Abstract
The quantity and activity of mitochondria vary dramatically in tissues and are modulated in response to changing cellular energy demands and environmental factors. The amount of mitochondrial DNA (mtDNA), which encodes essential subunits of the oxidative phosphorylation complexes required for cellular ATP production, is also tightly regulated, but by largely unknown mechanisms. Using murine T cells as a model system, we have addressed how specific signaling pathways influence mitochondrial biogenesis and mtDNA levels. T cell receptor (TCR) activation results in a large increase in mitochondrial mass and membrane potential and a corresponding increase of mtDNA copy number, indicating the vital role for mitochondrial function for the growth and proliferation of these cells. Independent activation of protein kinase C (via PMA) or calcium-related pathways (via ionomycin) had differential and sub-maximal effects on these mitochondrial parameters, as did activation of naïve T cells with proliferative cytokines. Thus, the robust mitochondrial biogenesis response observed upon TCR activation requires synergy of multiple downstream signaling pathways. One such pathway involves AMP-activated protein kinase (AMPK), which we show has an unprecedented role in negatively regulating mitochondrial biogenesis that is mammalian target of rapamycin (mTOR)-dependent. That is, inhibition of AMPK after TCR signaling commences results in excessive, but uncoordinated mitochondrial proliferation. We propose that mitochondrial biogenesis is not under control of a master regulatory circuit, but rather requires the convergence of multiple signaling pathways with distinct downstream consequences on the organelle’s structure, composition, and function.
Keywords: keywords: mitochondria, mtDNA, biogenesis, protein kinase C, calcium, signaling, AMP-activated protein kinase, mTOR, rapamycin, ionomycin, membrane potential, oxidative phosphorylation, T cell, cytokine, T cell receptor
1. Introduction
Mitochondria are essential organelles in eukaryotic cells that produce ATP via the process of oxidative phosphorylation (OXPHOS). In addition, mitochondria have many other key roles in metabolism and actively participate in important cellular processes such as apoptosis (Kroemer and Reed, 2000; Danial and Korsmeyer, 2004; Pinton et al., 2007), calcium homeostasis (Rizzuto et al., 2000; Winslow et al., 2003), signal transduction (Butow and Avadhani, 2004; Kelly and Scarpulla, 2004) and antiviral defenses (Seth et al., 2005; Yi et al., 2006). Critical for their function is the mitochondrial DNA (mtDNA), which in mammals is a ~16-kb circular molecule that encodes thirteen essential OXPHOS subunits, as well as twenty-two rRNAs and two rRNA required for their translation in the mitochondrial matrix (Bonawitz et al., 2006). However, the majority of the ~1,500 resident mitochondrial proteins is a product of nuclear genes that are translated by cytoplasmic ribosomes and imported into the organelles. These include not only the other metabolic enzymes and housekeeping proteins, but also the remaining ~70 subunits of the OXPHOS complexes (that assemble with those encoded by mtDNA) and factors required for expression and replication of the mitochondrial genome (Bonawitz et al., 2006). Therefore, the modulation of mitochondrial function in general and to meet tissue-specific demands requires signaling pathways that orchestrate crosstalk between the nucleus and mitochondria. Pathways that influence mitochondrial biogenesis and function have been described in yeast and more recently in mammalian cells (Wu et al., 2002; Butow and Avadhani, 2004; Kelly and Scarpulla, 2004), yet how specific signals alone or together modulate these important parameters remains largely uncharacterized. Given that mitochondria are increasingly implicated in common human disorders and age-related pathology (e.g. heart disease, Parkinson’s disease, and diabetes), understanding these pathways is critical in order to decipher the full impact of mitochondrial dysfunction in human disease and aging, which can result from mutations in mtDNA or in nuclear genes that influence mitochondrial function (Wallace, 1999, 2005).
Previous studies have implicated specific signaling pathways in the regulation of mitochondrial biogenesis in response to a variety of stimuli (Moyes et al., 1997; Wu et al., 1999; Wu et al., 2002; Zong et al., 2002; Butow and Avadhani, 2004; Akimoto et al., 2005; Scarpulla, 2006). For example, mitochondrial proliferation is increased by activation of AMP-activated kinase (AMPK) via energy depletion (Zong et al., 2002), c-MYC in response growth and cell cycle cues (Li et al., 2005), and by CaM kinase IV in response to chronic exercise (Wu et al., 2002). Ultimately, signaling from these upstream kinases usually converge on key downstream transcriptional activators and co-activators such as NRF-1, NRF-2, PGC-1α, PRC-1, and CREB that, in turn, activate transcription of nuclear genes encoding mitochondrial OXPHOS subunits and transcription factors (e.g. mtTFA, mtTFB1 and mtFB2), as well as other mitochondrial proteins needed to increase mitochondrial mass, gene expression and function (Kelly and Scarpulla, 2004; Wu et al., 2006). What remains unclear, however, is how specific signaling pathways coordinate the many individual aspects of the mitochondrial biogenesis response or whether there are different biogenesis scenarios that can be initiated to attain specific amounts and/or functional states of mitochondria to meet changing cellular needs. For example, the mitochondrial genome in mammals is usually present at 1000–10,000 copies per cell and is regulated in a tissue-specific manner, presumably representing one mechanism to tailor mitochondrial function to meet specialized cellular needs (Veltri et al., 1990; Moraes, 2001). While, in general, there is good correlation between the amount of mtDNA and the number of organelles in various cell types, whether mtDNA and overall mitochondrial biogenesis are under control of distinct signaling pathways and factors has not been addressed systematically. Although, in yeast and mammals modulation of mtDNA copy number can be achieved through alteration of the cellular dNTP pool via activation of the enzyme ribonucleotide reductase (Eaton et al. 2007; Lebedeva and Shadel, 2007; Taylor et al., 2005). Thus, it remains possible that altering mtDNA copy number or expression in the absence of a biogenesis response may represent a novel regulatory scenario in mammalian cells.
T cells of the immune system orchestrate adaptive responses against foreign agents such as viruses and bacteria. Quiescent T cells stimulated by an antigen via the T cell receptor (TCR) undergo a significant increase in cell size followed by a rapid rate of proliferation and differentiation. These processes require a tremendous amount of energy and hence, upon TCR activation, T cells immediately undergo a rapid increase in mitochondrial mass and OXPHOS (Frauwirth and Thompson, 2004) and then switch onto aerobic glycolysis for their energy needs (Fox et al., 2005). Accordingly, it has been shown that mitochondrial respiration is required for CD8+ T cell function and interestingly also for ERK phosphorylation after TCR stimulation (Yi et al., 2006). Several of the pathways downstream of the TCR are implicated in mitochondrial biogenesis and function, including the protein kinase C (PKC) pathway, the MAP kinase/ERK pathways, and calcium-related pathways such as those mediated by CAM kinases and calcineurin (Cantrell, 1996). Interestingly, in the case of PKC, this may even involve the translocation of isoforms of the enzyme into mitochondria (Wang et al., 2006). T cells also can respond to different cytokines via specific receptors. For example, proliferative cytokines like IL-2, IL-7 and IL-15 are important for homeostatic proliferation of T cells in the absence of antigen to maintain a constant T cell number, especially in the case of memory T cells (Geginat et al., 2001). The signaling pathways that are downstream of the TCR and cytokine receptors are relatively well-characterized and T cells purified from spleen readily undergo TCR- or cytokine-mediated activation, proliferation and differentiation ex vivo. Therefore, in this study, we used murine T cells as a model system to investigate signaling pathways that regulate mitochondrial biogenesis, activity, and mtDNA copy number and the degree to which these processes are coordinately controlled.
2. Methods
2.1 Preparation of mouse splenic T cells
Spleens were isolated from 8–12 week old C57BL/6 mice (purchased from The Jackson Laboratories). Using a wire mesh screen, spleens were disaggregated to generate a liquid suspension of splenocytes. Contaminating red blood cells were lysed osmotically in 0.83% NH4Cl buffer for 2 minutes. Splenocytes were then collected by centrifugation and resuspended in MACS buffer (0.5% BSA, 2 mM EDTA in 1x PBS) containing Thy1.2-labeled magnetic beads (100 μl beads/10 million splenocytes in 1 ml MACS buffer). This mixture was incubated at 4°C for 30 minutes to allow binding, washed with 20 volumes MACS buffer, resuspended at a concentration of 100 million spenocytes/500 μl MACS buffer, and then loaded onto a pre-equilibrated MACS column in a MACS magnetic separator. Unbound splenocytes were separated from magnetically bound Thy1.2 T cells by washing as described by the MACS protocol. The purified T cells were then eluted from the column and resuspended in RPMI 1640 supplemented with 10% FCS, 2 mM L-glutamine, penicillin-streptomycin, and 50 μM 2-β-mercaptoethanol. All preparations of T cell obtained by this protocol were 90–95% pure, as ascertained by FACS staining using FITC-labeled Thy1.2 antibodies.
2.2. Activation of mouse T cells in vitro and FACS analysis of mitochondrial mass and membrane potential
For anti-CD3/CD28 stimulation experiments, 96-well flat bottom plates were coated with 10 μg/ml anti-CD3 and anti-CD28 antibodies in 1x PBS overnight at 4°C. For all stimulations, 2 × 105 purified murine splenic T cells in 200 μl complete RPMI medium were seeded in each well. For stimulations with phorbol myristate acetate (PMA) and the Ca2+ ionophore, ionomycin (IONO), the drugs were added at 50 ng/ml and 500 ng/ml, respectively, in complete RPMI medium. IL-2, IL-7 and IL-15 were used at a concentration of 10 pg/ml for cytokine stimulations. The cytokines were added every 24 hours during the experiment. For inhibitor studies, compound C (AMPK inhibitor) and rapamycin (mTOR inhibitor) were used at 10 μM and 250 ng/ml, respectively. The drugs were added 12 hours post-stimulation of the murine T cells with anti-CD3/CD28 antibodies. For the no-inhibitor control samples, the drug vehicle (DMSO) was added at the same time at the maximum volume that was used for a single inhibitor in each experiment.
For FACS staining, 0.2–1×106 T cells were incubated with 100 nM or Mitotracker Green FM or Mitotracker Red (both from Molecular Probes) in complete RPMI medium and incubated at 37°C for 15 minutes. The cells were then washed three times with PBS containing 1% FCS, resuspended in 200 μl of 1x PBS containing 1% FCS. For surface staining of T cells with anti-CD25-FITC antibodies, cells were incubated with the antibody at 1:1000 dilution in FACS buffer (1% FCS, 1x PBS) for 20 minutes at 4°C, and washed three times with FACS buffer. Samples resuspended in FACS buffer were acquired on a FACS Calibur flow cytometer and analyzed using FloJo software (5,000–10,000 cells were acquired for analyses). For analyzing the FACS data, a “live” cell gate based on FSC vs. SSC plots was used. The median fluorescence intensity (MFI) values were determined for the cells in the live gate at all time points. The mean of the MFI of the unstimulated (or no-inhibitor) control samples was then used to determine the fold change in MFI during different stimulations and inhibitor treatments. Therefore, fold change in MFI refers to ratio of MFI of stimulated/inhibited sample to mean of MFI of the unstimulated or no-inhibitor control T cells at the same time point. All experiments were done in triplicate and carried out at least three times.
2.3. Analysis of cell proliferation by CFSE staining
Purified T cells were resuspended in 1x PBS containing 0.1% FCS at 1×107 cells/ml and stained with 1 μM CFSE (1 mM CFSE in DMSO) for 8 minutes in the dark at room temperature. Next, the cells were incubated at 37°C in equal volume of complete RPMI medium for 15 minutes, washed three times with complete RPMI medium, and resuspended in complete RPMI medium for FACS analysis.
2.4 Analysis of mtDNA copy number
Total cellular DNA from purified T cells was isolated as follows: 2 × 105 T cells were suspended in 250 μl of T cell extraction buffer (50 mM Tris-pH 8.0, 1 mM EDTA, 5 mM DTT, and 0.25% SDS), vortexed, boiled for 10 minutes, and then left to cool to room temperature in a water bath. Then 2.5 μl of RNaseA (10 mg/ml) was added and the sample was incubated at 37°C for 2 hours. Next, 2.5 μl of proteinase K (10 mg/ml) was added and the sample was incubated at 55°C for 12 hours. The sample was then boiled for 10 minutes, following which DNA was precipitated by adding 0.1 volume of 3M sodium acetate and 2 volumes of 100% ethanol and incubating for 12 hours. The DNA was pelleted in a micro-centrifuge, washed twice with 70% ethanol, dried, resuspended in 250 μl of ddH20, and incubated at room temperature overnight prior to performing the PCR reactions.
The ratio of mtDNA copy number per nuclear DNA was determined using real time, quantitative PCR analysis. For each sample analyzed, two independent PCR reactions were setup to amplify the COXI mtDNA target and the 18S rRNA nuclear DNA target. Twenty-five μl reaction volumes contained 14 μl of BioRad iQTM Sybr Green Supermix, 0.5 μL of a 25 μM stock of each primer, and 10 μl of template genomic DNA prepared as described above. DNA samples were diluted 10–20-fold and a 2-fold dilution series was run for each sample to insure accurate sample profiling and linearity. The PCR protocol consisted of 50°C for 2 minutes, 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds, 60°C annealing/extension for 1 minute with real-time data collection. Melt curves from 55°C to 95°C with 0.5°C increments every 10 seconds were included to confirm that a single PCR product was being analyzed, which was verified in early reactions by running products on 2% agarose gels. The primer sets used are as follows: 18S 1546 F: 5′-TAGAGGGACAAGTGGCGTTC-3′; 18S 1650 R: 5′-CGCTGAGCCAGTCAGTGT-3′; mouse COI F: 5′-GCCCCAGATATAGCATTCCC-3′; mouse COI R: 5′-GTTCATCCTGTTCCTGCTCC-3′. The reported fold changes in relative mtDNA copy number for each stimulation with or without inhibitor refers to ratio of relative mtDNA copy number for stimulation with or without inhibitor to relative mtDNA copy number of unstimulated cells, the latter of which was given a value of 1.0. All experiments were carried out in triplicate at all time point and repeated at least three times.
2.5 Analysis of markers of AMPK and mTOR pathway activation by Western immunoblotting
Whole cell protein extracts were prepared as follows: 1×107 cells were lysed in 50 μl cold lysis buffer [50mM Tris-HCL pH 8.8, 150 mM NaCl, 0.5% Tween-20, 0.5% Triton-X 100, 2 mM EDTA, 10% glycerol, 1X complete mini, EDTA free protease inhibitor cocktail (Roche Diagnostics), 1x phosphatase inhibitor cocktail II (AG Scientific) at 4°C for 30 minutes. The resulting cell lysate was centrifuged at 13,000 rpm for 10 minutes and the soluble extract was analysed after the protein concentration were determined using a BioRad protein assay kit.
Protein (10 μg) prepared as described above was separated on 8% SDS-polyacrylamide gels and then transferred electrophoretically to PVDF membrane. The membrane was then blocked with 5% BSA/TBST (10mM Tris-HCl pH 8.0, 150 mM NaCl, 0.05% Tween 20) for 30 minutes and then incubated with the desired primary antibody (in 5% BSA/TBST) overnight at 4°C. The blot was rinsed five times with 1X TBST for 5 minutes and then incubated with the corresponding secondary antibody (5% BSA/TBST) at 4°C for 1 hour. The blot was then washed five times for 10 minutes with TBST and the cross-reacting proteins were visualized using the western lighting TM chemiluminescence reagent plus kit (Perkin Elmer LAS, Inc) to expose x-ray film. Exposed films were developed and imaged with BioRad VersaDoc using Quantity One software. For serial western blots, the membrane was stripped using Restore TM western blot stripping buffer (Pierce) and reprobed as described above.
The antibodies used for the Western analysis were as follows: acetyl-CoA carboxylase or ACC (#3662), pSer79 ACC (#3661), p70 S6 kinase (#9202) and pThr389-p70 S6 kinase (Thr 389, #9205S) were purchased from Cell Signaling. Mouse monoclonal antibody to Tubulin alpha Ab-2 (DM1A) was obtained from Neomarkers. Hsp-60 (N-20) sc-1052 was a goat polyclonal IgG from Santa Cruz Biotechnology. All primary antibodies were used at 1:2000 dilution. HRP-conjugated secondary antibodies, donkey anti-rabbit IgG (sc-2313) and donkey anti-goat (sc-2020) were purchased from Santa Cruz Biotechnology and were used at 1:10,000 dilution.
3. Results
3.1 Activation of TCR signaling in purified murine T lymphocytes leads to a robust increase in mitochondrial biogenesis
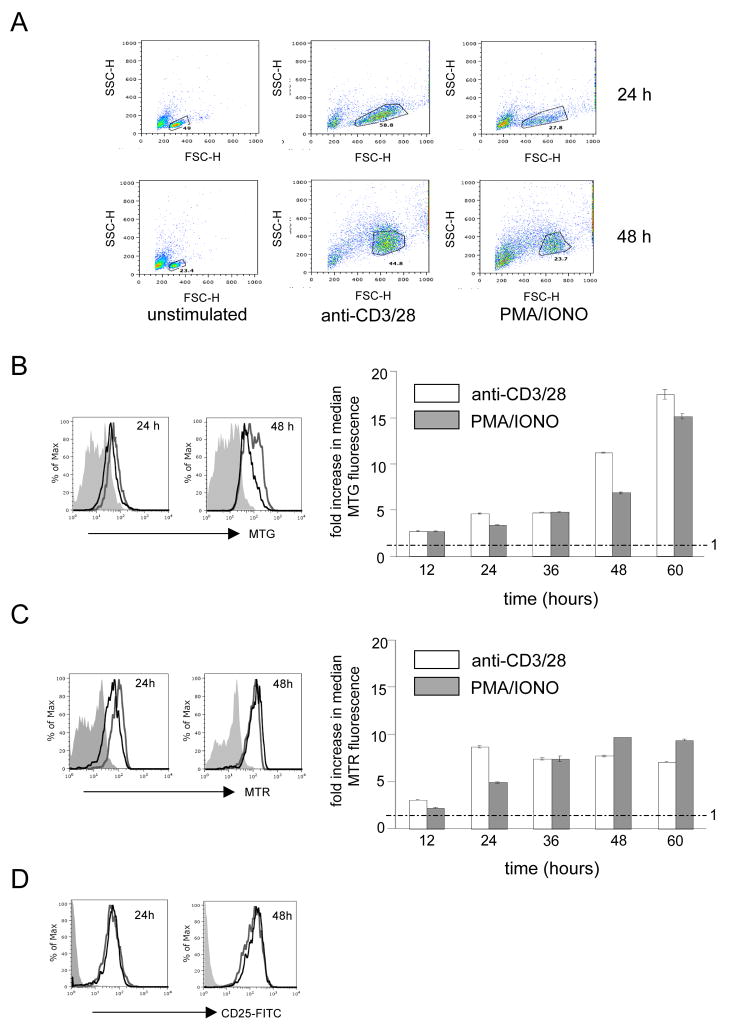
Activation of TCR signaling in T cells isolated from spleen is routinely achieved by cross-linking the CD3 and CD28 surface receptors using antibodies or by treating the cells with a combination of phorbol 12-myristate 13-acetate (PMA, a potent activator of protein kinase C) and the calcium ionophore ionomycin (IONO). These treatments mimic in many ways the activation of T cell proliferation and differentiation elicited by antigenic stimulation. Using these activation regimens we stimulated purified mouse splenic Thy1.2 T cells (CD4+ and CD8+) and measured the resulting changes in mitochondrial mass and membrane potential via FACS analysis after staining with Mitotracker Green FM and Mitotracker Red-CMXROS stains, respectively. In the case of anti-CD3/CD28 antibody-mediated stimulation, we observed typical cell blasting (i.e., increased cell size; Fig. 1A) by 24 hours and a steady increase in mitochondrial mass between 12–60 hours post-stimulation (Figure 1B), characterized by an early increase of ~2.5 fold after 12 hours of stimulation that eventually reached ~17-fold after 60 hours (Figure 1B). Mitochondrial membrane potential was increased by ~2.5 fold after 12 hours of stimulation and ~8-fold after 24 hours, but, unlike mitochondrial mass, did not show any further increase after 24-hours (Figure 1C). A very similar response, but with slightly delayed kinetics with regard to these mitochondrial parameters, was observed when the cells were stimulated by PMA/IONO (Figure 1A–1C). In both cases, the cells were activated (as judged by increased CD25 surface staining; Figure 1D) and proliferated between 24 and 48 hours (as measured by dilution of CFSE staining; Supplemental Figure 1).
Figure 1. Up-regulation of mitochondrial mass and mitochondrial potential during TCR stimulation of murine splenic T cells.
A. Representative FACS plots showing comparative FSC vs. SSC profiles of unstimulated, anti-CD3/28 and PMA/IONO stimulated T cells at 24 and 48 hours, respectively. The “live cell” gates used for analyses are shown. B. FACS analysis of Mitotracker green (MTG) staining for mitochondrial mass. C. FACS analysis of Mitotracker Red (MTR) staining for mitochondrial membrane potential. D. FACS analysis of CD25 staining, a marker for activated T cells. For B.–D. representative histograms comparing the fluorescence intensity (increasing from left to right as indicated by the arrow) of unstimulated (filled gray), anti-CD3/28 stimulated (open gray) and PMA/IONO stimulated (open black) cells at 24 and 48h are shown. For B. and C., a bar graph is also shown to the right that summarizes the fold-increase in the mean fluorescence intensity (MFI) during anti-CD3/28 (open) and PMA/IONO (filled gray) stimulations at 12, 24, 36, 48 and 60h relative to the unstimulated control sample. All plots show the mean+/− SD (brackets) calculated from triplicate samples. The dotted line across the graph denotes the mean MFI value of unstimulated cells, which was given a value of 1.0. Hence, a fold change of 1.0 indicates no effect.
3.2 Increasing intracellular calcium with ionomycin induces a moderate mitochondrial biogenesis response in the absence of cell proliferation, while activation of protein kinase C with PMA does not
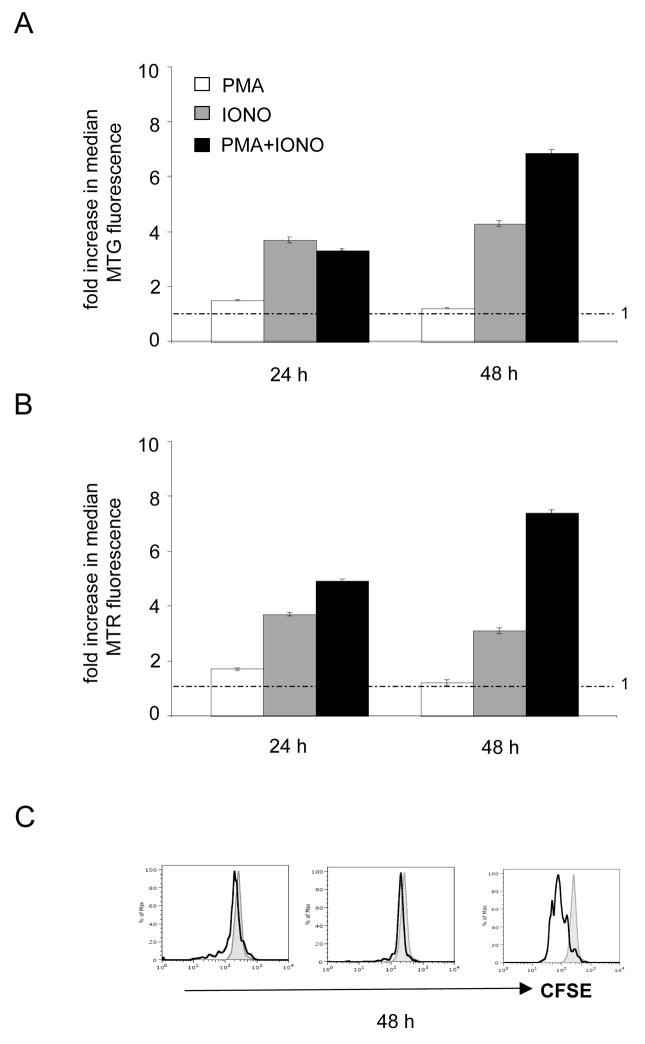
While the addition of PMA and IONO are synergistically required to obtain an activated and proliferating state of isolated T cell in vitro, these two stimuli activate largely separate signaling pathways. Therefore, we determined whether either of these drugs influences mitochondrial biogenesis or activity on their own in the absence of cell proliferation. The addition of PMA alone had no effect on mitochondrial biogenesis or membrane potential, however, IONO alone promoted a steady, but moderate (~2–4-fold) increase in mitochondrial mass (Figure 2A) and membrane potential (Figure 2B). As expected neither of the individual drug treatments resulted in cell proliferation, while addition of both did (Figure 2C). The fact that the increases provided by IONO alone was substantially lower than that observed when both drugs were added (Figures 1 and 2) suggests that, while PMA alone has no measurable effect, there is synergy between the two pathways and/or that cell proliferation is required to obtain a full mitochondrial biogenesis response.
Figure 2. Synergy between PMA-activated and ionomycin activated calcium-related signaling pathways is required for maximal mitochondrial biogenesis in response to T cell activation.
FACS analysis of T cells stimulated with PMA, ionomycin (IONO) or both is shown at the time point after stimulation indicated. A. Analysis of Mitotracker green (MTG) staining for mitochondrial mass. B. Analysis of Mitotracker green (MTG) staining for mitochondrial membrane potential. C. Analysis of CFSE staining at 72 hours post-stimulation. Reduced staining is an indicator of cell division.
3.3 Mitochondrial biogenesis and mtDNA copy number are coordinately regulated during T cell activation, but can be uncoupled
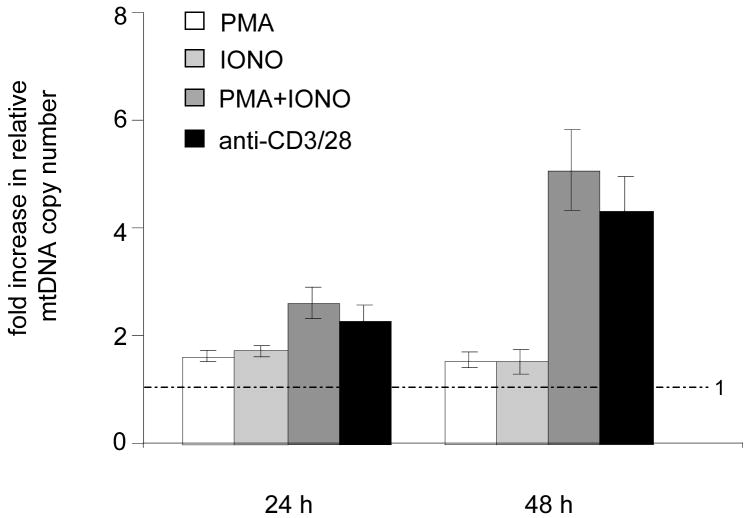
One of the primary questions we sought to address in this study is the degree to which mitochondrial biogenesis and mtDNA replication are coupled. Therefore, we measured mtDNA copy number using a real-time PCR assay under the various T cell activation scenarios described thus far. This assay measures the relative amount of mtDNA and nuclear DNA (see Materials and Methods). In response to anti-CD3/CD28 or PMA/IONO stimulation, we observed a 2-fold increase in mtDNA at 24 hours that increased further to ~4–5-fold at 48 hours (Figure 3), changes that are roughly proportional to those observed in mitochondrial mass under the same circumstances (Figures 1 and 2). Addition of PMA alone or IONO alone increased mtDNA copy number 1.5–2-fold at 24 hours, but they did not increase it further (Figure 3). The increase in mtDNA in the case of PMA is noteworthy given that there was not a corresponding increase in mitochondrial mass when it is added alone (Figure 2A). Even more striking, addition of IONO alone resulted in a significantly greater increase in mitochondrial mass (~4-fold), than in mtDNA copy number (1.5–2-fold). Taken together, these results indicate that mtDNA replication (and/or stability) is coordinately increased with overall mitochondrial biogenesis, but that these parameters can be uncoupled to a significant degree through the stimulation of individual signaling pathways (or by influencing cellular calcium levels).
Figure 3. Up-regulation of mtDNA copy number upon TCR stimulation also requires multiple synergistic pathways.
A real-time PCR assay that measures the amount of mtDNA relative to nuclear DNA was employed in activated mouse T cells. Plotted is the fold increase in the relative mtDNA copy number (mtDNA/nuclear DNA ratio) in T cells stimulated by PMA alone (open bars), ionomycin (IONO) alone (filled light gray), PMA and IONO (filled dark gray), and anti-CD3/CD28 (filled black) at 24 and 48 hours post-stimulation. All plots show mean +/− SD (brackets) calculated from triplicate samples. The dotted line across the graph denotes the mean relative mtDNA copy number of unstimulated control cells, which were given a value of 1.0.
3.4 Cytokine-stimulated T cell proliferation results in modest mitochondrial biogenesis and mtDNA copy number increases that are accentuated differentially in the presence ionomycin and PMA
Two questions arose from the results obtained this far. The first was, is cell proliferation per se the reason that both PMA and IONO are required for a full biogenesis response or are multiple signaling pathways downstream of the TCR needed for other reasons? And, the second was, does stimulation of T cell via proliferative cytokines result in a similar mitochondrial biogenesis response or does TCR activation represent a scenario where substantial up-regulation of mitochondrial biogenesis is needed? To address each of these questions, we stimulated T cells using proliferative cytokines instead of a TCR signal.
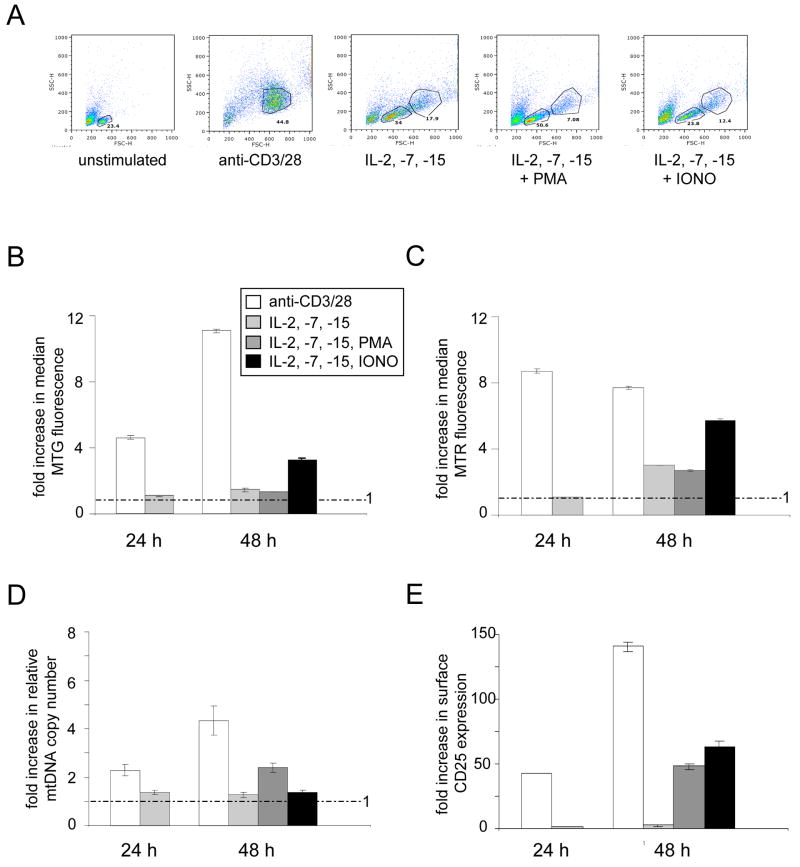
We added a cocktail of IL-2, IL-7 and IL-15 to naïve splenic T cells and analyzed those cells that were activated (blasted and stimulated to divide, Figure 4A) for mitochondrial mass, membrane potential, and mtDNA copy number. Cytokine stimulation resulted in modest increases in mitochondrial mass or membrane potential (~2–2.5 fold at 48 hours post stimulation; Figure 4B and 4C) and no significant increase in mtDNA (Figure 4D). Thus, cytokine-induced cell proliferation does not stimulate mitochondrial biogenesis or activity to nearly the same extent as TCR activation (also shown in Figure 4B–4D for comparison). We next wanted to determine if we could accentuate mitochondrial biogenesis in cytokine-stimulated cells by activating PKC with PMA or increasing cellular calcium via IONO. In this experiment, T cells were activated by adding the cytokine cocktail for 24-hours, then adding PMA or IONO, and measuring mitochondrial parameters after 24 more hours (i.e. the 48-hour time point). As was the case in purified T cells to which IONO alone was added (Figure 2), addition of IONO to the cytokine-stimulated T cells resulted in an increase in mitochondrial mass (Fig. 4A) and membrane potential (Figure 4B), but not mtDNA copy number (Fig. 4C). However, in contrast, addition of PMA alone to the cytokine-stimulated cells resulted in a 2-fold increase in mtDNA copy number (Figure 4C), but no significant changes in mitochondrial mass (Figure 4A) or membrane potential (Figure 4C). These results indicate that cell proliferation per se is not a major stimulus for mitochondrial biogenesis in T cells and that separate pathways or downstream targets influence mtDNA copy number and mass. Specifically they suggest that activation of the PKC pathway has a role in increasing mtDNA replication (and/or stability) and that calcium-related pathways influence changes in mitochondrial mass.
Figure 4. Stimulation of naïve T cell proliferation with cytokines results in minimal mitochondrial biogenesis that can be activated differentially by addition of PMA or ionomycin.
A. Representative FACS plots showing comparative FSC vs. SSC profiles of naïve T cells either unstimulated or stimulated for 48 hours as indicated with anti-CD3/CD28 or a cocktail of the cytokines IL-2, IL-7 and IL-15 (with or without PMA or IONO). PMA and IONO were added after 24 hours of stimulation with the cytokines, therefore, the 48-hour time point is 24 hours after drug addition. These were analyzed for mitochondrial biogenesis and activity and mtDNA copy number and are presented exactly as described Figures 1 and 3. The inset in B. also corresponds to the data presented in C–E. B. FACS analysis of Mitotracker green (MTG) staining for mitochondrial mass. C. FACS analysis of Mitotracker Red (MTR) staining for mitochondrial membrane potential. D. Analysis of relative mtDNA copy number. E. FACS analysis of CD25 staining, a marker for activated T cells.
3.5 Inhibition of AMPK results in mTOR-dependent over-proliferation of mitochondria without a concomitant increase in mtDNA
Thus far, our studies identified calcium-related and PKC as pathways downstream of the TCR that impact unique aspects of mitochondrial biogenesis. However, we wanted to also begin to examine the role of additional pathways in the regulation of this robust biogenesis response. One pathway that has been implicated in mitochondrial function according to cellular energy status is the AMP-activated protein kinase (AMPK) pathway (Zong et al., 2002). Since it was shown previously that AMPK is activated within minutes after TCR stimulation (Tamas et al., 2006), we first pre-incubated naïve T cells with the AMPK inhibitor compound C and activated TCR signaling. Inhibitor pre-treatment resulted in a complete lack of cell blasting and proliferation (presumably due to a G1/S cell cycle arrest, data not shown), which impeded further analysis of the role of this pathway in mitochondrial biogenesis in T cells. Western blot analyses confirmed that T cells inhibited with the concentrations of compound C used resulted in decreased phosphorylation of a known AMPK substrate acetyl-CoA carboxylase (ACC) (Supplementary Figure 2). Similar results were obtained with the mTOR inhibitor rapamycin and its substrate p70-S6 kinase (Supplementary Figure 2, and data not shown), which were analyzed in parallel given that mTOR is a known regulator of AMPK (Shaw et al. 2004; Arsham and Neufeld, 2006).
We next analyzed the kinetics of AMPK and mTOR activation over a longer time course of TCR activation by following the phosphorylation of ACC and p70-S6 kinase (Fig. 5A). Based on the analysis of these markers, there was a down-regulation of AMPK activation between 0–4 hours after TCR activation, followed by a period of increased activity that peaked between 12–18 hours (Fig. 5A; ACC pSer79 signal). In contrast, there was an immediate (within 2 hours) increase in mTOR activity that steadily increased to a peak that occurred between 12 and 18 hours after TCR stimulation (Fig. 5A; S6k pThr389 signal). Under these same conditions, there was also significant increase in Hsp60, a mitochondrial chaperone, by 8 hours after TCR activation, verifying the increase in mitochondrial biogenesis we had already documented using Mitotracker Green staining (Fig. 1).
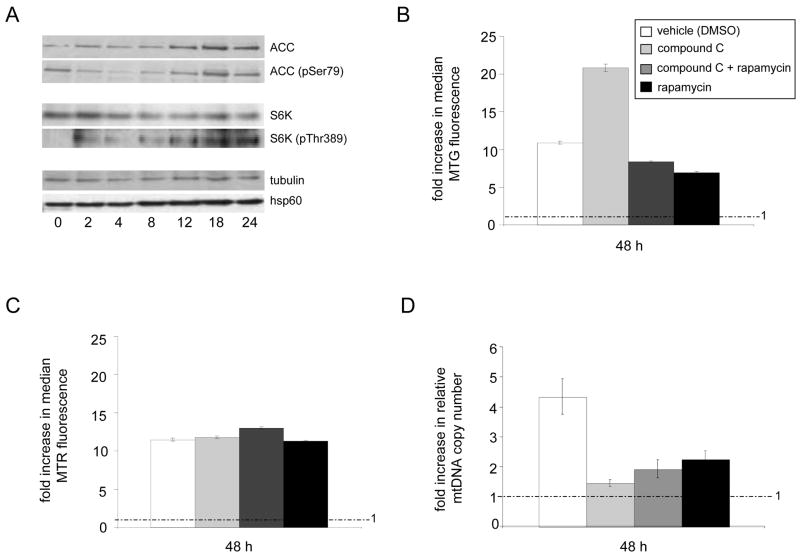
Figure 5. Differential and overlapping regulation of mitochondrial mass and mtDNA copy number by AMPK and mTOR.
A. Western blots of acetyl-CoA carboxylase (ACC; a AMPK phosphorylation substrate) and p70-S6K (S6K; a mTOR phosphorylation substrate) at the indicated time (in hours) after TCR stimulation using anti-CD3/CD28 antibodies. T cells were stimulated for 12 hours in the presence (+) or absence (−) of the AMPK inhibitor (compound C, 10 μM), the mTOR inhibitor (rapamycin, 250 nM), or both. Western blots were probed using antibodies specific for the non-phosphorylated (ACC or S6K) or specific phosphorylated (ACC-pSer79 or S6K-pThr389) forms of the target substrates. Tubulin was probed as a control for protein loading and normalization. Analysis of Hsp60, a mitochondrial chaperone and marker of total mitochondrial biogenesis, is also shown. B–D. Analysis of mitochondrial mass (B), membrane potential (C) and mtDNA copy number (D) of TCR-activated T cells in the presence (+) and absence (−) of compound C, rapamycin, or both is shown. The inset in B. also corresponds to the data presented in C. and D. The FACS and mtDNA copy number results are shown as bar graphs as described in Figures 1 and 3.
Based on the observed kinetics of AMPK and mTOR activation we determined in activated T cells (Fig. 5A), we employed a new strategy to examine the role of AMPK in mitochondrial biogenesis upon TCR activation. That is, instead of pre-incubating with the AMPK and mTOR inhibitors, we first stimulated the T cells with anti-CD3/CD28 antibodies for 12 hours, followed by addition of the inhibitors (alone or in combination) and analysis of mitochondrial parameters 36 hours later (i.e. at the 48-hour time point after TCR stimulation). Compared to drug-vehicle treated (uninhibited) control T cells, we observed a substantial over-accumulation of mitochondria (based on Mitotracker Green staining) without a concomitant increase in mitochondrial membrane potential when AMPK was inhibited ((Figure 5B and C). In addition, AMPK inhibition completely prevented the increase in mtDNA copy number normally observed in response to TCR stimulation (Figure 5D). Given that AMPK has been shown to negatively regulate the target of rapamycin (mTOR) pathway in other cell types (Shaw et al., 2004; Arsham and Neufeld, 2006), we determined whether the over-activation of mitochondrial biogenesis observed as the result of AMPK inhibition was affected by mTOR by adding rapamycin. This treatment completely abolished the over-accumulation of mitochondria caused by AMPK inhibition (Figure 5B). In addition, rapamycin alone inhibited the TCR-mediated increase in mtDNA copy number (Figure 5D) and mitochondrial mass (Figure 5B), but had no effect on membrane potential (Figure 5C). Altogether, these data demonstrate that the intersection of the AMPK and mTOR signaling pathways impinge on the mitochondrial biogenesis response of TCR-activated T cells and elucidate an unprecedented role for AMPK in negatively regulating mitochondrial biogenesis.
4. Discussion
In this study, we used mouse T cells as a model system to further define signaling pathways required for mitochondrial biogenesis and to understand the dynamic relationship between mitochondrial mass, membrane potential, and mtDNA copy number during a robust mitochondrial biogenesis response. The main conclusion we draw from our results is that mitochondrial biogenesis involves simultaneous inputs from multiple signaling pathways that each contribute to specific aspects of the response. This is best illustrated by our results showing that mitochondrial mass, membrane potential, and mtDNA copy number, while normally coordinately increased upon T cell activation, can be uncoupled from each other through the use of different stimulation regimens. Finally, our study illuminates individual and overlapping roles for AMPK and mTOR signaling in the regulation of mitochondrial mass and mtDNA copy number. The basis for these conclusions is discussed below.
Activating purified splenic T cells using either of two common methods, cross-linking the TCR and co-stimulatory receptors with anti-CD3 and anti-CD28 antibodies or addition of PMA/IONO, results in a significant increase in both mitochondrial mass and membrane potential (Figure 1B and 1C). In general, there is a good correlation between these two parameters, however, early in the response (24 hours post-stimulation) the membrane potential is activated to a greater extent than mitochondrial mass, which indicates that not only are there more mitochondria, but that they likely have increased OXPHOS activity. Interestingly, the cells continue to increase mitochondrial mass throughout the response (Figure 1A) even after the cells have begun to proliferate, while the membrane potential does not continue to increase as substantially (Figure 1C). Our interpretation of these results is that upon TCR stimulation, T cells first up-regulate both mitochondrial mass and OXPHOS activity to provide a sustained energy output needed to dramatically increase cell size and cytoplasmic content and to prepare for a high rate of cell proliferation. Later in the response it is postulated that these cells primarily generate ATP via glycolysis (Frauwirth and Thompson, 2004; Fox et al., 2005), which is perhaps why the membrane potential does not need to continue to increase as dramatically. However, mitochondrial biogenesis does continue and mitochondrial membrane potential is maintained, suggesting that there is a need for increased number of active organelles during the rapid proliferative state or for subsequent differentiation, perhaps for reasons in addition to OXPHOS-derived ATP production. Here it is noteworthy that mitochondrial electron transport has been implicated in ERK signaling in T cells under similar activation conditions (Yi et al., 2006), which might indicate that mitochondria are also playing a signaling role in this context. Finally, we observed a steady increase in mtDNA copy number in activated T cells (Figure 3), indicating a coordinate up-regulation of mitochondrial mass and mtDNA replication in response to TCR signaling.
To begin to dissect the pathways responsible for mitochondrial biogenesis we activated purified T cells with PMA or IONO alone, which activates PKC (and subsequently other downstream pathways like MAPK/ERK) or calcium-related pathways (e.g. calcineurin and CaMK pathways), respectively, but does not result in cell proliferation (Figure 2). We draw the following conclusions from these results. First, given that neither PMA nor IONO alone result in responses in any of the individual mitochondrial parameters to the same degree as adding both simultaneously, synergy of these pathways and signals is needed to achieve a full mitochondrial biogenesis response. Second, activation of PKC or calcium-related pathways can each increase mtDNA replication/stability to a small extent, but in isolation only the calcium-related signal can substantially increase mitochondrial mass. And third, that mtDNA copy number and mitochondrial mass can be uncoupled to a significant degree, as evidenced by an increase in mtDNA without a corresponding increase in mitochondrial mass in the case of PMA stimulation alone and a disproportional increase in mitochondrial mass compared to mtDNA in the case of IONO stimulation alone (Fig. 2A and 3).
To address whether the maximal increase in all mitochondrial parameters seen with anti-CD3/CD28 or PMA/IONO, but not with PMA or IONO alone, was due to cooperative downstream signaling from the TCR or the induction of cell proliferation, we stimulated naive T cells with proliferative cytokines to induce similar changes in cell size and proliferation in the absence of a TCR-based signal. Based on the fact that there is minimal, if any increase in mitochondrial mass (Figure 4B) or mtDNA copy number (Figure 4D) under these conditions, we conclude that cell proliferation per se is not a major stimulus for overall mitochondrial biogenesis in T cells. However, the 2–3 fold increase in mitochondrial membrane potential is consistent with the heightened energy requirements for cell division being met at, least in part, by increasing mitochondrial OXPHOS activity independently of an increase the number of organelles. These results suggest that it is indeed synergy between the signaling pathways downstream of the TCR that lead to the full biogenesis response in the anti-CD3/CD28 and PMA/IONO stimulations. This conclusion is bolstered by the observation that, when added to cytokine-stimulated cells, IONO results in an increase in mitochondrial mass (Figure 4B) and PMA results in an increase in mtDNA (Figure 4D), and each increased mitochondrial potential (Figure 4C). However, again none of these treatments resulted in increases of same magnitude as a TCR response, which suggests that maximal synergy of these pathways is only achieved through TCR signaling. Since cytokines act as important differentiation and survival signals for naïve, activated, and memory T cells in vivo, our results raise the intriguing possibility that the degree of mitochondrial biogenesis and activity, which is markedly different in TCR-activated versus cytokine-activated T cells, is an important determinant of T cell fate and longevity.
Our results with PMA and IONO support a role for collaboration of PKC activation and calcium-related pathways in promoting mitochondrial biogenesis. However, other signaling pathways have also been implicated in mitochondrial biogenesis, including those mediated by AMPK, which senses cellular energy charge and activates mitochondrial biogenesis under certain conditions (Zong et al., 2002). Furthermore, AMPK is involved in early TCR signaling events in T cells that bridge calcium-related pathway with the nutrient-sensing mTOR pathway (Shaw et al., 2004; Tamas et al., 2006). Therefore, we also examined the potential involvement of AMPK in regulating the mitochondrial biogenesis response by inhibiting it in the context of TCR activation. Here, our results were perhaps surprising. That is, we observed a marked over-accumulation of mitochondria (Figure 5A), suggesting that AMPK has a negative influence on mitochondrial biogenesis. This heightened (or unabated) biogenesis response was completely dependent on mTOR kinase activity (Figure 5B), consistent with mTOR being downstream of AMPK in this pathway. Despite the enhanced mitochondrial biogenesis, AMPK inhibition did not result in greater membrane potential (Figure 5C) and completely inhibited the typical ~4-fold increase in mtDNA copy number induced by anti-CD3/CD28 (Figure 5D). Thus, not only does AMPK have a negative role in the control of mitochondrial mass, it has an opposing positive influence on mtDNA replication/stability. Furthermore, these results firmly establish that mitochondrial mass and mtDNA copy number can be completely uncoupled from each other, indicating that different pathways or downstream effectors modulate these two parameters. Altogether, we interpret these results to indicate that AMPK normally tempers the mitochondrial biogenesis response though its influence on mTOR signaling and also has a positive role in enhancing mtDNA replication in conjunction with increased mitochondrial mass. The fact that inhibition of mTOR signaling alone results in inhibition of mitochondrial mass and mtDNA copy number (Figure 5B and 5D) is consistent with mTOR having a positive influence on these parameters in the context of TCR activation that is negatively regulated by AMPK. However, the fact that AMPK alone prevents up-regulation of mtDNA copy number suggests that this effect is not through inhibition of mTOR or that AMPK has both positive and negative influences on mitochondrial biogenesis as described by others (Zong et al., 2002).
It is important to note that we observe maximal activation of AMPK 18 hours after TCR activation (Fig. 5A), which is well after the initial wave of AMPK activation that occurs within minutes (Tamas et al., 2006). Since we added the inhibitors after 12 hours of TCR stimulation, it follows that it is inhibition of this “second wave” of AMPK activation that results in over-accumulation of mitochondria. This leaves open the interesting possibility that AMPK may both initiate a mitochondrial biogenesis response early during TCR activation and provide a negative signal to cap mitochondrial biogenesis later in the response.
In conclusion, based on our results in T cells, we propose that mitochondrial biogenesis is not under control of a single “master” regulatory circuit, but rather is a multi-faceted response involving many signals and overlapping pathways. This is evident not only from this study, but many others that have implicated a multitude conserved signaling pathways that activate mitochondrial biogenesis and activity in different cell types (Moyes et al., 1997; Wu et al., 1999; Moraes, 2001; Wu et al., 2002; Zong et al., 2002; Butow and Avadhani, 2004; Kelly and Scarpulla, 2004; Akimoto et al., 2005; Li et al., 2005; Scarpulla, 2006; Wu et al., 2006). During T cell activation this not only involves synergistic influence from PKC and calcium-related pathways, but also complex regulation by the AMPK and mTOR pathways. Our results also indicate that separate downstream targets increase mitochondrial mass and mtDNA copy number. This is logical given that the building blocks required for these processes are quite distinct. We recently demonstrated that modulation of cellular deoxynucleotide pools via the enzyme ribonucleotide reductase is a key parameter that influences mtDNA copy number in yeast and mammals (Eaton et al. 2007; Lebedeva and Shadel, 2007; Taylor et al., 2005). Whether expression or activity of this enzyme is modulated by the signaling pathways described herein to influence mtDNA copy number remains in important area of future investigation. That the mRNAs for ribonucleotide reductase subunits are potential targets of multiple signaling pathways including PKC/ERK is interesting in this regard (Burton et al., 2003). It is also clear that mitochondrial biogenesis usually involves the up-regulation of nuclear transcription factors and co-activators such as NRF1, NRF2, PGC1-α, and PRC-1 (Scarpulla, 2006), which in turn increase the expression and activity of key nucleus-encoded factors required for mitochondrial OXPHOS, gene expression, and mtDNA replication (Bonawitz et al., 2006). Almost certainly, a subset of these factors is targeted by the signaling pathways that are influencing mitochondrial parameters in T cells delineated in this study. However, given that the signaling pathways that control mitochondrial biogenesis and activity are likely cell-type specific, it will of great interest to determine precisely which downstream modulators of mitochondrial function are activated in T cells to evoke such a robust mitochondrial biogenesis response. Understanding this complex process in multiple systems will be necessary to understand how mitochondrial dysfunction results in its stereotypical tissue-specific pathology and how these pathways might be modulated as a therapeutic strategy for mitochondrial diseases and age-related pathology. The study of the exceptional mitochondrial biogenesis response in murine T lymphocytes should continue to be a useful and genetically amenable model system in this regard.
Supplementary Material
Acknowledgments
This work was supported by Program Project Grant ES-011163 awarded to G.S.S. We wish to thank Drs. Bing Su and Gerald Shulman for providing antibodies used in this study, Weiming Ouyang and Rich Resnick for helpful advice, and Dr. Sharen McKay for many useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–93. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–97. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–25. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Burton TR, Kashour T, Wright JA, Amara FM. Cellular signaling pathways affect the function of ribonucleotide reductase mRNA binding proteins: mRNA stabilization, drug resistance, and malignancy (Review) Int J Oncol. 2003;22:21–31. [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Cantrell DA. T cell antigen receptor signal transduction pathways. Cancer Surv. 1996;27:165–75. [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Eaton JS, Lin Z, Sartorelli AC, Bonawitz ND, Shadel GS. J Clinical Investigation. 2007 doi: 10.1172/JCI31604. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–52. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, Thompson CB. Regulation of T lymphocyte metabolism. J Immunol. 2004;172:4661–5. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- Geginat J, Sallusto F, Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4(+) T cells. J Exp Med. 2001;194:1711–9. doi: 10.1084/jem.194.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–68. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Reed JC. Mitochondrial control of cell death. Nat Med. 2000;6:513–9. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- Lebedeva M, Shadel GS. Cell cycle- and ribonucleotide reductase-driven changes in mtDNA copy number influence mtDNA inheritance without compromising mitochondrial gene expression. Cell Cycle. 2007;6 doi: 10.4161/cc.6.16.4572. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- Moyes CD, Mathieu-Costello OA, Tsuchiya N, Filburn C, Hansford RG. Mitochondrial biogenesis during cellular differentiation. Am J Physiol. 1997;272:C1345–51. doi: 10.1152/ajpcell.1997.272.4.C1345. [DOI] [PubMed] [Google Scholar]
- Pinton P, Rimessi A, Marchi S, Orsini F, Migliaccio E, Giorgio M, Contursi C, Minucci S, Mantovani F, Wieckowski MR, Del Sal G, Pelicci PG, Rizzuto R. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the lifespan determinant p66Shc. Science. 2007;315:659–63. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Bernardi P, Pozzan T. Mitochondria as all-round players of the calcium game. J Physiol 529 Pt. 2000;1:37–47. doi: 10.1111/j.1469-7793.2000.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97:673–83. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–9. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Tamas P, Hawley SA, Clarke RG, Mustard KJ, Green K, Hardie DG, Cantrell DA. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J Exp Med. 2006;203:1665–70. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O’Rourke TW, Siede W, Shadel GS. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:3010–8. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri KL, Espiritu M, Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 1990;143:160–4. doi: 10.1002/jcp.1041430122. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–8. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Biswas G, Prabu SK, Avadhani NG. Modulation of mitochondrial metabolic function by phorbol 12-myristate 13-acetate through increased mitochondrial translocation of protein kinase Calpha in C2C12 myocytes. Biochem Pharmacol. 2006;72:881–92. doi: 10.1016/j.bcp.2006.06.032. [DOI] [PubMed] [Google Scholar]
- Winslow MM, Neilson JR, Crabtree GR. Calcium signalling in lymphocytes. Curr Opin Immunol. 2003;15:299–307. doi: 10.1016/s0952-7915(03)00050-5. [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–52. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- Wu Z, Huang X, Feng Y, Handschin C, Feng Y, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci U S A. 2006;103:14379–84. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Yi JS, Holbrook BC, Michalek RD, Laniewski NG, Grayson JM. Electron transport complex I is required for CD8+ T cell function. J Immunol. 2006;177:852–62. doi: 10.4049/jimmunol.177.2.852. [DOI] [PubMed] [Google Scholar]
- Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–7. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.