Abstract
We have previously shown that environmental enrichment decreases the activating and rewarding effects of the psych psychostimulant cocaine and increases resistance to the neurotoxic effect of the Parkinson-inducing drug MPTP. These effects were accompanied by an increase in the striatal expression of the neurotrophin BDNF, an increase in the striatal levels of delta- Fos B and by a decrease in striatal levels of the dopamine transporter, the main molecular target for cocaine and MPTP. Here, we used cDNA arrays to investigate the effects of rearing mice in enriched environments from weaning to adulthood on the profile of expression of genes in the striatum focusing on genes involved in intracellular signalling and functioning. We found that mice reared in an enriched environment show several alterations in the levels of mRNA coding for proteins involved in cell proliferation, cell differentiation, signal transduction, transcription and translation, cell structure and meta metabolism. Several of these findings were further confirmed by real-time quantitative PCR and, in the case of protein kinase C lambda, also by western blot. These findings are the first description of alterations in striatal gene expression by an enriched environment. The striatal gene expression regulation by environment that we report here may play a role in the resistance to the effects of drugs of abuse and dopaminergic neurotoxins previously reported.
Keywords: cDNA array, Real time-PCR, Western blot, PKC, Epigenetic, Dopamine, Environmental enrichment
1. Introduction
The term enriched environment (EE), refers to housing conditions that enhance sensory, cognitive and motor stimulation. EE usually consists of larger cages containing several objects, often including a running wheel, which vary in composition, shape size, texture and color (Nithianantharajah and Hannan, 2006; van Praag et al., 2000). The positive effects of EE on animal learning and memory were first described in the 1940's by Hebb who allowed laboratory rats to roam freely in his house and observed behavioural improvements in these rats compared to ratshoused in standard laboratory conditions(Hebb, 1947). Later on, Rosenzweig and colleagues (Rosenzweig, 1966) described EE as a combination of complex inanimate and social stimulation. Since then, many studies in laboratory settings have confirmed and extended the notion that rodents reared in EE show improved learning and memory performance (Iuvone et al., 1996; Young et al., 1999). In addition, exposure to an EE has been shown to induce biochemical and structural changes in several brain regions in particular in the cortex and the hippocampus (Diamond et al., 1976; Kempermann et al., 1997; Nithianantharajah and Hannan, 2006; van Praag et al., 2000). Alterations include increased brain weight and size (Bennett et al., 1969), increased neurotransmitter levels such as acetylcholine (Rosenzweig and Bennett, 1969), serotonin and noradrenaline (Chaouloff, 1989), as well as enhanced gliogenesis (Diamond et al., 1966) and neurogenesis (Kempermann et al., 1997). These alterations may be due, at least in part, to changes in gene expression produced by environmental stimulation. Indeed, previous studies have shown that, in the hippocampus, EE induces alterations in the expression of several genes involved in synaptic function and cellular plasticity of wild-type as well as transgenic mice lacking the NMDA receptor 1 subunit (Li et al., 2007; Rampon et al., 2000a,b).
The positive effects of EE are not limited to learning and hippocampal structures but may be extended to motivational and motor functions and to striatal structures. In fact, environmental enrichment was shown to be beneficial in several psychiatric and neurodegenerative disorders implicating monoamine systems where it can (i) compensate for impairments in animal models of schizophrenia, Huntington's, and Parkinson's diseases; (ii) increase resistance to the addictive properties of psychostimulant drugs; (iii) level-out the consequences of prenatal stress in animal models of depression (Laviola et al., in press). For instance, we have previously shown that mice reared in EE were protected against the neurotoxic effects of a pro-parkinsonian neurotoxin, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) (Bezard et al., 2003; Faherty et al., 2005), and against the stimulation of locomotor activity by an acute injection of cocaine (Bezard et al., 2003). In addition, we have recently found that the rewarding and sensitizing effects of cocaine are also reduced in EE mice and that this reduction is associated with reduced reactivity of striatal neurons to the effects of cocaine but not with changes in cocaine-induced dopamine release in the nucleus accumbens (Solinas et al., in press). The resistance of enriched mice to dopaminergic drugs may be due, in part, to the reduced levels of DAT, the main molecular target of cocaine and MPTP, and to increased levels of BDNF and delta-Fos B in the striatum (Bezard et al., 2003; Solinas et al., in press). Nevertheless, the positive influence of stimulation by EE against the effects of MPTP and cocaine is likely to be also due to other EE-induced changes in gene expression in striatal neurons. In order to test this hypothesis we used cDNA array analysis techniques to identify more global and complex influence of EE on the striatum. The cDNA array chips contained sequences targeting genes involved in several intracellular functions such as signal transduction, cell proliferation and differentiation, transcription and translation, structural rearrangements and metabolism that were chosen because little is known on the effects of EE on these genes (Tanaka et al., 2000). Some of the array data were further confirmed by real-time PCR. Effects of EE were finally verified by western blot analysis for protein kinase lambda. A special emphasis was placed on this gene because PKCs are known to participate in several cellular processes including synaptic plasticity.
2. Results
2.1. Several families of genes are regulated by environmental stimulation
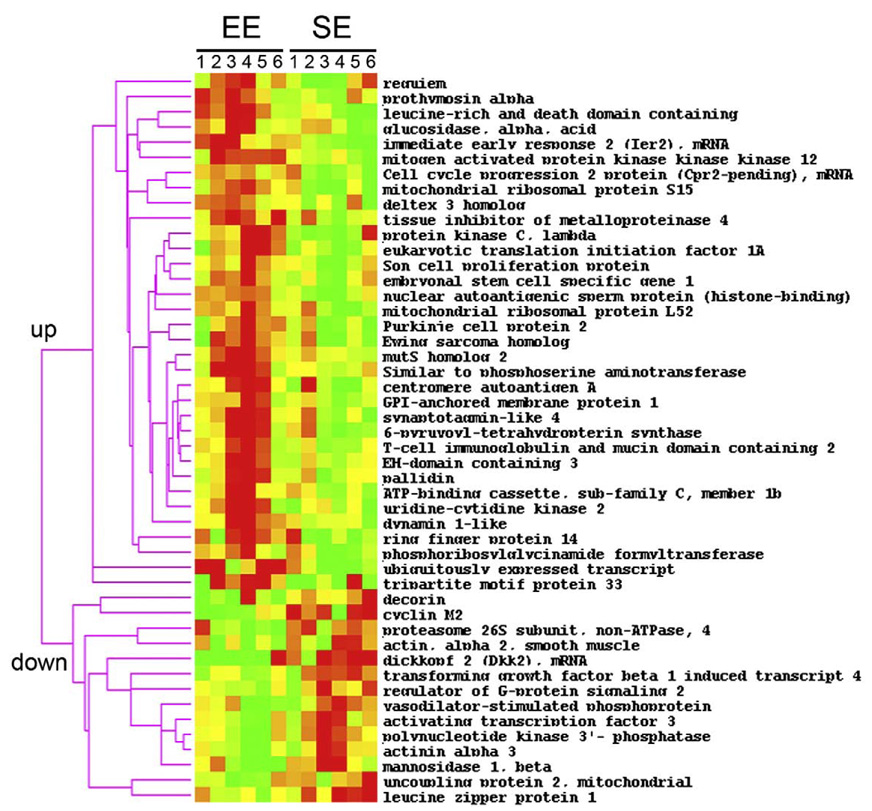
To identify possible molecular adaptations induced by enriched housing conditions in comparison to standard environments (SE) conditions, we analysed the transcriptional responses in striatal tissue by cDNA arrays. Our analysis revealed that out of 16896 transcripts, 48 were differentially regulated (p<0.05). We applied hierarchical cluster analysis to profile these transcripts based on similarities in the patterns of expression (Fig. 1). Two differential expression profiles were obtained showing that 34 genes were significantly up-regulated whereas 14 genes were down-regulated after exposure to environmental stimulation from weaning to adulthood. These genes belong to several classes of genes that play a role in cell proliferation and differentiation, intracellular signalling including transcription and translation, structural changes, and cell metabolism (Table 1). Among the genes that code for proteins involved in cell proliferation, prothymosin alpha (Ptma), which is regulated by spatial discrimination learning (Robles et al., 2003), was up-regulated whereas cyclin M2 was down-regulated. Genes belonging to the Notch pathway (dtx3, esg1 and dkk2) involved in cell fate and differentiation were regulated as well. Given the role of this pathway in learning and memory (Costa et al., 2005), these changes could be involved in the cognitive enhancing effects of EE (van Praag et al., 2000). EE down-regulated several members of another important family of protein involved in cell structure (Actn3, Dec, Acta2, except for Sytl4) and these changes might be involved in causing EE-induced architectural changes in the brain. Several other genes regulated by EE may be involved in remodelling/axonal connection (Gpiap1, Timp4, Vasp). Other regulated genes participate in gene expression (Ier2 and Atf3) and protein synthesis or degradation (Mrpl52, Mrps15, Psmd4, Rnf4, Uck2-pending). Dynamin 1-like (Dnm1l), a gene involved in endocytosis of activated growth factor receptors (Harrison-Findik et al., 2001) was found to be decreased in EE mice. Finally, genes that belong to cellular signalling such as the protein kinase PKC lambda and mitogen-activated protein kinase kinase kinase 12 (MAP3K12) were also regulated by enriched environment. We have mostly focused on these later genes because protein kinases and MAP kinases are known to play a crucial role in the effects of stimulant drugs and may be responsible for the differential response to drugs in EE mice in comparison to SE mice that others and we have previously reported (Bardo et al., 2001; Bezard et al., 2003; Green et al., 2002; Solinas et al., in press).
Fig. 1.
Hierarchical cluster analysis of gene expression profiles in the striatum of SE and EE mice. The levels of expression of 48 transcripts were changed by EE. The modifications are in two opposite directions (increased or decreased). For each gene, the expression in SE or EE is shown for each of the 6 animals in both SE and EE groups. Each row shows changes in the expression of a single gene whereas each column represents the expression of the genes within a particular animal. Color scale, the expression level of each gene in a single sample relative to its median abundance across all samples; red, expression level above the median; yellow,median expression; green, expression level below the median; color saturation shows the magnitude of deviation from the median.
Table 1.
Transcripts whose expression was changed in the striatum of mice raised in EE in comparison to mice raised in SE
| Gene name (gene symbol) | Unigene | Diffrential expression (mean EE-mean ES) | z-ratio | p-values | Changes |
|---|---|---|---|---|---|
| Cell proliferation | |||||
| Cell cycle progression 2 protein (Cpr2) | Mm.196027 | 0.133 | 1.59 | 0.01 | + |
| Phosphoribosylglycinamide formyltransferase (Gart) | Mm.4505 | 0.151 | 1.81 | 0.04 | + |
| Prothymosin alpha (Ptma) | Mm.19187 | 0.142 | 1.70 | 0.01 | + |
| Nuclear autoantigenic sperm protein (Nasp) | Mm.7516 | 0.152 | 1.82 | 0.00 | + |
| Son cell proliferation protein (Son) | Mm.46401 | 0.141 | 1.69 | 0.02 | + |
| Cyclin in M2 (Cnnm2) | Mm.38506 | −0.258 | −3.09 | 0.03 | − |
| Cell differentiation | |||||
| Purkinje cell protein 2 (Pcp2) | Mm.41456 | 0.161 | 1.93 | 0.02 | + |
| Deltex 3 homolog (Dtx) | Mm.29197 | 0.129 | 1.54 | 0.02 | + |
| Embryonal stem cell specific gene 1 (Esg1) | Mm.139314 | 0.125 | 1.50 | 0.03 | + |
| Dickkopf 2 (Dkk2) | Mm.103593 | −0.550 | −6.59 | 0.03 | − |
| Signal transduction | |||||
| Eukaryotic translation initiation factor 1A (Eif1a) | Mm.143141 | 0.221 | 2.64 | 0.01 | + |
| Protein kinase C, lambda (Prkcl) | Mm.23823 | 0.212 | 2.53 | 0.01 | + |
| Mitogen-activated protein kinase kinase kinase 12 (Map3k12) | Mm.4358 | 0.182 | 2.18 | 0.00 | + |
| Leucine-rich and death domain containing (Lrdd) | Mm.158086 | 0.281 | 3.37 | 0.00 | + |
| Regulator of G-protein signaling 2 (Rgs2) | Mm.28262 | −0.130 | −1.56 | 0.02 | − |
| Transforming growth factor beta 1 induced transcript 4 (Tgfb1i4) | Mm.20927 | −0.132 | −1.58 | 0.00 | − |
| Polynucleotide kinase 3′- phosphatase (Pnkp) | Mm.29545 | −0.200 | −2.39 | 0.00 | − |
| Transcription/translation | |||||
| Immediate early response 2 (Ier2) | Mm.399 | 0.413 | 4.95 | 0.01 | + |
| Mitochondrial ribosomal protein L52 (Mrpl52) | Mm.30240 | 0.175 | 2.09 | 0.02 | + |
| Mitochondrial ribosomal protein S15 (Mprs15) | Mm.25143 | 0.198 | 2.37 | 0.00 | + |
| Proteasome 26S subunit, non-ATPase, 4 (Psmd4) | Mm.2261 | −0.223 | −2.67 | 0.02 | − |
| Activating transcription factor 3 (Atf3) | Mm.2706 | −0.156 | −1.86 | 0.05 | − |
| Structural proteins | |||||
| Synaptotagmin-like 4 (Sytl4) | Mm.38674 | 0.197 | 2.36 | 0.02 | + |
| Ubiquitously expressed transcript (Uxt) | Mm.34779 | 0.542 | 6.49 | 0.01 | + |
| Tissue inhibitor of metalloproteinase 4 (Timp4) | Mm.36851 | 0.154 | 1.84 | 0.01 | + |
| GPI-anchored membrane protein 1 (Gpiap1) | Mm.1098 | 0.133 | 1.59 | 0.03 | + |
| Vasodilator stimulated phosphoprotein (Vasp) | Mm.9684 | −0.139 | −1.66 | 0.02 | − |
| Actinin alpha 3 (Actn3) | Mm.5316 | −0.177 | −2.12 | 0.01 | − |
| Decorin (Dcn) | Mm.1987 | −0.152 | −1.82 | 0.04 | − |
| Actin, alpha 2, smooth muscle (Acta2) | Mm.16537 | −0.136 | −1.63 | 0.05 | − |
| Metabolism | |||||
| 6- pyruvoyl-tetrahydropterin synthase (Pts) | Mm.35856 | 0.196 | 2.34 | 0.03 | + |
| Glucosidase, alpha, acid (Gaa) | Mm.4793 | 0.127 | 1.52 | 0.05 | + |
| Similar to phosphoserine aminotransferase | Mm.29902 | 0.176 | 2.11 | 0.02 | + |
| Ring finger protein 14 (Rnf14) | Mm.22086 | 0.202 | 2.42 | 0.03 | + |
| Uridine-cytidine kinase 2 (Ck2-pending) | Mm.25309 | 0.195 | 2.33 | 0.01 | + |
| Mannosidase 1, beta (Man1b) | Mm.4389 | −0.570 | −6.82 | 0.01 | − |
| Others | |||||
| Requiem (Req) | Mm.2651 | 0.153 | 1.84 | 0.04 | + |
| MutS homolog 2 (Msh2) | Mm.4619 | 0.149 | 1.79 | 0.01 | + |
| T- cell immunoglobulin and mucin domain containing 2 (Timd2) | Mm.29550 | 0.159 | 1.90 | 0.02 | + |
| Tripartite motif protein 33 (trim33) | Mm.31048 | 0.365 | 4.37 | 0.05 | + |
| Centromere autoantigen A (Cenpa) | Mm.6579 | 0.206 | 2.47 | 0.04 | + |
| Ewing sarcoma homolog (Ewsh) | Mm.142822 | 0.169 | 2.02 | 0.04 | + |
| EH-domain containing 3 (Ehd3) | Mm.18526 | 0.147 | 1.76 | 0.04 | + |
| Pallidin (Pldn) | Mm.148195 | 0.129 | 1.55 | 0.04 | + |
| Dynamin 1-like (Dnm1l) | Mm.140013 | 0.168 | 2.01 | 0.01 | + |
| ATP-binding cassette, sub-family C, member 1b (Abcc1b) | Mm.196634 | 0.160 | 1.92 | 0.04 | + |
| Uncoupling protein 2, mitochondrial (ucp2) | Mm.144413 | −0.130 | −1.55 | 0.03 | − |
| Leucine zipper protein 1 (Luzp1) | Mm.92659 | −0.173 | −2.07 | 0.01 | − |
2.2. Effects of environments on striatal mRNA levels of PKC lambda, MAP3K12, DEC, PSDM4 and DNM1L
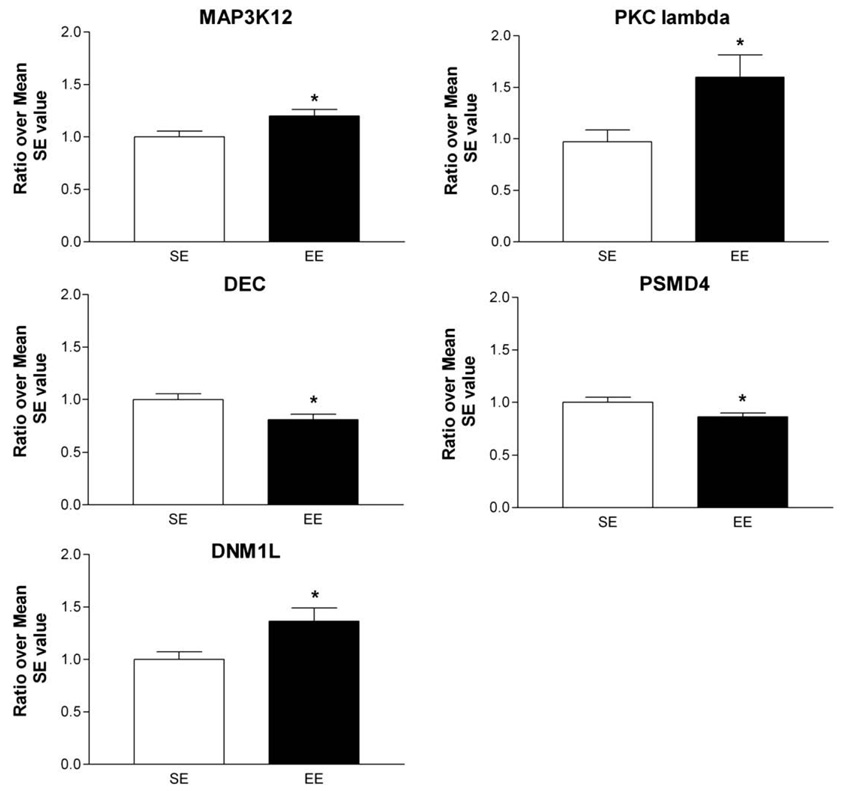
We used quantitative RT-PCR to confirm the changes in transcripts belonging to various families of genes involved in either signalling (PKC lambda, MAP3K12), cell architecture (DEC), transcription and translation (PSDM4) as well as Dynamin 1-like gene (DNM1L) a member of the dynamin superfamily (Praefcke and McMahon, 2004). Consistent with cDNA arrays findings, significant changes were found in the same direction than observed with cDNA arrays (Fig. 2). Specifically, EE caused increased expression of PKC lambda (+60%; Student t-test: DF=15, t-value=−2.38; p<0.05), of MAP3K12 (+20%; Student t-test: DF=15, t-value=−2.35; p<0.05) and of DNM1L (+40%; Student t-test: DF=12, t-value=−2.69; p<0.05). On the other hand, the expressions of DEC and PSDM4 were decreased by EE (−20% and −15%, respectively; Student t-test: for DEC, DF=15, t-value=2.46; p<0.05 and for PSDM4, DF=12, t-value=2.45; p<0.05).
Fig. 2.
Quantitative RT-PCR analysis of some transcripts affected by EE. The expression of genes, selected from various families of genes for their potential involvement in EE's effects on striatal plasticity, was confirmed by RT-PCR. Selected genes were: Mitogen-activated protein kinase kinase kinase 12 (MAP3K12), Protein kinase C lambda (PKC lambda), Decorin (DEC), Proteasome 26S subunit non-ATPase 4 (PSMD4), and Dynamin 1-like (DNM1L). For all genes examined, statistically significant changes were observed in the same direction as arrays results. The data were obtained from 8 mice per group and determined individually. The amount of each product was normalized by GAPDH value and then a ratio over mean SE value was obtained. Values represent means±SEM. Student t-test, *p< 0.05 different from SE control.
2.3. Effects of environments on striatal protein levels of PKC lambda
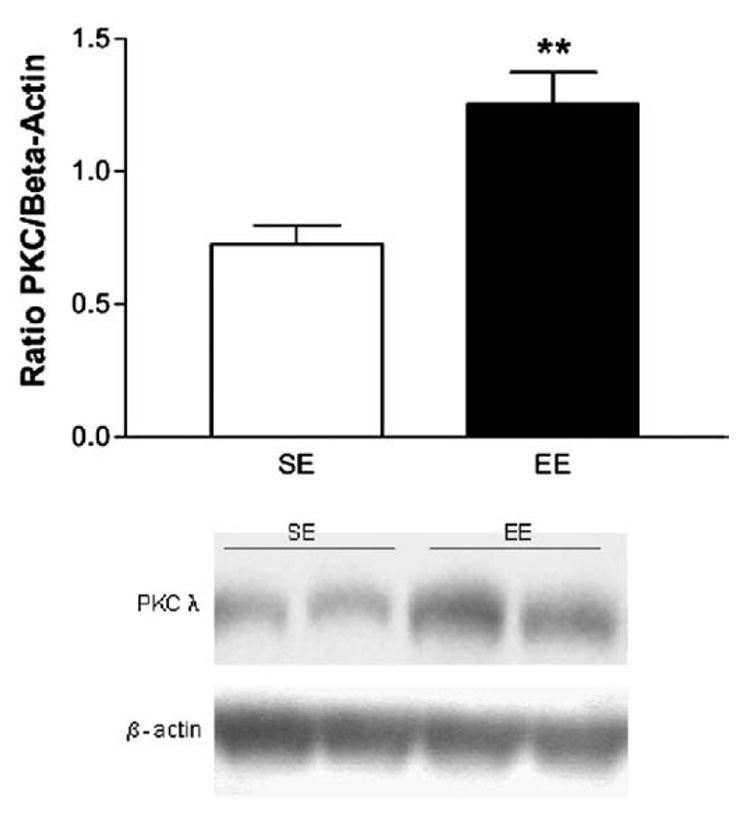
Since mRNA changes are not always correlated by changes at the protein level, we measured the PKC lambda protein levels in the striatum of SE and EE by western blot (Fig. 3). In agreement with what was found at the mRNA levels, we observed that EE increased the level PKC lambda protein (+80%; Student t-test: DF=15, t-value=−3.74; p<0.01).
Fig. 3.
Levels of PKC lambda protein in the striatum of mice reared in standard or enriched environments. EE mice show higher basal levels of PKC lambda than SE mice. Results are expressed as level of PKC lambda over level of beta-actin. Values represent means±SEM. Student T test, **p<0.01 different from SE control.
3. Discussion
We have used a cDNA arrays approach to identify the differential profile of transcription in the striatum induced by rearing mice in enriched or standard conditions.Mice were placed in enriched or standard conditions right after weaning and for 2 months to expose them to these environments during the entire duration of their adolescence, a critical period for neuro-developmental plasticity (Crews et al., 2007; Sisk and Zehr, 2005). We found that indeed EE caused a complex profile of gene expression which consisted of the regulation of 48 genes that are implicated in various functions including cell proliferation and differentiation, intracellular signalling, transcription and translation, as well as structural changes and cell metabolism. In what follows, we will discuss the possible role of some of these changes in modifying striatal functions observed after EE exposure and their potential implication in the resistance to drugs of abuse and neurotoxins that was previously reported (Bezard et al., 2003; Nithianantharajah and Hannan, 2006; Solinas et al., in press).
EE has been previously shown to produce up- or down-regulation of some genes and proteins in several regions. Particularly relevant for the present work, some studies have used cDNA arrays to investigate the effects of enriched environments on gene expression profiles in models of neurode-generative diseases and ischemia (Costa et al., 2007; Lazarov et al., 2005; Ronnback et al., 2005). In all these studies, EE were found to ameliorate cognitive impairments and to produce a number of changes in gene expression in the hippocampus (Costa et al., 2007; Lazarov et al., 2005; Ronnback et al., 2005). In particular, Ronnback et al. (2005) found that in rat models of ischemia, the effects of EE in the hippocampus were concentrated on the 3 main families of genes (signal transduction, cell metabolism and cytoskeletal proteins) that we also found to be altered by EE in the striatum. On the other hand, the other two studies that used genetic mouse model of Alzheimer disease found that EE stimulate expression of genes associated with learning, memory and neurotrophic actions in the hippocampus (Costa et al., 2007; Lazarov et al., 2005). Concerning the striatum, no large-scale study has been described in the literature. However, two genes known to participate in neuroplasticity, namely nerve growth factor 1-A (NGF1-A) and Activity-regulated cytoskeleton (Arc) are over-expressed after EE exposure (Pinaud et al., 2001, 2002). Also, our laboratory has previously described EE-mediated down-regulation of the dopamine transporter and the up-regulation of the trophic factor BDNF (Bezard et al., 2003) and increases in the striatal levels of the transcription factor delta-Fos B (Solinas et al., in press). However, we estimated that it was unlikely that these few genes could, by themselves, account for all the plastic changes induced by EE in the striatum. Indeed, our array analysis was able to identify additional families of genes that participate in the control of cell proliferation and differentiation, cell signalling and cell architecture. The results of the present study are consistent with those of another study that reported that training in an EE for 3 h, 6 h, 12 days or 14 days changes the expression of about 100 genes in the cortex of adult mice (Rampon et al., 2000a). Specifically, the genes whose expression levels were changed at 14 days belong to the same families of genes that those found in the present study that focused on the striatum. Therefore, it appears that the effects of EE do indeed involve a complex pattern of transcriptional changes in the brain.
Neuroplasticity is characterized by changes in synaptic organization and transmission. We have found changes in the expression of genes involved in cell signalling including genes that are involved in the regulation of the MAP kinase pathway. For example, a gene regulated by EE in the striatum was MAP3K12 (also known as DLK, MUK, Zpk). MAP3K12 belongs to the MAP kinase–kinase–kinase (MAPKKK) class of protein kinases and regulates MAP kinases c-Jun amino-terminal kinase (JNK) via MAPKK7, which belongs to the MAP kinase–kinase class of protein kinases (Gallo and Johnson, 2002). MAP3K appears to be involved in neurodevelopment in mice (Hirai et al., 2005) as well as axon growth and neuronal migration (Hirai et al., 2006). In a previous paper, expression of MAP3K12 mRNA was detected in the brain of adult mice, but not in any other tissue tested. In situ hybridization analysis of mouse brain sections revealed specific association of MAP3K12 mRNA with neuronal cell populations, primarily in the hippocampus, the cerebral cortex, and the Purkinje cell layer of the cerebellum (Blouin et al., 1996). Our results indicate that MAP3K12 is also expressed in the striatum and that its expression can be regulated by environmental manipulations.
Another interesting gene that we found to be regulated by EE is Dynamin 1-like (DNM1L) which belongs to the dynamin superfamily (Praefcke and McMahon, 2004) that has been shown to be involved in endocytosis of activated growth factor receptors (Harrison-Findik et al., 2001). The effects of EE are believed to be due, at least in part, to increases in neurotrophic factors (van Praag et al., 2000) and, indeed, we have previously shown that BDNF levels are increased in the striatum of mice reared in EE (Bezard et al., 2003). Thus it is possible that changes in DNM1L are related to increases in BDNF levels associated with enrichment. Importantly, it has been recently shown that levels of Dynamin 1 are increased in the brain of old mice (Poon et al., 2006). Given the well-known anti-aging effects of EE (Mora et al., 2007), our data support the hypothesis that increases in DNM1L may be a part of the brain compensatory reactions to counteract cellular aging.
Among the other genes that were regulated in our study, we focused on PKC lambda because PKCs are known to participate in several cellular processes including synaptic plasticity. PKC lambda is a newly discovered form of serine/threonine PKC that belongs to anatypical family class of PKC (Blouin et al., 1996). In a previous study, PKC lambda mRNA was found to be expressed mainly in the cortex and the hippocampus but not in the striatum of C57/BL6 mice (Oster et al., 2004). Here we report the expression, and regulation by enriched environments, of PKC lambda in the striatum and the cDNA arrays results were confirmed, at the mRNA level, by real time quantitative PCR and, at the protein level, by western blot. The negative results by Oster et al. (2004) might have been due to lower sensitivity of the in situ hybridization techniques compared to the techniques used in this study. This possibility is supported by findings that PKC lambda/zeta protein is indeed present in the nucleus accumbens of rats (Chen et al., 2007) and the authors hypothesised that this protein might be involved in cocaine addiction (Chen et al., 2007). The suggestion that atypical PKCs lambda/iota/zeta may be involved in neurodegenerative diseases such as Alzheimer and Parkinson diseases also provide support for the proposal that these kinases might be involved in plastic changes in the brain (Crary et al., 2006) and that their expression can be affected by EE (Nithianantharajah and Hannan, 2006). Thus, PKC lambda could play an important role in neuronal plasticity or neuroprotection induced by EE. Interestingly, Regala and collaborators have shown a regulation of Erk pathway by PKC lambda (Regala et al., 2005). Thus, PKC lambda and MAP3K12 converge on and participate to the regulation of the same MAP kinase pathway. Given the role of Erk pathway in cocaine addiction(Girault et al., 2007; Lu et al., 2006), it is possible that EE modulates the reactivity to drugs through regulation of the expression of PKC lambda and MAP3K12.
Another interesting result of our study is the regulation of genes involved in cell differentiation and architectural changes that could be more directly involved in the neuroplastic changes induced by EE and in the ability of EE to reduce the effects of drugs and toxins. Indeed, we have found the regulation of three members of the Notch pathway, namely deltex homolog 3, embryonal stem cell specific gene 1 and dickkopf 2. Given the involvement of this pathway in cell fate and differentiation and in the regulation of neurite growth and adult neurogenesis (Berezovska et al., 1999; Costa et al., 2005), it is likely that these genes participate in the synaptic morphological changes induced by EE in the striatum (Comery et al., 1996; Kolb et al., 2003a,b). EE down-regulated the expression of several other genes that encode for proteins involved in cell structure such as synaptotagmin-like 4, Actinin alpha 3, Decorin, Actin alpha 2. Other genes regulated by EE may be involved in remodelling axonal connections (GPI-anchored membrane protein 1, Tissue inhibitor 4, Vasodilator-stimulated phosphoprotein). Altogether, these results support the proposition that EE induces a profound structural changes and reorganization of striatal neurons (Bezard et al., 2003; Nygren et al., 2006; Spires et al., 2004). For example, it has been shown EE induces long-lasting adaptations such as increases in spine density and arborisation in the striatum of rats (Comery et al., 1996; Kolb et al., 2003a,b). The striatum is one of the main terminals of dopamine neurons and it is a brain region that plays a pivotal role in drug addiction and in voluntary motor behaviour especially its dorsal part (Everitt and Robbins, 2005; Jaber et al., 1996). As a matter of fact, several behavioral consequences have been described after EE. For example, we have found that EE produces faster adaptation to novelty and reduces the locomotor activity (Bezard et al., 2003) as well as conditioned place preference and behavioral sensitization induced by cocaine (Solinas et al., in press).We and others have also found that EE protects against the neurotoxic effects of the Parkinson-inducing drug MPTP (Bezard et al., 2003; Faherty et al., 2005) while several findings from the group of Hannan have shown that EE ameliorates several symptoms in a mice model of Huntington disease (Nithianantharajah and Hannan, 2006). Since there is a high level of correlation between transcript expression and protein expression in murine tissue (Kislinger et al., 2006), and although it is difficult to single out a gene responsible for these in vivo effects, it is possible and even likely that the changes reported here played a role in the resistance to cocaine and MPTP previously reported (Bezard et al., 2003; Faherty et al., 2005).
In conclusion, we found that rearing mice in an enriched environment during adolescence induces a variety of changes in the expression of genes in the striatum, an area that plays a pivotal role in motor and motivated behaviours. The pattern of gene expression is compatible with several studies that have reported dramatic morphological changes induced by EE in the striatum. In addition, the regulation of genes involved in cell signalling is compatible with our recent results indicating that effects of cocaine and MPTP are reduced by EE.
4. Experimental procedures
4.1. Subjects
Male C57Bl/6J mice were housed in a temperature-controlled environment on a 12-h light/12-h dark cycle with the lights on from 7:00 a.m. to 7:00 p.m. and had ad libitum access to food and water. All experimentation was conducted during the light period. Experiments were carried out in accordance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) for the care of laboratory animals.
4.2. Housing environmental conditions
After weaning (3 weeks of age), mice were randomly divided in groups of 4 in either a standard environment (SE) or an enriched environment (EE). SE consisted of common housing cage (25×20×15 cm). The EE consisted of a larger (60× 38×20 cm) cage containing constantly a running wheel and a small house and four-five toys that were changed once a week with new toys of different shape and color. Mice were housed in either SE or EE for at least 2 months, throughout the adolescence period and until adulthood, before the start of the experiments. All experiments were conducted at adulthood.
4.3. Tissue and RNA preparation
After 2 months in either SE or EE, mice (n=8 per group) were decapitated and brains were removed. Striatum (including caudate and accumbens nuclei) were isolated by freehand dissection as previously performed (Thiriet et al., 2005), immediately frozen on dry ice and stored at −80 °C until use. RNAs were isolated using RNeasy Mini kit (Qiagen). RNA integrity was assessed by electrophoresis in a denaturating 1% agarose-formaldehyde gel and concentration was determined by measuring OD at 260 nm. In EE and SE groups, the 6 samples from each group having the best RNA quality were used for cDNA arrays (n=6). All RNA samples were used for real-time PCR (n=8) experiments.
4.4. CDNA array analysis
Arrays experiments were performed using single channel labelling 33P nylon membrane-based cDNA arrays containing 16896 features, which include 12341 unique mouse genes and expressed sequence tags (ESTs). These membranes were kindly provided by the Gene expression and Genomics Unit, National Institute of Aging, NIH (Baltimore, USA). Information about the cDNA array used can be found at the Gene Expression and Genomics Unit websites (http://www.daf.jhmi.edu/microarray/index.htm and http://lgsun.grc.nia.nih.gov). Protocols on array printing are available at this website (http://www.daf.jhmi.edu/microarray/protocols.htm). Each mouse's striatal RNA was processed and run on a separated array membrane. Five µg of total RNA from each sample was reverse-transcribed into 33P-labeled individual cDNA samples using the Superscript II reverse transcriptase (Invitrogen) and purified on biospin P-30 spin columns (BioRad). Probes were then used for overnight hybridization at 55 °C onto cDNA arrays membranes. Detailed hybridization protocol can be found at the following website (http://www.daf.jhmi.edu/microarray/protocols/protocol4.pdf). The arrays were exposed to a low-energy phosphor screen (Molecular Dynamics) for 5 days, the screen was scanned using a Phosphorimager 860 (Molecular Dynamics), and pixel intensities quantified using the software ImageQuant.
4.5. Analysis of cDNA arrays and hierarchical clustering
Array data analysis was supervised by the Gene Expression and Genomics Unit (NIA/NIH, Baltimore, USA). To minimize possible bias due to individual background differences, 6 mice were used to represent each group. Due to potential experimental variations, non specific uniform background across entire arrays was normalized by Excel software using global normalization. The data value for each spot on each membrane was divided by the median intensity value of that membrane to obtain a normalized intensity value. Intensity data for each gene were logarithmically transformed. To eliminate noise from low level expression, we filtered out all genes whose average normalized intensity between the two groups of mice was less than zero. The cDNA arrays data sets were analyzed using the z-score transformation normalization method (Cheadle et al., 2003), in which log-transformed and normalized hybridization intensity values provide the basis for p-value-based significance calculations by a z-test in Excel. The Z scores were calculated on the overall mean of the transcripts before any filtering or selection, using the following formula:
Two-tailed p-values were used to identify transcripts with decreased or increased expression, respectively. Individual z-ratio data represent a normalized ratio between the 2 groups (EE and SE) of animals. A gene expression was regarded as up-regulated if the z-score value of its transcript was ≥1.5 times higher in the enriched conditions in comparison to the standard conditions. Agene was regarded as down-regulated if the z-score value of this transcript was ≤0.5 times smaller in the enriched conditions in comparison to the standard conditions. Only differences with a p≤0.05 were regarded as significant. The choice of an appropriate fold-change and a non-stringent P cutoff has been based on recent reports that this approach can be successful in identifying reproducible gene lists (Shi et al., 2006). Functional grouping analysis for significantly changed genes was performed using Amigo (http://www.geneontology.org/) and supported through literatures search. Hierarchical clustering analysis was performed using TreeView and Cluster software downloaded from Eisenlab website (http://rana.lbl.gov/EisenSoftware.htm).
4.6. Real-time reverse transcription-polymerase chain reaction (real-time RT-PCR)
We confirmed cDNA array results by using the real-time RT-PCR technique for 5 genes picked in different families, i.e. 2 kinases involved in cell signalling (PKC lambda and MAP3K12); decorin (DEC) a gene involved in cellular architecture, Proteasome 26S subunit, non-ATPase 4 (PSMD4) a gene involved in transcription and translation and a member of the dynamin family, Dynamin 1-like (DNM1L),which is involved in several processes including budding of transport vesicles, division of organelles, cytokinesis and pathogen resistance (Praefcke and McMahon, 2004). Total RNA previously extracted from striatum (n=8) was reverse-transcribed using the Superscript II reverse transcriptase (Invitrogen) (Thiriet et al., 2005, 2002). PCR experiments were then performed using LightCycler technology and the DyNAmo™ Capillary SYBR® Green qPCR kit (Ozyme, France). HPLC-purified and gene-specific primers were obtained from the Synthesis and Sequencing Facility of Johns Hopkins University (Baltimore, USA). The sequences of primers used were: TTT GGA CCA AGT TGG TGA (upstream) and CAT TGC ATA AAT GCG ATC T (downstream) for PKC lambda; CTA GAA GAG GAA CTG GTG A (upstream) and TGA GCA ATT CCC TCT CTT T (downstream) for MAP3K12; TTG TCA TAG AAC TGG GCG (upstream) and GCA CTT CAG TGA GAG AAG TA (downstream) for DEC; TGG TGA AAC TAG CTA AAC G (upstream) and GAG GCA CTG TCA CTA GA (downstream) for PSMD4; TAG GTG GCC TTA ACA CTA T (upstream) and AAT GAT CCT CTG CAT CTC (downstream) for DNM1L; ATG GTG AAG GTC GGT GTG (upstream) and ACT CCA CGA CAT ACT CAG (downstream) for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The reactions were performed as follows: DNA Master SYBR Green I mix (containing Taq DNA polymerase, dNTP, MgCl2, and SYBR Green I dye) was incubated with primers (20 µM) and cDNA template. The amplification program consisted of 1 cycle of 95 °C with 10 min hold (“hot start”) followed by 40 cycles of denaturation (95 °C, 10 s), annealing (56 °C, 20 s) and extension (72 °C, 15 s). Fluorescence data collection was performed at the end of each extension phase. Amplification was followed by melting curve analysis using the program run for 1 cycle 95 °C (0 s), 65 °C (10 s), and 95 °C (0 s) in order to confirm the amplification specificity. A negative control without cDNA template was run with every assay to assess the overall specificity and to verify that no primer-dimer was generated. The relative standard curve was established with serial dilution of a cDNA solution with an unknown concentration,which corresponds to a mix of 5 samples randomly picked. Template concentrations using the relative standard curve were given arbitrary values. The mean concentration of GAPDH was used to control for input RNA because it is considered a stable housekeeping gene. The mean GAPDH concentration was determined once for each cDNA sample and used to normalize all other genes tested from the same cDNA sample. The relative change in gene expression was recorded as the ratio of normalized data over the mean value for each gene in SE mice.
4.7. Western blot
For determinations of PKC lambda (78 kDa) protein levels in the striatum, 8 mice that were either reared in SE or EE were sacrificed by decapitation, their brains were removed and striata were immediately dissected as described above. Proteins were extracted according to the protocol of PKC lambda antibody manufacturer (BD Biosciences). Briefly, brain tissues were homogenized in boiling lysis buffer (350 µl for 25mg), containing SDS 1%, sodium ortho-vanadate 1 mM, Tris 10 mM pH 7,4 in distilled water, with 0.05 U aprotinin A, 1 µM pepstatin, 1 µM leupeptin, 0.1 mM benzamidine, 10 nM chloroquin, 10 nM Soybean Trypsin inhibitor, 0.1 mM Nα-Tosyl-l-lysine chloro-methyl ketone hydrochloride, 100 µg/ml Nα-p-Tosyl-l-arginine methyl ester hydrochloride and 0.1 mM PMSF protease inhibitors. After homogenization using ultra turrax for 15 s (IMLAB Laboratories technology, France), samples were boiled for 3 min and centrifuged at full speed for 10 min. Supernatant were collected and stored at −20°C until analysis. Protein samples (about 30 µg) were subjected to SDS-polyacrylamide gel electrophoresis (10% acrylamide/0.27% N,N′-methylenebisacrylamide resolving gel), for 120 min at 90 V. Proteins were transferred to PVDF membranes at 20 V overnight and the blots were blocked for 1 h in 5% non-fat dry milk in TBS-Tween 0.1%. Primary antibody incubation was then performed with rabbit anti-PKC lambda IgG (1:1000) (BD Biosciences) in blocking buffer (5% milk in TBS-Tween 0.1%) overnight. The blots were then washed 3×10 min in blocking buffer and incubated for 120 min in blocking buffer with goat anti-mouse antibody (1:1000) conjugated to horse-radish peroxidase (Bio-Rad Laboratories, France). The blots were washed 2×10 min in blocking buffer and 10 min in TBS, then developed with an enhanced chemiluminescence system (Pierce, France) before being exposed to a Kodak MS Film (VWR, France) for 1min. The blots were subsequently incubated with an anti-beta-actin antibody (1/2000, Sigma-Aldrich Chimie, FRANCE SIGMA). Levels of band density were quantified by densitometry using the ViosioLab 2000 analyser (BIOCOM) and for each sample the results are expressed as level of PKC lambda/level of beta-actin to adjust results according to protein loaded in each well. Care was taken to stay within the linear range of the film.
4.8. Statistics
For real-time RT-PCR and western blot, we used a Student T test to compare SE and EE mice. All analyses were done using the Statview 4.02 program (SAS Institute, Cary, NC). The null hypothesis was rejected at p<0.05.
Acknowledgments
This work was supported by the CNRS, Poitiers University, Fondation pour la Recherche Médicale (2003), MILDT-INSERM (2006–2007), Région Poitou Charentes (2006), and the Intramural Research programs of the DHHS/NIH NIA and NIDA.
Abbreviations
- EE
Enriched environments
- SE
Standard Environment
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- BDNF
Brain-derived neurotrophic factor
- cDNA
complementary DNA
- MAPK
mitogen-activated protein kinase
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- SDS
c-Jun amino-terminal kinase (JNK)
REFERENCES
- Bardo MT, Klebaur JE, Valone JM, Deaton C. Environmental enrichment decreases intravenous self-administration of amphetamine in female and male rats. Psychopharmacology (Berl) 2001;155:278–284. doi: 10.1007/s002130100720. [DOI] [PubMed] [Google Scholar]
- Bennett EL, Rosenzweig MR, Diamond MC. Rat brain: effects of environmental enrichment on wet and dry weights. Science. 1969;163:825–826. doi: 10.1126/science.163.3869.825. [DOI] [PubMed] [Google Scholar]
- Berezovska O, McLean P, Knowles R, Frosh M, Lu FM, Lux SE, Hyman BT. Notch1 inhibits neurite outgrowth in postmitotic primary neurons. Neuroscience. 1999;93:433–439. doi: 10.1016/s0306-4522(99)00157-8. [DOI] [PubMed] [Google Scholar]
- Bezard E, Dovero S, Belin D, Duconger S, Jackson-Lewis V, Przedborski S, Piazza PV, Gross CE, Jaber M. Enriched environment confers resistance to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine and cocaine: involvement of dopamine transporter and trophic factors. J. Neurosci. 2003;23:10999–11007. doi: 10.1523/JNEUROSCI.23-35-10999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin R, Beaudoin J, Bergeron P, Nadeau A, Grondin G. Cell-specific expression of the ZPK gene in adult mouse tissues. DNA Cell Biol. 1996;15:631–642. doi: 10.1089/dna.1996.15.631. [DOI] [PubMed] [Google Scholar]
- Chaouloff F. Physical exercise and brain monoamines: a review. Acta Physiol. Scand. 1989;137:1–13. doi: 10.1111/j.1748-1716.1989.tb08715.x. [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J. Mol. Diagnostics. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lee TH, Wetsel WC, Sun QA, Liu Y, Davidson C, Xiong X, Ellinwood EH, Zhang X. Reversal of cocaine sensitization-induced behavioral sensitization normalizes GAD67 and GABAA receptor alpha2 subunit expression, and PKC zeta activity. Biochem. Biophys. Res. Commun. 2007;356:733–738. doi: 10.1016/j.bbrc.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Stamoudis CX, Irwin SA, Greenough WT. Increased density of multiple-head dendritic spines on medium-sized spiny neurons of the striatum in rats reared in a complex environment. Neurobiol. Learn. Mem. 1996;66:93–96. doi: 10.1006/nlme.1996.0049. [DOI] [PubMed] [Google Scholar]
- Costa RM, Drew C, Silva AJ. Notch to remember. Trends Neurosci. 2005;28:429–435. doi: 10.1016/j.tins.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Costa DA, Cracchiolo JR, Bachstetter AD, Hughes TF, Bales KR, Paul SM, Mervis RF, Arendash GW, Potter H. Enrichment improves cognition in AD mice by amyloid-related and unrelated mechanisms. Neurobiol. Aging. 2007;28:831–844. doi: 10.1016/j.neurobiolaging.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Crary JF, Shao CY, Mirra SS, Hernandez AI, Sacktor TC. Atypical protein kinase C in neurodegenerative disease I: PKMzeta aggregates with limbic neurofibrillary tangles and AMPA receptors in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2006;65:319–326. doi: 10.1097/01.jnen.0000218442.07664.04. [DOI] [PubMed] [Google Scholar]
- Crews F, He J, Hodge C. Adolescent cortical development: a critical period of vulnerability for addiction. Pharmacol. Biochem. Behav. 2007;86:189–199. doi: 10.1016/j.pbb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Diamond MC, Law F, Rhodes H, Lindner B, Rosenzweig MR, Krech D, Bennett EL. Increases in cortical depth and glia numbers in rats subjected to enriched environment. J. Comp. Neurol. 1966;128:117–126. doi: 10.1002/cne.901280110. [DOI] [PubMed] [Google Scholar]
- Diamond MC, Ingham CA, Johnson RE, Bennett EL, Rosenzweig MR. Effects of environment on morphology of rat cerebral cortex and hippocampus. J. Neurobiol. 1976;7:75–85. doi: 10.1002/neu.480070108. [DOI] [PubMed] [Google Scholar]
- Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat. Neurosci. 2005;8:1481–1489. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- Faherty CJ, Raviie Shepherd K, Herasimtschuk A, Smeyne RJ. Environmental enrichment in adulthood eliminates neuronal death in experimental Parkinsonism. Brain Res. Mol. Brain Res. 2005;134:170–179. doi: 10.1016/j.molbrainres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat. Rev., Mol. Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- Girault JA, Valjent E, Caboche J, Herve D. ERK2: a logical AND gate critical for drug-induced plasticity? Curr. Opin. Pharmacol. 2007;7:77–85. doi: 10.1016/j.coph.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Green TA, Gehrke BJ, Bardo MT. Environmental enrichment decreases intravenous amphetamine self-administration in rats: dose-response functions for fixed- and progressive-ratio schedules. Psychopharmacology (Berl) 2002;162:373–378. doi: 10.1007/s00213-002-1134-y. [DOI] [PubMed] [Google Scholar]
- Harrison-Findik D, Misra S, Jain SK, Keeler ML, Powell KA, Malladi CS, Varticovski L, Robinson PJ. Dynamin inhibits phosphatidylinositol 3-kinase in hematopoietic cells. Biochim. Biophys. Acta. 2001;1538:10–19. doi: 10.1016/s0167-4889(00)00130-0. [DOI] [PubMed] [Google Scholar]
- Hebb D. he effects of early experience on problem-solving at maturity. Am. Psychol. 1947;2:306–307. [Google Scholar]
- Hirai S, Kawaguchi A, Suenaga J, Ono M, Cui DF, Ohno S. Expression of MUK/DLK/ZPK, an activator of the JNK pathway, in the nervous systems of the developing mouse embryo. Gene Expr. Patterns. 2005;5:517–523. doi: 10.1016/j.modgep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Hirai S, Cui de F, Miyata T, Ogawa M, Kiyonari H, Suda Y, Aizawa S, Banba Y, Ohno S. The c-Jun N-terminal kinase activator dual leucine zipper kinase regulates axon growth and neuronal migration in the developing cerebral cortex. J. Neurosci. 2006;26:11992–12002. doi: 10.1523/JNEUROSCI.2272-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuvone L, Geloso MC, Dell'Anna E. Changes in open field behavior, spatial memory, and hippocampal parvalbumin immunoreactivity following enrichment in rats exposed to neonatal anoxia. Exp. Neurol. 1996;139:25–33. doi: 10.1006/exnr.1996.0077. [DOI] [PubMed] [Google Scholar]
- Jaber M, Robinson SW, Missale C, Caron MG. Dopamine receptors and brain function. Neuropharmacology. 1996;35:1503–1519. doi: 10.1016/s0028-3908(96)00100-1. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386:493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Kolb B, Gorny G, Li Y, Samaha AN, Robinson TE. Amphetamine or cocaine limits the ability of later experience to promote structural plasticity in the neocortex and nucleus accumbens. Proc. Natl. Acad. Sci. U. S. A. 2003a;100:10523–10528. doi: 10.1073/pnas.1834271100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb B, Gorny G, Soderpalm AH, Robinson TE. Environmental complexity has different effects on the structure of neurons in the prefrontal cortex versus the parietal cortex or nucleus accumbens. Synapse. 2003b;48:149–153. doi: 10.1002/syn.10196. [DOI] [PubMed] [Google Scholar]
- Laviola G, Hannan A, Macrì S, Solinas M, Jaber M. Effects of enriched environment on animal models of neurodegenerative diseases and psychiatric disorders. Neurobiol Brain Dis. (Advance online publication) doi: 10.1016/j.nbd.2008.05.001. in press. [DOI] [PubMed] [Google Scholar]
- Lazarov O, Robinson J, Tang YP, Hairston IS, Korade-Mirnics Z, Lee VM, Hersh LB, Sapolsky RM, Mirnics K, Sisodia SS. Environmental enrichment reduces Abeta levels and amyloid deposition in transgenic mice. Cell. 2005;120:701–713. doi: 10.1016/j.cell.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Li C, Niu W, Jiang CH, Hu Y. Effects of enriched environment on gene expression and signal pathways in cortex of hippocampal CA1 specific NMDAR1 knockout mice. Brain Res. Bull. 2007;71:568–577. doi: 10.1016/j.brainresbull.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Lu L, Koya E, Zhai H, Hope BT, Shaham Y. Role of ERK in cocaine addiction. Trends Neurosci. 2006;29:695–703. doi: 10.1016/j.tins.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Mora F, Segovia G, del Arco A. Aging, plasticity and environmental enrichment: structural changes and neurotransmitter dynamics in several areas of the brain. Brain Res. Rev. 2007;55:78–88. doi: 10.1016/j.brainresrev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Nithianantharajah J, Hannan AJ. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat. Rev. Neurosci. 2006;7:697–709. doi: 10.1038/nrn1970. [DOI] [PubMed] [Google Scholar]
- Nygren J, Wieloch T, Pesic J, Brundin P, Deierborg T. Enriched environment attenuates cell genesis in subventricular zone after focal ischemia in mice and decreases migration of newborn cells to the striatum. Stroke. 2006;37:2824–2829. doi: 10.1161/01.STR.0000244769.39952.90. [DOI] [PubMed] [Google Scholar]
- Oster H, Eichele G, Leitges M. Differential expression of atypical PKCs in the adult mouse brain. Brain Res. Mol. Brain Res. 2004;127:79–88. doi: 10.1016/j.molbrainres.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Pinaud R, Penner MR, Robertson HA, Currie RW. Upregulation of the immediate early gene arc in the brains of rats exposed to environmental enrichment: implications for molecular plasticity. Brain Res. Mol. Brain Res. 2001;91:50–56. doi: 10.1016/s0169-328x(01)00121-8. [DOI] [PubMed] [Google Scholar]
- Pinaud R, Tremere LA, Penner MR, Hess FF, Robertson HA, Currie RW. Complexity of sensory environment drives the expression of candidate-plasticity gene, nerve growth factor induced-A. Neuroscience. 2002;112:573–582. doi: 10.1016/s0306-4522(02)00094-5. [DOI] [PubMed] [Google Scholar]
- Poon HF, Vaishnav RA, Getchell TV, Getchell ML, Butterfield DA. Quantitative proteomics analysis of differential protein expression and oxidative modification of specific proteins in the brains of old mice. Neurobiol. Aging. 2006;27:1010–1019. doi: 10.1016/j.neurobiolaging.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev., Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Rampon C, Jiang CH, Dong H, Tang YP, Lockhart DJ, Schultz PG, Tsien JZ, Hu Y. Effects of environmental enrichment on gene expression in the brain. Proc. Natl. Acad. Sci. U. S. A. 2000a;97:12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C, Tang YP, Goodhouse J, Shimizu E, Kyin M, Tsien JZ. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat. Neurosci. 2000b;3:238–244. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Copland JA, Thompson EA, Fields AP. Atypical protein kinase Ciota plays a critical role in human lung cancer cell growth and tumorigenicity. J. Biol. Chem. 2005;280:31109–31115. doi: 10.1074/jbc.M505402200. [DOI] [PubMed] [Google Scholar]
- Robles Y, Vivas-Mejia PE, Ortiz-Zuazaga HG, Felix J, Ramos X, Pena de Ortiz S. Hippocampal gene expression profiling in spatial discrimination learning. Neurobiol. Learn. Mem. 2003;80:80–95. doi: 10.1016/s1074-7427(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Ronnback A, Dahlqvist P, Svensson PA, Jernas M, Carlsson B, Carlsson LM, Olsson T. Gene expression profiling of the rat hippocampus one month after focal cerebral ischemia followed by enriched environment. Neurosci. Lett. 2005;385:173–178. doi: 10.1016/j.neulet.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Rosenzweig MR. Environmental complexity, cerebral change, and behavior. Am. Psychol. 1966;21:321–332. doi: 10.1037/h0023555. [DOI] [PubMed] [Google Scholar]
- Rosenzweig MR, Bennett EL. Effects of differential environments on brain weights and enzyme activities in gerbils, rats, and mice. Dev. Psychobiol. 1969;2:87–95. doi: 10.1002/dev.420020208. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisk CL, Zehr JL. Pubertal hormones organize the adolescent brain and behavior. Front. Neuroendocrinol. 2005;26:163–174. doi: 10.1016/j.yfrne.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Solinas M, Thiriet N, El Rawas R, Lardeux V, Jaber M. Environmental enrichment during early stages of life reduces the behavioral, neurochemical and molecular effects of cocaine. Neuropsychopharmacology. (Advance online publication) doi: 10.1038/npp.2008.51. in press. [DOI] [PubMed] [Google Scholar]
- Spires TL, Grote HE, Varshney NK, Cordery PM, van Dellen A, Blakemore C, Hannan AJ. Environmental enrichment rescues protein deficits in a mouse model of Huntington's disease, indicating a possible disease mechanism. J. Neurosci. 2004;24:2270–2276. doi: 10.1523/JNEUROSCI.1658-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, III, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet N, Ladenheim B, McCoy MT, Cadet JL. Analysis of ecstasy (MDMA)-induced transcriptional responses in the rat cortex. Faseb J. 2002;16:1887–1894. doi: 10.1096/fj.02-0502com. [DOI] [PubMed] [Google Scholar]
- Thiriet N, Deng X, Solinas M, Ladenheim B, Curtis W, Goldberg SR, Palmiter RD, Cadet JL. Neuropeptide Y protects against methamphetamine-induced neuronal apoptosis in the mouse striatum. J. Neurosci. 2005;25:5273–5279. doi: 10.1523/JNEUROSCI.4893-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH. Neural consequences of environmental enrichment. Nat. Rev. Neurosci. 2000;1:191–198. doi: 10.1038/35044558. [DOI] [PubMed] [Google Scholar]
- Young D, Lawlor PA, Leone P, Dragunow M, During MJ. Environmental enrichment inhibits spontaneous apoptosis, prevents seizures and is neuroprotective. Nat. Med. 1999;5:448–453. doi: 10.1038/7449. [DOI] [PubMed] [Google Scholar]