MUBA following electrophoresis of intestinal microsomal proteins on native PAGE gels (see Fig. 2B, inset). In contrast, staining of the native gel demonstrated a substantial amount of hCE2 protein in the intestinal microsomes.
For pools of either rat intestinal microsomes (5 rats, 1 pool) or rat intestinal cytosol (5 rats, 1 pool) the rates of trans-permethrin hydrolysis were comparable, with microsomes and cytosol exhibiting specific activities of 0.42 ± (0.07) and 0.29 ± (0.02) nmol/min/mg protein, respectively. However, the pool of human intestinal microsomes (5 individuals) was ~4–5-fold more active than was the pool of rat intestinal microsomes (Fig. 2B and 2C). Hydrolysis of deltamethrin was not detected following incubation with either rat intestinal microsomes or cytosol (Fig. 2C). Rat intestinal microsomes and cytosol appear to possess at least one CES2-like esterase based on similar migration properties to hCE2 in native PAGE gels (Fig. 2C, inset).
3.2. Hydrolysis of pyrethroids by human and rat liver subcellular fractions
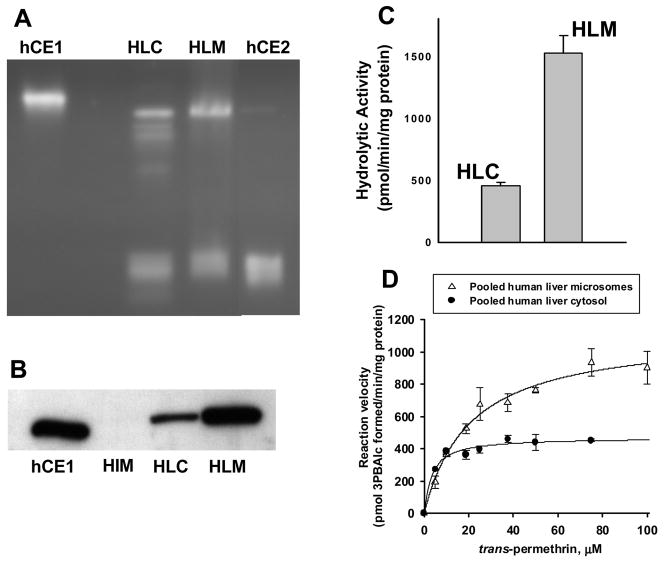
When human hepatic microsomes and cytosol were examined by native PAGE, hCE1 and hCE2 were clearly present in both fractions (see Fig. 3A). It appears that native hCE1 protein in each tissue fraction migrates slightly faster than the pure recombinant protein, perhaps reflecting differences in glycosylation state. When both subcellular fractions were examined by western blotting, hCE1 protein was expressed at significantly higher levels in the pooled human liver microsomes than in pooled cytosol (see Fig. 3B). (The immunoblot in Fig. 3B also demonstrated that hCE1 was not expressed in the intestinal microsomes, which is consistent with the data in Fig. 2). The greater hCE1 expression in hepatic microsomes compared to cytosol was reflected in the ability of each subcellular fraction to hydrolyze trans-permethrin; e.g., the specific activity for the pooled human liver microsomes was 3.4-fold higher than the pooled human liver cytosol (Fig. 3C) and Vmax was >2-fold higher for the microsomes compared to cytosol (Fig. 3D).
Fig. 3.
Distribution of CE enzymes in a pool of human liver microsomes or cytosol. A, Native PAGE gel stained with 4-MUBA. hCE1, pure recombinant human carboxylesterase 1 protein; HLC, pooled human liver cytosol; HLM, pooled human liver microsomes; hCE2, human carboxylesterase 2. Equal amounts of cytosolic and microsomal protein (30 μg) and approximately 0.5–1 μg each of the recombinant CE proteins were loaded on the gel. B, Immunoblot of pooled human liver cytosol (HLC), microsomes (HLM), and intestinal microsomes (HIM). Equal amounts of protein were loaded (0.5 μg per lane) for each hepatic fraction. The membrane was probed using an anti-hCE1 antibody. C, Comparison of hydrolytic activities of a pool (n=1) of human liver microsomes (HLM) and a pool (n=1) of cytosols (HLC) using trans-permethrin as the substrate (at 50 μM concentration). D, Substrate-velocity plots of trans-permethrin hydrolysis catalyzed by pooled human liver microsomes and cytosols. The kinetic parameters that were derived are as follows: microsomes, Vmax = 1120 (± 50) pmol/min/mg, Km = 20.7 (± 3.0) μM; cytosol, Vmax = 469 (± 18) pmol/min/mg, Km = 3.4 (± 0.8) μM. Each symbol and error bar is the mean and S.D. of two determinations at each substrate concentration.
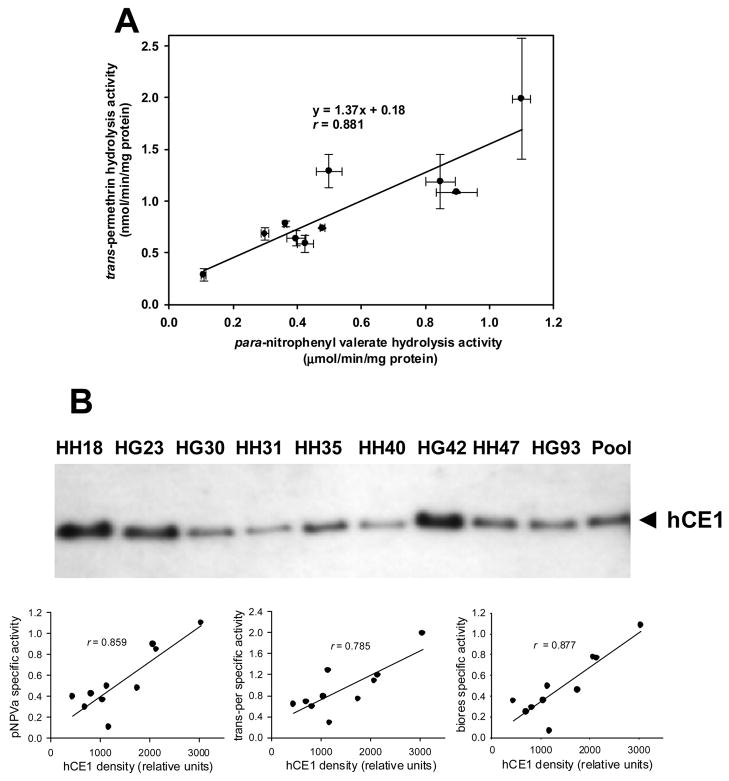
Nonetheless, a substantial fraction of hCE1 protein in liver appears to be present in the cytosolic fraction. Thus to address the role of soluble esterases in the hydrolysis of esterified compounds, the activities of ten individual human liver cytosolic samples were determined using pNPV and trans-permethrin as ester substrates. Substantial variation in hydrolysis rates was observed for both substrates with interindividual rates varying as much as 10-fold (Fig. 4A). It was also found that hydrolysis rates for the individual cytosols toward pNPV and trans-permethrin correlated reasonably well (r = 0.881), suggesting that both substrates were hydrolyzed by the same CE isoforms(s).
Fig. 4.
Hydrolysis of ester substrates catalyzed by individual human liver cytosols. A, Correlation of rates of para-nitrophenyl valerate hydrolysis and trans-permethrin hydrolysis catalyzed by individual human liver cytosols. Symbols are mean values and error bars are S.D. (n = 3 replicates). B, Immunoblot of individual human liver cytosols. The membranes were probed using an anti-hCE1 antibody. The density of the hCE1 immunoreactive band in each cytosolic sample was plotted against the pNPV, trans-permethrin, and bioresmethrin hydrolysis activities.
We next examined the level of hCE1 protein in each cytosolic sample by western blotting. In contrast to the non-variable levels of hCE1 protein observed in eleven individual human liver microsomes (Ross et al., 2006), variable amounts of hCE1 protein expression were seen in the cytosolic samples (Fig. 4B). Moreover, the specific activities of the cytosolic samples for pNPV, trans-permethrin and bioresmethrin appeared to correlate with the level of immunoreactive hCE1 protein present in each sample (r = 0.859, 0.785, and 0.877 for pNPV, trans-permethrin and bioresmethrin, respectively), which suggested that hCE1 was the primary esterase responsible for hydrolyzing these esters.
Electrophoretic separation of a representative human cytosolic sample on native PAGE gels and subsequent 4-MUBA staining allowed the distribution of esterases in cytosol to be determined (see Fig. 5). Both hCE1 and hCE2 were detected in the native gels. Furthermore, an esterase that migrates midway between the hCE1 and hCE2 bands was also observed. The identity of this esterase is likely Esterase D, which is also known as S-formyl glutathione hydrolase (Gonzalez et al., 2006). A minor band designated as monomeric hCE1 was also observed to migrate just below the trimeric form of hCE1 (Fig. 5). A similar monomeric band for hCE1 in native PAGE gels was observed by Imai (2006). Moreover, hCE1 (both monomeric and trimeric forms) and hCE2 were found to be sensitive to the inhibitory effects of 1-μM chlorpyrifos oxon (CPO); in contrast, Esterase D was not inhibited by CPO (see Fig. 5). These results are consistent with the known OP sensitivity of hCE1 and hCE2, and the lack of sensitivity of Esterase D to OP-mediated inhibition.
Fig. 5.
Distribution and organophosphate-mediated inhibition of esterases in human liver cytosol. A representative human liver cytosol sample (individual HG42) was treated with increasing concentrations (0–1000 nM) of chlorpyrifos oxon (CPO) for 10 min before electrophoresis on a native PAGE gel and subsequent staining with 4-MUBA. The identities of hCE1 (mono) and Esterase D were aided by staining native gels with a mixture of α- and β-naphthylacetate since Esterase D is not stained by α- or β-naphthylacetate, but the monomeric and trimeric forms of hCE1 are (data not shown).
A comparison of hydrolytic kinetic parameters for rat and human hepatic microsomes (Ross et al., 2006) and cytosol (this study) toward trans-permethrin is shown in Table 1. The data demonstrate that the microsomal Vmax and Km values are very similar for each species. For cytosol, the human and rat Vmax values were also comparable, but the human Km value was much lower than the rat Km. The rat appears to be a reasonable model of human hepatic pyrethroid metabolism, at least for trans-permethrin, although the apparent Km in human liver cytosol is markedly lower than that for rat liver cytosol.
Table 1.
Kinetic parameters of trans-permethrin hydrolysis catalyzed by human and rat hepatic subcellular fractions (microsomes and cytosols)a
|
trans-permethrin |
|||
|---|---|---|---|
| Species (fraction) | Km (μM) | Vmax (nmol/min/mg) | CLintd (nmol/min/mg/mM) |
| rat (microsomes) | 19.2 ± 3.2 | 1.3 ± 0.3 | 67.7 |
| rat (cytosol) | 24.6 | 0.56 | 22.8 |
| human (microsomesb) | 20.7 ± 3.0 | 1.1 ± 0.1 | 53.1 |
| human (cytosolc) | 3.4 ± 0.8 | 0.47 ± 0.02 | 138.2 |
Rat microsomes and cytosol (pooled) were prepared from another study (Ross et al., 2006). Values (± S.E.) were obtained by non-linear regression of kinetic plots. The microsomal kinetic data are from Ross et al. (2006). The cytosol kinetic data are from this study.
Pooled sample (n = 18 individuals)
Pooled sample (n = 20 individuals)
CLint = Vmax/Km
3.3. Serum carboxylesterases: Whole serum and purified enzyme
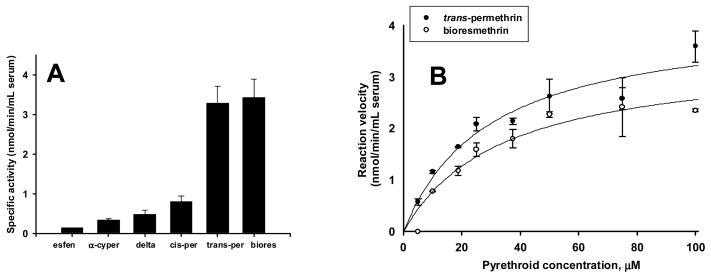
We also investigated the kinetics of pyrethroid metabolism by rat serum in order to estimate the total amount of hydrolytic metabolism that occurs in blood relative to that of the liver. Of the six pyrethroid compounds we studied, bioresmethrin and trans-permethrin were hydrolyzed the fastest and esfenvalerate the slowest in rat serum (Fig. 6A). When concentration-velocity studies were performed using rat serum, the type 1 pyrethroids bioresmethrin and trans-permethrin were hydrolyzed at comparable rates with apparent Michaelis-Menten kinetics (Fig. 6B). In contrast, the type 2 pyrethroid deltamethrin was hydrolyzed more slowly and did not exhibit hyperbolic enzyme kinetics (Godin et al., personal communication). For trans-permethrin (utilizing the specific activities measured at 50 μM) we estimated that serum possesses 4% of the total hydrolytic capacity in the rat. Liver weight and serum volume (per kg body weight) were estimated using allometric data (Lindstedt and Schaeffer, 2002). In contrast, human serum was unable to hydrolyze either deltamethrin or trans-permethrin (data not shown). Not surprisingly purified human butyrylcholinesterase does not hydrolyze trans-permethrin (Ross et al., 2006) or deltamethrin (Ross MK, unpublished data).
Fig. 6.
Hydrolysis of pyrethroids by whole rat serum. A, Specific activity of a pool (n=1) of rat serum following hydrolysis of six pyrethroids (esfen, esfenvalerate; α-cyper, alpha-cypermethrin; delta, deltamethrin; cis-per, cis-permethrin; trans-per, trans-permethrin; biores, bioresmethrin). Hydrolysis rates were determined at a single concentration of pyrethroid substrate (50 μM). Whole serum activity data was normalized to the volume (mL) of serum. Bars represent means and error bars represent S.D. (n = 3 replicates of the pooled serum sample). B, Substrate concentration-velocity kinetic plots for trans-permethrin and bioresmethrin in whole serum. The kinetic parameters for trans-permethrin and bioresmethrin, respectively, are: Vmax (nmol/min/mL serum): 4.2 (± 0.5) and 3.4 (± 0.5); Km (μM): 29.4 (± 8.3) and 33.9 (± 11.1).
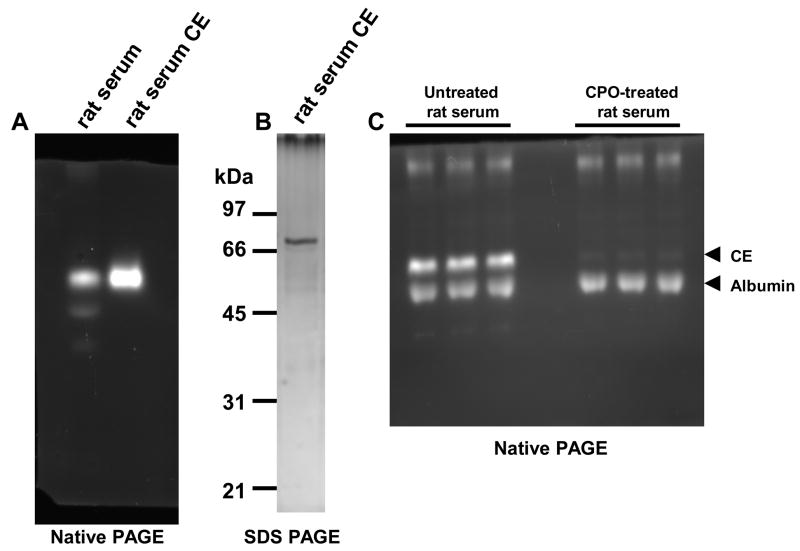
We next purified the OP-sensitive rat serum CE to homogeneity to further investigate the substrate specificity and kinetics of pyrethroid hydrolysis (Fig. 7A–C). When the hydrolysis rates of the six pyrethroids by rat serum CE were examined at a single concentration of substrate (50 μM), a similar trend was observed as seen with whole serum (compare Fig. 7D with Fig. 6A). Bioresmethrin and trans-permethrin were again hydrolyzed the fastest by the serum CE, as seen in whole serum, while the type 2 pyrethroids were hydrolyzed much less efficiently. Upon further kinetic study of trans-permethrin and bioresmethrin hydrolysis by rat serum CE the following parameters were obtained, respectively: kcat = 1.30 and 1.65 min−1, Km = 23.6 and 16.2 μM, and kcat/Km = 55.1 and 102 min−1mM−1 (see Fig. 7E). Thus, rat serum CE is nearly twofold more efficient at hydrolyzing bioresmethrin than trans-permethrin.
Fig. 7.
Purification of rat serum CE and pyrethroid hydrolysis. A, Native PAGE gel of whole rat serum and purified rat serum CE following staining with 4-MUBA. B, SDS-PAGE gel of purified rat serum CE protein stained with silver. The estimated MW of the pure protein is ~70 kDa. C, Native PAGE gel of untreated rat serum or CPO-treated (5 μM) rat serum after staining with 4-MUBA. The identity of albumin in the serum was aided by the migration behavior of pure albumin in native PAGE gels (data not shown), since it is known that albumin is stained by bromophenol blue present in gel loading buffers (Li et al., 2005), and because the 4-MUBA hydrolyzing activity of albumin is not inhibited by treatment with CPO. D, Specific activity of pyrethroid hydrolysis by pure rat serum CE. Hydrolysis rates were determined at a single concentration of pyrethroid substrate (50 μM). E, Substrate concentration-velocity plot for trans-permethrin and bioresmethrin hydrolysis catalyzed by pure serum CE.
3.4. Lipases and pyrethroid hydrolysis
In an effort to characterize other hydrolytic enzymes that may metabolize pyrethroid compounds, we have also examined lipases. Specifically, we investigated the activity of a porcine pancreatic lipase with two different pyrethroids (bioresmethrin and trans-permethrin), both in the absence and presence of the bile acids deoxycholic or cholic acid. However, we did not detect any pyrethroid hydrolytic activity with this lipase when either substrate was used, which suggested that it does not metabolize pyrethroids (data not shown). We did find, however, that the pancreatic lipase could hydrolyze the water-soluble substrate 4-MUBA and the water-insoluble 4-MUBS (data not shown), demonstrating that the enzyme was active. Furthermore, we also used a commercially available Pseudomonas lipase to attempt to cleave trans-permethrin but again did not detect any hydrolytic activity (data not shown).
4. Discussion
Biotransformation of pyrethroid insecticides by CEs accounts for a significant fraction of the clearance of these compounds from the body (Godin et al., 2006; Nishi et al., 2006; Ross et al., 2006). Further studies that examine the functional activities of intestinal, liver and serum CEs are important to understand better the systemic availability and disposition of pyrethroid insecticides in exposed mammals. Moreover, physiologically based pharmacokinetic (PBPK) models of pyrethroids may require inclusion of esterase activities that are present in the intestinal tract, liver and serum if it is shown that these compounds are appreciably hydrolyzed within these tissues. Such data will help to improve the accuracy of PBPK models and thus enable better predictions of tissue and blood concentrations following exposure to these insecticides.
To examine the distribution of CE isozymes in rat and human tissue preparations, we used native PAGE and in-gel hydrolysis of 4-MUBA, which stains esterases in tissue fractions. We used quantitative immunoblotting to determine hCE1 protein abundance in human hepatic cytosols. Furthermore, we used in vitro biochemical assays with standard esterified substrates and pyrethroid insecticides to study the activities of esterases in subcellular fractions and whole sera. We also measured the pyrethroid hydrolytic activity of three purified enzymes. These studies extend our previous work, which examined the roles of human hepatic CEs in pyrethroid metabolism (Ross et al., 2006; Godin et al., 2006), and further characterize the role of CEs in pyrethroid clearance. Our results show that hCE2, which is abundant in human intestine, has a significant role in the metabolism and detoxication of trans-permethrin, but has a more limited role in the metabolism of bioresmethrin and deltamethrin. The presence of hCE2 in the gastrointestinal tract may protect humans following oral exposures to permethrin, which is one of the most extensively used pyrethroid insecticides in agricultural practices (Wheelock et al., 2003). However, since deltamethrin and bioresmethrin are not effectively hydrolyzed by the intestinal microsomes, the efficiency of pyrethroid metabolism in the intestines will obviously depend on the structure of the pyrethroid encountered in the gut. Compared to human intestinal hydrolysis rates for trans-permethrin, the rat intestinal microsomes exhibited 4.5-fold lower activity. For deltamethrin, little hydrolysis was detected using human intestinal microsomes, and no hydrolysis was detected using rat intestinal microsomes. Therefore, serum concentrations of trans-permethrin in humans following oral doses will likely be lower than in rats, while we would predict that serum concentrations of deltamethrin will be comparable for each species. Differences between rats and humans exist in the CE isozymes that are expressed in intestinal tissue. Rat small intestine expresses a CES1 class isozyme (ES3) and two CES2 class isozymes (AY034877 and AB010632) based on Northern blots (Sanghani et al., 2002; Imai, 2006), while only hCE2 is expressed in the human small intestine. Our native gel data (Fig. 2C) also suggests that multiple CES2-like isozymes are found in rat intestine. The substrate specificity of the individual rat intestinal CE isozymes toward pyrethroids is unknown. However, rat ES3 exhibits a high degree of sequence homology with Hydrolase B, which does not hydrolyze trans-permethrin, bioresmethrin or deltamethrin very effectively (Ross et al., 2006; Godin et al., 2006). Therefore, ES3 is unlikely to contribute significantly to pyrethroid hydrolysis. While it is likely that intestinal metabolism of esterified compounds by CEs contributes to their clearance from the body, we estimate that intestinal hydrolysis accounts for just 2.5% of the total hydrolytic metabolism of trans-permethrin that occurs in the rat.
The highest content of CE protein in the body is found in the microsomal fraction of the liver. It is generally assumed that this subcellular fraction is responsible for most of the esterolytic metabolism in the whole animal. However, a relatively high proportion of CE protein and activity is also found in the hepatic cytosolic fraction (see Fig. 3). Indeed, we estimate that ~40% of the esteratic metabolism of trans-permethrin in both human and rat liver is due to cytosolic esterases. Therefore, the hepatic cytosol should be considered in metabolism and kinetic studies of esters such as pyrethroids. Consistent with our findings, Hodgson and co-workers previously found that human liver cytosol possessed significant hydrolytic activity toward trans-permethrin when studied by the parent compound depletion approach (Choi et al, 2002; Choi et al., 2004). Thus our present findings, which utilized a product appearance kinetic approach to study permethrin hydrolysis, confirm this previous result. Rat liver cytosol also has a similar amount of esterase activity toward trans-permethrin as human liver cytosol (Table 1), which is likely attributable to the presence of cytosolic CEs (Natarajan et al., 1996). Therefore, toxicokinetic studies that examine esterified compounds should use whole tissue homogenates (or ‘S9’ fraction) when assessing overall esterase activity.
Another finding in our study was the variable levels of hCE1 protein in ten individual human liver cytosols (coefficient of variation, CV = 56%; Fig. 4). This is in marked contrast to the lack of variable hCE1 expression previously observed in eleven individual human microsomal samples (CV = 9%; Ross et al., 2006). The reason for the variable hCE1 expression in cytosols within this small human population is unknown. The presence of hCE1 in cytosol is probably a result of the partial solublization of proteins during liver hepatic microsomal preparation and thus represents an artifact of the separation method. However, two possible physiological scenarios could account for the presence of hCE1 in both cytosol and microsomes. Newly synthesized hCE1 protein possesses an N-terminal signal peptide that directs it into the endoplasmic reticulum lumen (Satoh and Hosokawa, 1998), whereupon the 18-amino acid long signal peptide is clipped off. Perhaps the cytosolic pool of CEs is due in part to the presence of immature hCE1 protein that has not yet been trafficked into the ER and processed. Alternatively, a fraction of the hCE1 in the hepatocyte may reside in the cytoplasm directed there by some yet to be elucidated mechanism. The C-terminal sequence KDEL is responsible for the retention of CEs in the lumen of the endoplasmic reticulum through its interaction with the KDEL receptor (Robbi and Beaufay, 1991). If hCE1 present in the cytosol lacks both the signal peptide and the C-terminal KDEL tetrapeptide, it would suggest the existence of a previously undescribed pool of hCE1 targeted to the cytoplasm. Future in situ localization studies could confirm the presence of hCE1 in the cytoplasm of hepatocytes. The lack of an adequate number of commercially available individually-matched cytosolic and microsomal samples (i.e., derived from the same liver) precludes a definitive explanation here, although future studies will examine this issue.
Rat serum has esterase activity in the form of a serum CE, which actively hydrolyzes pyrethroids. While bioresmethrin and trans-permethrin were hydrolyzed in whole rat serum equally well, deltamethrin was hydrolyzed more slowly. To study pure serum CE-mediated hydrolysis of pyrethroids in more detail, we examined the kinetics of trans-permethrin hydrolysis (Fig. 7E). The estimated Km value for rat serum CE-catalyzed hydrolysis of trans-permethrin was similar to that obtained when trans-permethrin was hydrolyzed by whole rat serum (Fig. 6B). This suggested that a single enzyme in rat serum was catalyzing the hydrolysis of trans-permethrin, i.e., rat serum CE. The kcat value for trans-permethrin hydrolysis by the serum CE is 1.8-fold lower than that seen with Hydrolase A (Ross et al., 2006), which is the most abundant CE isozyme expressed in rat liver (Morgan et al., 1994). It is approximately the same, however, as the second most abundant rat hepatic isozyme, which is termed Hydrolase B (Morgan et al, 1994). However, the Km value for the serum CE was ~2–4-fold higher than the Km for Hydrolases A and B, suggesting a lower affinity of trans-permethrin for the serum CE compared to the liver CEs. Thus, based on the kinetic parameters for the pure liver CEs and the greater abundance of Hydrolases A and B than serum CE, the hepatic CEs have a greater capacity to clear trans-permethrin than the serum CE. These results highlight a significant species difference between rats and humans with respect to pyrethroid metabolism in the blood because no detectable metabolism (hydrolysis) of pyrethroids occurs in human blood. The metabolism of pyrethroids in blood is likely important in rodents, but not in humans because of the lack of CEs in human blood.
Another goal of this study was to determine whether lipases, which are abundantly found in adipose tissue and the gastrointestinal tract (two locations where pyrethroid compounds are likely found), contribute to the hydrolytic clearance of the pyrethroids. Small, water-soluble esters are preferentially hydrolyzed by carboxylesterases, while large hydrophobic substrates are hydrolyzed by lipases. However, there is overlap in the substrate specificities of these two classes of hydrolytic enzymes (Gilham and Lehner, 2005). Our limited results, using both a porcine pancreatic lipase and a bacterial lipase (from Pseudomonas), suggest that lipases will not significantly contribute to the metabolism of pyrethroids in vivo. Moreover, the carboxylesterases are likely the most important esterases that need to be considered in the formulation of models that examine the disposition and pharmacokinetics of pyrethroid insecticides.
In conclusion, human and rat intestinal microsomes effectively hydrolyze trans-permethrin, but not deltamethrin or bioresmethrin, which corresponds to the known substrate specificity of the major human intestinal esterase hCE2 (Ross et al., 2006) and possibly the CES2-like esterase(s) in rat intestine. Similar results were found for rat intestinal cytosol. Human liver cytosols were shown to have lower levels of hCE1 protein and hydrolytic activity when compared to the hepatic microsomal fraction. However, the level of hCE1 in human liver cytosol is not insignificant. Moreover, significant interindividual variability in the abundance of hCE1 protein was observed in ten individual hepatic cytosols. Thus, the contribution of the hepatic cytosolic fraction to hydrolytic metabolism of pyrethroids and the variable amounts of hCE1 expressed in this subcellular fraction warrants consideration in toxicokinetic studies. Lastly, the pyrethroid substrate specificity of purified rat serum CE was shown to match very closely that of whole rat serum, which suggests this enzyme is the major esterase in rat serum that metabolizes pyrethroids. While the hepatic CEs undoubtedly contribute the bulk of pyrethroid hydrolysis, the serum CE in rats may contribute to an appreciable extent at low serum concentrations. This is a major difference between rats and humans when attempting to extrapolate animal studies of pyrethroid exposure because humans lack detectable CEs in their blood. For pyrethroids such as bioresmethrin and trans-permethrin, which are rapidly hydrolyzed by serum CEs, the rat is probably a poor surrogate animal for human health risk assessment, especially at environmentally relevant low level exposures. Transgenic rodent models that do not express serum CEs (Morton et al., 2005) may represent a better model for predicting the pharmacokinetics of such compounds in humans.
Acknowledgments
The project described was supported by Grant Number P20RR017661 (J.A.C. and M.K.R.) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. P.M Potter was supported by NIH CA76202, CA79763, CA108775, CA98468, a Cancer Center Core Grant P30 CA-21765 and the American Lebanese Syrian Associated Charities. We gratefully acknowledge the gift of CE antibodies from Dr. M. Hosokawa (Chiba University, Japan). We are also grateful to Drs. Nick Filipov, Rogelio Tornero-Velez, and Steven Gwaltney for their critical reading of the manuscript.
List of abbreviations
- hCE1
human carboxylesterase 1
- hCE2
human carboxylesterase 2
- 3PBAlc
3-phenoxybenzyl alcohol
- pNPA
para-nitrophenyl acetate
- pNPV
para-nitrophenyl valerate
- 4-MUBA
4-methylumbelliferyl acetate
- 4-MUBS
4-methylumbelliferyl stearate
Footnotes
Conflict of Interest Statement. The authors declare there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand SS, Bruckner JV, Haines WT, Muralidhara S, Fisher JW, Padilla S. Characterization of deltamethrin metabolism by rat plasma and liver microsomes. Toxicol Appl Pharmacol. 2006;212:156–166. doi: 10.1016/j.taap.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Choi J, Hodgson E, Rose RL. In vitro human metabolism of permethrin: the role of human alcohol and aldehyde dehydrogenases. Pest Biochem Physiol. 2002;73:117–128. [Google Scholar]
- Choi J, Hodgson E, Rose RL. Inhibition of trans-permethrin hydrolysis in human liver fractions by chloropyrifos oxon and carbaryl. Drug Metabol Drug Interact. 2004;20:233–246. doi: 10.1515/dmdi.2004.20.4.233. [DOI] [PubMed] [Google Scholar]
- Cygler M, Schrag JD, Sussman JL, Harel M, Silman I, Gentry MK, Doctor BP. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RA, Zhang J, Brzezinski MR, Bosron WF. Tissue distribution of cocaine methyl esterase and ethyl transferase activities: correlation with carboxylesterase protein. J Pharmacol Exp Ther. 1995;275:965–971. [PubMed] [Google Scholar]
- Dolinsky VW, Gilham D, Alam M, Vance DE, Lehner R. Triacylglycerol hydrolase: role in intracellular lipid metabolism. Cell Mol Life Sci. 2004;61:1633–1651. doi: 10.1007/s00018-004-3426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughan LC, Engel JL, Casida JE. Pesticide Interactions: Effects of organophophorus pesticides on the metabolism, toxicity, and persistence of selected pyrethroid insecticides. Pest Biochem Physiol. 1980;14:81–85. [Google Scholar]
- Gilham D, Lehner R. Techniques to measure lipase and esterase activity in vitro. Methods. 2005;36:139–147. doi: 10.1016/j.ymeth.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Godin SJ, Scollon EJ, Hughes MF, Potter PM, DeVito MJ, Ross MK. Species differences in the in vitro metabolism of deltamethrin and esfenvalerate: Differential oxidative and hydrolytic metabolism by humans and rats. Drug Metab Dispos. 2006;34:1764–1771. doi: 10.1124/dmd.106.010058. [DOI] [PubMed] [Google Scholar]
- Gonzalez CF, Proudfoot M, Brown G, Korniyenko Y, Mori H, Savchenko AV, Yakunin AF. Molecular basis of formaldehyde detoxification. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J Biol Chem. 2006;281:14514–14522. doi: 10.1074/jbc.M600996200. [DOI] [PubMed] [Google Scholar]
- Hashinotsume M, Higashino K, Hada T, Yamamura Y. Purification and enzymatic properties of rat serum carboxylesterase. J Biochem. 1978;84:1325–1333. doi: 10.1093/oxfordjournals.jbchem.a132254. [DOI] [PubMed] [Google Scholar]
- Hodgson E, Levi PE. Pesticides: an important but underused model for the environmental health sciences. Environ Health Perspect. 1996;104(Suppl 1):97–106. doi: 10.1289/ehp.96104s197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DY, Howles PN. Carboxyl ester lipase: structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis. J Lipid Res. 2002;43:2017–2030. doi: 10.1194/jlr.r200013-jlr200. [DOI] [PubMed] [Google Scholar]
- Imai T. Human carboxylesterase isozymes: catalytic properties and rational drug design. Drug Metab Pharmacokinet. 2006;21:173–185. doi: 10.2133/dmpk.21.173. [DOI] [PubMed] [Google Scholar]
- Imai T, Taketani M, Shii M, Hosokawa M, Chiba K. Substrate specificity of carboxylesterase isozymes and their contribution to hydrolase activity in human liver and small intestine. Drug Metab Dispos. 2006;34:1734–1741. doi: 10.1124/dmd.106.009381. [DOI] [PubMed] [Google Scholar]
- Li B, Sedlacek M, Manoharan I, Boopathy R, Duysen EG, Masson P, Lockridge O. Butyrylcholinesterase, paraoxonase, and albumin esterase, but not carboxylesterase, are present in human plasma. Biochem Pharmacol. 2005;70:1673–1684. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Lindstedt SL, Schaeffer PJ. Use of allometry in predicting anatomical and physiological parameters of mammals. Laboratory Animals. 2002;36:1–19. doi: 10.1258/0023677021911731. [DOI] [PubMed] [Google Scholar]
- Morgan EW, Yan B, Greenway D, Petersen DR, Parkinson A. Purification and characterization of two rat liver microsomal carboxylesterases (hydrolase A and B) Arch Biochem Biophys. 1994;315:495–512. doi: 10.1006/abbi.1994.1531. [DOI] [PubMed] [Google Scholar]
- Morton CL, Potter PM. Comparison of Escherichia coli, Saccharomyces cerevisiae, Pichia pastoris, Spodoptera frugiperda, and COS7 cells for recombinant gene expression. Application to a rabbit liver carboxylesterase. Mol Biotechnol. 2000;16:193–202. doi: 10.1385/MB:16:3:193. [DOI] [PubMed] [Google Scholar]
- Morton CL, Iacono L, Hyatt JL, Taylor KR, Cheshire PJ, Houghton PJ, Danks MK, Stewart CF, Potter PM. Activation and antitumor activity of CPT-11 in plasma esterase-deficient mice. Cancer Chemother Pharmacol. 2005;56:629–636. doi: 10.1007/s00280-005-1027-y. [DOI] [PubMed] [Google Scholar]
- Natarajan R, Ghosh S, Grogan W. Catalytic properties of the purified rat hepatic cytosolic cholesteryl ester hydrolase. Biochem Biophys Res Commun. 1996;225:413–419. doi: 10.1006/bbrc.1996.1188. [DOI] [PubMed] [Google Scholar]
- Nishi K, Huang H, Kamita SG, Kim IH, Morisseau C, Hammock BD. Characterization of pyrethroid hydrolysis by the human liver carboxylesterases hCE-1 and hCE-2. Arch Biochem Biophys. 2006;445:115–123. doi: 10.1016/j.abb.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter PM, Wadkins RM. Carboxylesterases: detoxifying enzymes and targets for drug therapy. Curr Med Chem. 2006;13:1045–1054. doi: 10.2174/092986706776360969. [DOI] [PubMed] [Google Scholar]
- Robbi M, Beaufay H. The COOH terminus of several liver carboxylesterases targets these enzymes to the lumen of the endoplasmic reticulum. J Biol Chem. 1991;266:20498–20503. [PubMed] [Google Scholar]
- Ross MK, Pegram RA. In vitro biotransformation and genotoxicity of the drinking water disinfection byproduct bromodichloromethane: DNA binding mediated by glutathione transferase theta 1–1. Toxicol Appl Pharmacol. 2004;195:166–181. doi: 10.1016/j.taap.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Ross MK, Borazjani A, Edwards CC, Potter PM. Hydrolytic metabolism of pyrethroids by human and other mammalian carboxylesterases. Biochem Pharmacol. 2006;71:657–669. doi: 10.1016/j.bcp.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Sanghani SP, Davis WI, Dumaual NG, Mahrenholz A, Bosron WF. Identification of microsomal rat liver carboxylesterases and their activity with retinyl palmitate. FEBS. 2002;269:4387–4398. doi: 10.1046/j.1432-1033.2002.03121.x. [DOI] [PubMed] [Google Scholar]
- Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu Rev Pharmacol Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- Satoh T, Taylor P, Bosron WF, Sanghani SP, Hosokawa M, La Du BN. Current progress on esterases: from molecular structure to function. Drug Metab Dispos. 2002;30:488–493. doi: 10.1124/dmd.30.5.488. [DOI] [PubMed] [Google Scholar]
- Schwer H, Langmann T, Daig R, Becker A, Aslanidis C, Schmitz G. Molecular cloning and characterization of a novel putative carboxylesterase, present in human intestine and liver. Biochem Biophys Res Commun. 1997;233:117–120. doi: 10.1006/bbrc.1997.6413. [DOI] [PubMed] [Google Scholar]
- Soderlund DM, Casida JE. Effects of pyrethroid structure on rates of hydrolysis and oxidation by mouse liver microsomal enzymes. Pest Biochem Physiol. 1977;7:391–401. [Google Scholar]
- Soderlund DM, Abdel-Aal YAI, Helmuth DW. Selective inhibition of separate esterases in rat and mouse liver microsomes hydrolyzing malathion, trans-permethrin, and cis-permethrin. Pest Biochem Physiol. 1982;17:162–169. [Google Scholar]
- Soderlund DM. Metabolic considerations in pyrethroid design. Xenobiotica. 1992;22:1185–1194. doi: 10.3109/00498259209051872. [DOI] [PubMed] [Google Scholar]
- Testa B, Mayer JM. Hydrolysis in Drug and Prodrug Metabolism: Chemistry, Biochemistry, and Enzymology. Verlag Chimica Acta; Zurich, Switzerland: 2003. [Google Scholar]
- Takken W. Do insecticide-treated bednets have an effect on malaria vectors? Trop Med Int Health. 2002;7:1022–1030. doi: 10.1046/j.1365-3156.2002.00983.x. [DOI] [PubMed] [Google Scholar]
- Wadkins RM, Hyatt JL, Wei X, Yoon KJ, Wierdl M, Edwards CC, et al. Identification and characterization of novel benzil (diphenylethane-1,2-dione)analogues as inhibitors of mammalian carboxylesterases. J Med Chem. 2005;48:2906–2915. doi: 10.1021/jm049011j. [DOI] [PubMed] [Google Scholar]
- Wheelock CE, Wheelock AM, Zhang R, Stok JE, Morisseau C, Le Valley SE, et al. Evaluation of alpha-cyanoesters as fluorescent substrates for examining interindividual variation in general and pyrethroid-selective esterases in human liver microsomes. Anal Biochem. 2003;315:208–222. doi: 10.1016/s0003-2697(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Yan B, Yang D, Bullock P, Parkinson A. Rat serum carboxylesterase. Cloning, expression, regulation, and evidence of secretion from liver. J Biol Chem. 1995;270:19128–19134. doi: 10.1074/jbc.270.32.19128. [DOI] [PubMed] [Google Scholar]
- Zhao B, Fisher BJ, St Clair RW, Rudel LL, Ghosh S. Redistribution of macrophage cholesteryl ester hydrolase from cytoplasm to lipid droplets upon lipid loading. J Lipid Res. 2005;46:2114–2121. doi: 10.1194/jlr.M500207-JLR200. [DOI] [PubMed] [Google Scholar]