Abstract
Nutritional genomics offers a way to optimize human health and the quality of life. It is an attractive endeavor, but one with substantial challenges. It encompasses almost all known aspects of science, ranging from the genomes of humans, plants and microorganisms, to the highest levels of food science, analytical science, computing and statistics of large systems, as well as human behavior. The underlying biochemistry that is targeted by the principal issues in nutritional genomics is described and entails genomics, transcriptomics, proteomics and metabolomics. A major feature relevant to nutritional genomics is the single nucleotide polymorphisms in genes that interact with nutrients and other bioactive food components. These genetic changes may lead to alterations in absorption, metabolism and functional responses to bioactive nutritional factors. Bioactive food components may also regulate gene expression at the transcriptome, protein abundance and/or protein turnover levels. Even if all of these variables are known, additional variables to be taken into account include the nutritional variability of the food (unprocessed and processed), the amount that is actually eaten, and the eating-related behaviors of those consuming the food. These challenges are explored within the context of soy intake. Finally, the importance of international co-operation in nutritional genomics research is presented.
Keywords: nutritional genomics, nutritional genomics, human nutrition, gene variant, genetic variation, dietetics
Introduction
Although many people have become familiar with the term genomics, being the flagship of science at the National Institutes of Heath at the transition into the 21st century, other –omics have also been introduced in our vocabulary. Perhaps naïvely, it was assumed by many in the 1980s and 1990s that if the human genes could be defined, then the causes of diseases and syndromes could be understood sufficiently well to develop strategies (gene replacement/repair, optimal therapeutics or lifestyle changes involving what we eat) that would improve human health. When the sequencing approaches adopted by the National Human Genome Research Institute (1) and latterly Celera (2) only yielded 20,000–24,000 genes instead of the 80,000–100,00 that were expected, it caused many investigators to rethink how the cell really operates. Besides genomics, many other –omics, transcriptomics, metabolomics, physiological genomics, proteomics, epigenomics, and now nutritional genomics, have been born. Most recently, data from the ENCODE (ENCyclopedia Of DNA Elements) project have suggested that the concept of a gene and the resulting transcriptome may require substantial revision (3,4). The goal of the ENCODE project is to identify all the functional elements in the human genome sequence. Results reported in June 2007 revealed that the majority of the genome is transcribed, including non-protein encoding regions, and that genes extensively overlap each other (3).
This review describes the underlying biochemistry, introduces the variation in the human genome and the role of nutrition associated with nutritional genomics, and presents the role of timing of specific dietary components on which parts of the genome are expressed. As an example, the role of dietary polyphenols in health is discussed from a nutritional genomics and dietetics point of view. Finally, the importance of the national and international efforts that are in progress to deliver the fruits of this new field are briefly discussed. For overviews of the impact of nutritional genomics on dietetics, see Afman and Müller (5) and Trujillo et al. (6).
The flow of biochemical information
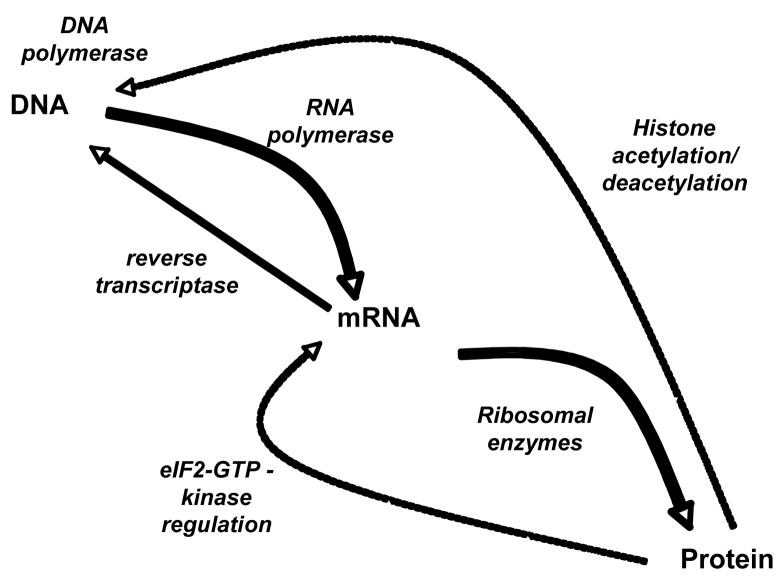
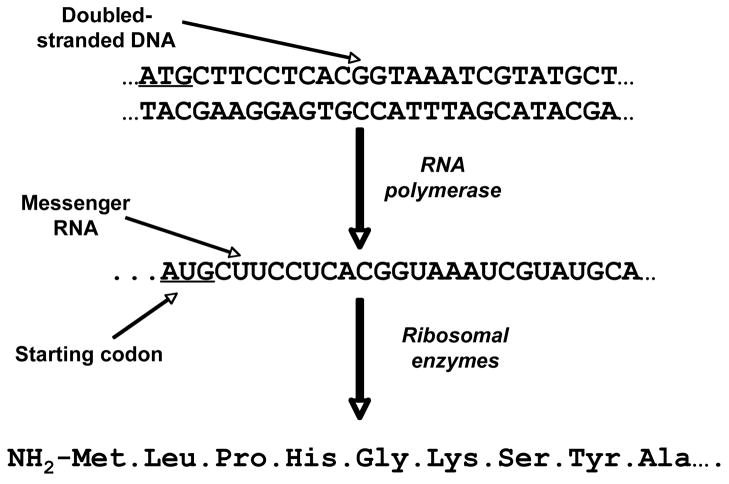
In the classical biochemical paradigm, genetic information encoded in genes in DNA (deoxyribonucleic acid) is transcribed by RNA (ribonucleic acid) polymerases to form messenger RNAs (the transcriptome) (Fig. 1). Recent research in the ENCODE project has revealed that, rather than information being drawn from one gene, the transcribed RNA may be a product of more than one adjacent gene (7). The mRNAs are exported from the nucleus and form complexes with ribosomes; these ribonuclear proteins synthesize polypeptides using the mRNAs as templates (Fig. 1). The triplet codon sequence of the mRNA is translated into amino acids one at a time to form polypeptides (Fig. 2). The polypeptides fold to form proteins with a wide variety of properties that are essential for life. These include cytoskeletal structures (actin, tubulin, etc.), membrane transporters, and enzymes that utilize externally derived compounds (glucose, amino acids, fats) to generate energy to power the cell and to synthesize biochemically important intermediates (e.g., amino acids, Coenzyme A thioesters, deoxyribonucleotides and ribonucleotides) needed for the synthesis of the materials required for cell division (that include DNA and RNA) (Fig. 1). Although for multicellular organisms the total available genes are the same for each cell, only a fraction of them is expressed to create the cellular phenotype (the effective genome of that cell type over the life of the cell). Of these expressed genes, while many are converted to proteins (structural proteins, enzymes for intermediary metabolism) that are used throughout the lifetime of a cell, others that are needed to control the timing of the cell cycle are only present transiently. The set of proteins recovered at any moment in the life of a cell is termed the proteome.
Figure 1. Information flow between nucleic acids and proteins.
The principal pathway is from DNA to mRNA to proteins (the bold lines) and is mediated by a bevy of proteins including RNA polymerase and those of the translational apparatus. However, it is well appreciated that enzymes control the synthesis of both DNA and RNA (their control is represented by the dotted lines). Information flows from RNA to DNA by the action of reverse transcriptase. Kinases regulate the phosphorylation status of the elongation-initiation factor 2-guanosine triphosphate (eIF2-GTP) complex. Large scale regulation of the genes that are expressed in a cell at a particular time in our lifetime is determined by the post-translational modification of histones, basic proteins that bind tightly to DNA. Deacetylation of histones leads to the suppression of gene expression.
Figure 2. Translation of DNA sequence information via mRNA to synthesis of a specific polypeptide.
DNA is double stranded, each strand being the mirror image of the other [the image is created by matching cytosine (C) with guanosine (G) and adenosine (A) with thymidine (T) and vice versa]. The information in the double stranded DNA is transcribed using RNA polymerase to form a mRNA copy [note that while the C/G, G/C and T/A copies are upheld as in the DNA pairs, T is transcribed to uridine (U)]. The AUG codon on mRNA is the start codon (it also encodes methionine). The mRNA nucleotide sequence is converted to an amino acid sequence by “reading” of a series of contiguous triplet codons (three successive nucleotides). Most, but not all, amino acids are encoded by more than one triplet codon.
A complex set of small molecules in a cell represents its metabolome. The metabolome can be measured in a cell, in tissues (brain, heart, kidney, liver, muscle, ovary, testis, etc.), or in biological fluids (serum/plasma, urine, bile, etc.). It is a function of the genes available in the genome, the expressed genes in the transcriptome, the transporters that move extracellular and intracellular compounds across the cell membrane, and the catalytic activity and organization of the enzymes within a cell. The metabolome is constantly changing. Maintaining the elements of the metabolome within certain ranges is called homeostasis. For some compounds the range that is normal is very narrow, e.g., intracellular ATP (adenosine triphosphate). For others, e.g., plasma glucose, it is broader. However, for glucose there is a lower limit at which point various mechanisms are called into play to supply glucose and maintain homeostasis. As glucose levels rise after a meal, signals from insulin released from the pancreas serve to increase glucose transport into cells and to store it as glycogen and fat. Persistent high levels of plasma glucose lead to the complications of diabetes mellitus due to low levels of insulin secretion or failure to respond to insulin.
Humans use food as the fuel to run their metabolic engines–the food keeps the lights on and allows the metabolic system to create the pieces that sustain a cell so it can perform its necessary functions, and for undifferentiated cells to undergo division. Is there a particular set of foods needed to sustain metabolism? Fortunately, the answer is no. Foods consist of many different polymers of carbohydrates, fats and proteins. These are broken down to their sugar, fatty acid and amino acid monomeric units through hydrolysis by saccharidases, lipases, and proteases released into the intestine from the pancreas. The value of a food can vary. In order to be optimal, the food should generate sufficient glucose and provide essential fatty acids and amino acids. It is unusual for one food to satisfy all these requirements; thus a diet built on a variety of food sources is usually necessary. For some foods, external fermentation (pre-digestion by microorganisms) is used to make a food more bioavailable or palatable. Milk is converted to cheese and yogurt, whereas soybeans are converted to soy sauce, miso, tempeh and soy paste. An advantage of fermentation can be a shift of the amino acid content towards a more optimal value due to the addition of proteins from the microorganisms to the food.
Besides saccharides, fatty acids and amino acids, the human body has requirements for other essential compounds in the diet including water, vitamins and minerals. Other bioactive food components, many of which are phytochemicals (plant chemicals), are being identified as important to human health. Thus, the complete diet is a complex matrix of food components often supplemented with herbals, botanicals and dietary supplements containing bioactive components and vitamins/minerals. Delivering that complex matrix is the challenge for the dietitian who not only determines the nutrients that need to be combined to make a healthy diet, but also formulates methods based on knowledge of several sciences that ensure that the individual foods are safe and palatable (within the standards of the community in which they operate).
Genetic variation in human beings
So, if humans have a given set of genes, why aren’t we all the same? And if we all ate a healthy diet, why aren’t we all uniformly healthy? The answer to both questions is that, although Homo sapiens is a discrete species, there are differences in our individual genomes. In the case of well-described single-gene disorders, there are distinct mutations, deletions and additions in certain genes that lead to the absence or dysfunction of the proteins derived from them. Since each parent contributes one copy of every gene (an allele) to their children, in most cases (but not always) both alleles have to be dysfunctional in order for the associated condition to become manifest.
In addition, there are other site-specific differences that occur throughout the genome, on average every 300 bases. These are termed single nucleotide polymorphisms (SNPs). Even if they are in the open-reading frame that is converted to messenger RNA and translated into a protein, the differences may be silent if they do not lead to a change in the amino acid that is translated from each codon. For instance, if the codon for glycine shown in Figure 2 (GGU) is mutated at the third nucleotide to form GGA, GGC or GGG, all the mutations will still be translated as glycine. On the other hand, for lysine (AAA in Figure 2), while mutation to AAG has no effect, mutation to AAU or AAC results in a change in translation to asparagine. This amino acid remains hydrophilic, but it loses the charge possible on the lysine residue. The outcome may be to alter rather than destroy the enzyme activity of the protein. Mutations of the first or second nucleotides in a triplet codon almost always cause a change in the amino acid that is translated. Gene polymorphisms may also occur in non-coding DNA regions that regulate expression of a gene. If the mutation stops gene expression, the outcome is severe and may result in classification as a genetic disease. However, a mutation that modulates (i.e., causing non-zero gene expression) the change in expression of a gene may be much harder to detect since its effects may fall within what is regarded as physiologically normal.
Proteins have a wide range of sizes from ~50 amino acids (e.g., insulin) to >2,500 (e.g., fatty acid synthase – accession number P49327 at http://www.expasy.org). These proteins are encoded by genes having 150 to >7,500 nucleotides. Accordingly, some genes have no SNPs, whereas others have several. For individuals with SNPs in multiple genes, the number of SNP combinations is restricted – the observed blocks of SNPs are termed haplotypes, combinations of alleles at multiple linked loci that are transmitted together. The pattern of SNPs is a reflection of genetic heritance, and certain haplotypes may be typical of those located within a particular region. Since the Y-chromosome in males is present in only one copy, it is not subject to shuffling via recombination and the pattern of SNPs therein establishes the paternal bloodline and the geographic origins of an individual’s patrilineal ancestors.
As noted above, SNPs typically lead to altered function of the protein product rather than severe impairment or total loss of function. Therefore, the population of a country may carry a high preponderance of a particular SNP. Of course, with the intermixing of populations that has occurred from migration, new SNPs may be introduced into a population. An example of a gene with a well-known SNP relevant to nutrition and disease is the gene that encodes the enzyme methylenetetrahydrofolate reductase (MTHFR) (accession number P42898 at http://www.expasy.org). This gene has a polymorphism at residue 677 (C or T) resulting in an alanine (677C) or valine (677T). Thus, an individual may carry a CC, CT/TC, or TT genotype. The TT genotype is common in northern China (20%), southern Italy (26%) and Mexico (32%) (8). Its frequency is low in those of African ancestry (9). MTHFR is important in the remethylation of homocysteine to methionine. It catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. Unlike many of the other mutations in MTHFR, the 677TT genotype lowers, but does not abolish, enzymatic activity. Thus, individuals with this genotype have a mild form of hyperhomocysteinemia since they have less efficient conversion of homocysteine to methionine than the common genotype. Alcohol consumption also increases total plasma homocysteine. Both the MTHFR 677TT genotype and alcohol effects on plasma homocysteine can be offset by increasing the intake of folate (10). Interestingly, 677TT individuals have a lowered risk of colon cancer if they have a folate-supplemented diet (11). This polymorphism exemplifies how knowledge of the disadvantages of a specific genotype can be prevented by adjustment in nutrient intake. This association, and the subsequent interventions, is described as nutrient-gene interaction and is part of the field of study of nutritional genomics. It provides an explanation for why a nutritional recommendation that is optimal for a large group, may not benefit an individual in the group.
The nutrient-dependent transcriptome
In the above scenario, the mutations in the coding regions of the gene of interest are the dominant issue. However, as noted in Fig. 1, there are feedback mechanisms from the distal part of the gene-transcript-protein pathway. A mutant protein may lead to alteration of the way the expression of genetic information is regulated. Similarly, small molecules produced by concerted enzyme action in the various pathways of the cell can feed back to regulate the activity of the protein and also gene expression via hormonally-sensitive receptors. For example, endogenously produced steroids and eicosanoids interact with a variety of receptors that, once activated, alter the expression of a large number of relevant genes. Receptor activation is not restricted to endogenous compounds; some ligands are derived directly from the diet. Plant-derived estrogens (phytoestrogens), such as genistein, coumestrol and zearalenone bind to the estrogen receptor (12) and may switch on a similar set of genes such as 17β-estradiol, the physiologic estrogen (13–15). Thus the diet becomes an important regulator of gene expression with the potential to offset the effects of the body of SNPs that an individual may have. The matrix in which the phytoestrogen is delivered may also have a role in gene expression. Su et al. (16) showed that the effects of soy protein isolate (containing the isoflavones daidzein, genistein and glycitein and their β-glycosides as well as other soy matrix components) and genistein on gene expression in rat mammary epithelial cells were essentially independent. Therefore, some of the benefits of a phytochemical may be lost if it is removed from its usual matrix.
Microarray analysis is used to assess changes in the transcriptome. While microarray technology has improved substantially over the past 10 years, it still suffers from being too expensive to overcome the major limitation of its use – the small number of replicates versus the number of parameters (genes) being tested. Under the best analytical circumstances, the number of expected differences between a control and treatment group under the null hypothesis (i.e., there is no difference) with α set at 0.05 for an array where there are 10,000 features (genes) is 500. This issue is not peculiar to microarray analysis – it also applies to proteomic and metabolomic analyses. Early publications of DNA microarray data focused on genes whose expression changed (up or down) 2-fold. These parameters may have led to selecting for genes whose expression changes considerably, but without biological importance (17). The key issue is to have sufficient biological replicates that the variances can be calculated. Another way of looking at microarray data is to determine whether a whole pathway is affected, which may help to identify where a critical gene or its gene product is located.
Another way to sort out likely true-positives is to map the observed changes onto the metabolic, synthetic and signaling pathways in the cell. A group of similar changes in the same pathway may be reasonably regarded as significant. An important feature of microarray research is the standardization and recording of every aspect of an experiment (18). By following guidelines laid down for such research (the MIAME [Minimum Information About a Microarray Experiment] standards) (19), investigators can download publicly available microarray data (National Center for Biotechnology Information Gene Omnibus) (20) from experiments that are similar to the ones they are planning in order to calculate the statistical power needed for their experiment. Similarly, statistical tests of the genes and pathways changed by a nutrient can be strengthened by combining data sets. This approach has not yet come into practice in proteomics and metabolomics, but presumably this will occur just as it has for microarray analysis.
Timing of the nutrient-gene interaction
A feature of growth and development is the programmed switching on and switching off of sets of genes that are necessary at different stages of life. Not all genes in a cell are “on” at any one time. When they are switched off, the intention is often that they stay that way. The on/off expression of a gene is regulated by the extent to which it is bound to chromatin in the nucleus. Nuclear DNA is packaged by histone proteins into a complex. The “openness” of the complex (to allow RNA polymerase to transcribe a gene) is regulated by acetylation (to relax chromatin) and methylation (to bind the DNA more tightly). Since there are several modification sites on histones, the concept of a histone code to augment that of gene transcription and translation has been proposed (21). Recent evidence suggests that nutrients/phytochemicals can alter the degree of histone acetylation. Administration of the sulphoraphane present in Brassica (broccoli) to human embryonic kidney cells and to human colorectal tumor cells inhibited histone deacetylase (22). Importantly, sulphoraphane was not active itself, rather its metabolites formed after reaction with glutathione. Sulphoraphane has also been shown to have histone deacetylase activity in human subjects, albeit transiently (23,24). This type of modulation of gene expression is termed epigenetics (changing expression without altering the DNA nucleotide sequence) and may turn out to be crucial in terms of the benefit of the diet in preventing chronic disease.
Dietary polyphenols and nutritional genomics
Polyphenols enter the diet from a wide variety of edible plants. As part of the activities of the University of Alabama at Birmingham’s Center for Nutrient-Gene Interaction, three polyphenols (the isoflavone genistein from soy, the stilbene trans-resveratrol from grapes and epigallocatechin-3-gallate from green tea) are being investigated for their roles in the prevention of breast cancer. Evidence from rodent studies suggests that the chemopreventive effects of genistein (25) and proanthocyanidin-rich grape seed extract (26) depend on the animals being exposed to genistein or soy at the time of weaning and puberty. Epidemiological data suggest exposure to soy (in the form of tofu) in adolescence is critical for beneficial effect of soy in lowering breast cancer risk (27,28). Similar to genistein, trans-resveratrol administered in the diet (1000 ppm) from birth to weaning in rats and then from 100 days of age caused a 50% reduction in the number of mammary tumors induced by carcinogens (29). In contrast, epigallocatechin-3-gallate administered in the drinking water had no effect in this model.
Polyphenol variability and disease prevention
Do polyphenolics have a significant effect in preventing chronic diseases such as cancer? Many of the variables mentioned earlier that constitute nutritional genomics come into play and are not limited to the human subject. Even the soybean produces variable amounts of isoflavones. This has a genetic basis that is a function of the soybean strain, an environmental basis (temperature, humidity, altitude and sunlight) (30,31), and the time of harvesting (32). Thus a gram (or ounce) of a soybean or a soy food is not a unit of amount of isoflavones and therefore food frequency questionnaires should be augmented by analysis of what is being eaten by the subject group under study (33). Furthermore, the processing that takes place in the production of a soy food alters the composition of the isoflavones (Fig. 3). As examples, many of the Asian-style forms of soy foods (soy paste, miso, tempeh, soy sauce) are fermented products (34). Fermentation removes the glycosidic moiety that is chemically bound to isoflavones in soybeans. The unconjugated isoflavone is rapidly absorbed in the upper gut (35). In addition, fermentation leads to altered chemistry of the isoflavone, typically hydroxylation of the A-ring. Soy sauce can also be prepared by chemical treatment of soybeans. However, this leads to almost total loss of isoflavones. Most American-style forms of soy involve recovery of the soy protein fraction from soybeans without fermentation and tend to preserve the glycosidic conjugates; indeed, heating (toasting) of the soy protein can convert the isoflavone-6″-O-malonylglucoside to the 6″-O-acetylglucoside (36,37). This form must enter the colon before it can be hydrolyzed to unconjugated isoflavones, which leads to a more rapid conversion to secondary bacterial metabolites of isoflavones (dihydrodaidzein, O-desmethylangolensin and S-equol). Even the colonic bacteria are a variable, as these are a function of what we eat and drink. Approximately one in three individuals is an equol-producer. Some have suggested that equol production is tightly linked to the beneficial effects of a soy diet (38–41). This is believed to be due to the interaction of S-equol with the estrogen receptor beta (42). Perimenopausal women who are equol producers retain bone better than non-equol producers (38). In a recent randomized, controlled, parallel study design of 62 adults with hypercholesterolemia on a step II diet containing 80 g of pasta with or without soy germ isoflavones, equol producers had improved in serum lipid and other cardiovascular parameters compared to non-producers (43)
Figure 3. Effect of processing of soybeans into different soy food products on the isoflavone composition.
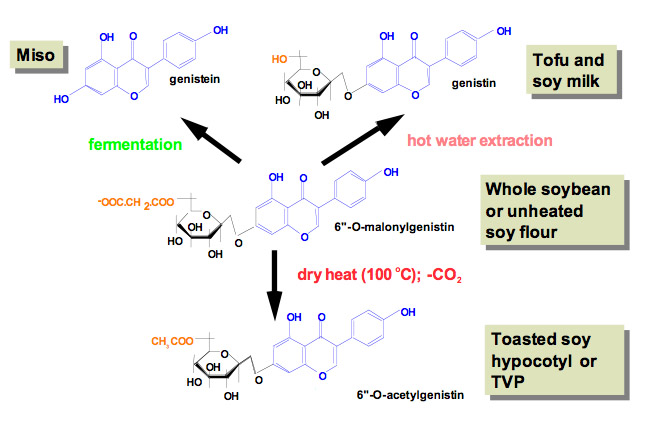
Soybeans consist mostly of 6″-O-malonylglucosides of daidzein, genistein and glycitein (genistein is shown in this example). Fermentation leads to the cleavage of the entire glucose moiety as well as undergoing hydroxylation at the 6-and 8-positions. Extraction of crushed soybeans with n-hexane to recover the oils does not alter the isoflavone composition. However, heating to toast the soy flour causes a loss of CO2 from the malonyl group that converts the isoflavone to a 6″-O-acetylglucoside. This also occurs for soy products formed by hot extrusion methods such as those for textured vegetable protein (TVP) or roasted products (SoyLife™). Hot pressurized extraction of soybeans with water to make soy milk leads to hydrolysis of the malonyl group, converting the isoflavones to simple glucosides. Products made from full fat soy milk have this composition. Some soy products are designed to be very bland and are made from soy flour that has been treated with hot aqueous ethanol. This solvent extracts the isoflavones, so these products are essentially isoflavone-free.
Polyphenols and dietetics
A polyphenol-rich diet is consistent with the recommendations for 8–10 servings per day of fruits and vegetables, as recommended by both the Centers for Disease Control and Prevention (44) and the National Heart, Lung and Blood Institute (45). Isoflavones are the major polyphenols in soybeans and soy foods; proanthocyanidins are prominent in apples, grapes and many berries; catechins (flavanols) are in teas, particularly green tea; resveratrol is in peanuts and grape skin. Integrating these foods into a healthy and tasty diet is attainable. In 2007 fast food outlets and schools began to provide fruit and salad alternatives to the ubiquitous French fry. This may be in response to increasing awareness of obesity and changes in eating-related behavior by the public.
Conclusions and Challenges in Nutritional Genomics
What becomes clear as the description of nutritional genomics unfolds is that it consists of numerous dimensions – the genome, the complement of genes and their mutations that are available to be expressed, the transcriptome (both protein-encoding and non-protein encoding RNAs), the proteome (primary polypeptides, their posttranslational modifications and protein complexes), the epigenome (methylated DNA and methylated and acetylated histones), the metabolome (the small endogenous molecules that create energy and are used for the synthesis of complex bioactive intermediates including lipids, complex carbohydrates, proteins, and nucleic acids), the nutribiome (food-derived xenobiotics) and the xenobiome (other compounds derived from sources outside of the body –including pollutants). Analyzing all of these variables is a substantial challenge to the investigator.
As for all studies of populations, their genes and the factors from the diet that interact with them, a major hurdle to surmount is the need to have sufficient statistical power to be able to draw firm conclusions. Researchers need a large enough study group to satisfy the statistical requirements and sufficient grant support to carry out such large studies. The ability to conduct suitably powered clinical studies is a problem even in the USA and at the most prestigious institutions. Koushik et al. (46), using data gathered from the Nurses’ Health Study and the Health Professionals Follow-up Study cohorts, were able to show in a study of 376 men and women with colorectal cancer and 849 control subjects that patients with the MTHFR 677TT genotype had a reduced risk of colorectal cancer (odds ratio 0.66; 95% confidence interval, 0.43–1.00), but could not discern a relationship with dietary methyl status. They concluded that their work lacked the requisite statistical power.
Nutrient-gene interaction is becoming an important part of public health policy throughout the world as each country strives to most efficiently ensure the health and productivity of its peoples through prevention strategies. Accordingly, a group of investigators with expertise in nutritional genomics from 21 different countries and 5 continents called for an international alliance to address the most important questions (47). Coordination of research in nutritional genomics is provided by the European Nutritional Genomics Organization (48) and the Nutritional Genomics Society (49). What is envisaged by these groups is cooperative, multinational studies of the major questions in nutritional genomics. This was discussed in depth at a 2007 Conference entitled, “Who We Are and What We Eat: The Role of Metabolomics and Nutritional Genomics in Creating Healthful Foods and Healthy Lives” (50). This cooperation is analogous to the advantages provided by binocular vision or large array radio telescopes. However, it depends on international standards for how nutritional genomics projects are to be conceived, executed and analyzed. For now, it is work in progress.
Acknowledgments
Support for the UAB Center for Nutrient-Gene Interaction in Cancer Prevention is provided by a grant-in-aid (U54 CA100949, S. Barnes, PI) from the National Cancer Institute. Support for research on botanicals and dietary supplements at the Purdue University-University of Alabama at Birmingham Botanical Center for Age-related Disease is provided by a grant (P50 AT00477, Connie M. Weaver, PI) from the National Center for Complementary and Alternative Medicine and the NIH Office of Dietary Supplements. The editorial advice provided by Ruth DeBusk during the writing of the manuscript is warmly appreciated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 4.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 5.Afman L, Müller M. Nutritional genomics: from molecular nutrition to prevention of disease. J Am Diet Assoc. 2006;106:569–576. doi: 10.1016/j.jada.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Trujillo E, Davis C, Milner J. Nutritional genomics, proteomics, metabolomics, and the practice of dietetics. J Am Diet Assoc. 2006;106:403–413. doi: 10.1016/j.jada.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, Dike S, Wyss C, Henrichsen CN, Holroyd N, Dickson MC, Taylor R, Hance Z, Foissac S, Myers RM, Rogers J, Hubbard T, Harrow J, Guigo R, Gingeras TR, Antonarakis SE, Reymond A. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilcken B, Bamforth FLZ, Zhu H, Ritvanen A, Redlund M, Stoll C, Alembik Y, Dott B, Czeizel AE, Gelman-Kohan Z, Scarano G, et al. Geographical and ethnic variation of the 677C-T allele of 5,10 methylenetetrahydrofolate reductase (MTHFR): findings from over 7000 newborns from 16 areas world wide. J Med Genet. 2003;40:619–625. doi: 10.1136/jmg.40.8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAndrew PE, Brandt JT, Pearl DK, Prior TW. The incidence of the gene for thermolabile methylene tetrahydrofolate reductase in African Americans. Thromb Res. 1996;83:195–198. doi: 10.1016/0049-3848(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 10.Chiuve SE, Giovannucci EL, Hankinson SE, Hunter DJ, Stampfer MJ, Willett WC, Rimm EB. Alcohol intake and methylenetetrahydrofolate reductase polymorphism modify the relation of folate intake to plasma homocysteine. Am J Clin Nutr. 2005;82:155–162. doi: 10.1093/ajcn.82.1.155. [DOI] [PubMed] [Google Scholar]
- 11.Le Marchand L, Wilkens LR, Laurence N, Kolonel LN, Henderson BE. The MTHFR C677T Polymorphism and Colorectal Cancer: The Multiethnic Cohort Study. Cancer Epidemiol Biomark Prev. 2005;14:1198–1203. doi: 10.1158/1055-9965.EPI-04-0840. [DOI] [PubMed] [Google Scholar]
- 12.Martin PM, Horwitz KB, Ryan DS, McGuire WL. Phytoestrogen interaction with estrogen receptors in human breast cancer cells. Endocrinology. 1978;103:1860–1867. doi: 10.1210/endo-103-5-1860. [DOI] [PubMed] [Google Scholar]
- 13.Naciff JM, Jump ML, Torontali SM, Carr GJ, Tiesman JP, Overmann GJ, Daston GP. Gene expression profile induced by 17alpha-ethynyl estradiol, bisphenol A, and genistein in the developing female reproductive system of the rat. Toxicol Sci. 2002;68:184–199. doi: 10.1093/toxsci/68.1.184. [DOI] [PubMed] [Google Scholar]
- 14.Moggs JG, Ashby J, Tinwell H, Lim FL, Moore DJ, Kimber I, Orphanides G. The need to decide if all estrogens are intrinsically similar. Environ Health Perspect. 2004;112:1137–1142. doi: 10.1289/ehp.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ise R, Han D, Takahashi Y, Terasaka S, Inoue A, Tanji M, Kiyama R. Expression profiling of the estrogen responsive genes in response to phytoestrogens using a customized DNA microarray. FEBS Lett. 2005;579:1732–1740. doi: 10.1016/j.febslet.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 16.Su Y, Simmen FA, Xiao R, Simmen RC. Expression profiling of rat mammary epithelial cells reveals candidate signaling pathways in dietary protection from mammary tumors. Physiol Genomics. 2007;30:8–16. doi: 10.1152/physiolgenomics.00023.2007. [DOI] [PubMed] [Google Scholar]
- 17.Barnes S, Allison D. B. Excitement and realities in microarray analysis of the biological effects of polyphenols. Pharmaceut Biol. 2004;42(suppl):94–101. [Google Scholar]
- 18.Barnes S, Allison DB, Page GP, Carpenter M, Gadbury GL, Meleth S, Horn-Ross P, Kim H, Lamartinere CA, Grubbs CJ. Genistein and Polyphenols in the Study of Cancer Prevention: Chemistry, Biology, Statistics, and Experimental Design. In: Kaput J, Rodriguez R, editors. Nutritional Genomics: Discovering the Path to Personalized Nutrition. Wiley and Sons, Inc.; NY: 2006. pp. 55–82. [Google Scholar]
- 19.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 20.Edgar R, Barrett T. NCBI GEO standards and services for microarray data. Nat Biotechnol. 2006;24:1471–1472. doi: 10.1038/nbt1206-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc Natl Acad Sci USA. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 23.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007;232:227–234. [PMC free article] [PubMed] [Google Scholar]
- 24.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: From cells to mice to man. Semin Cancer Biol. 2007 May 5; doi: 10.1016/j.semcancer.2007.04.001. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamartiniere CA. Timing of exposure and mammary cancer risk. J Mammary Gland Biol Neoplasia. 2002;7:67–76. doi: 10.1023/a:1015722507237. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Hall P, Smith M, Kirk M, Prasain JK, Barnes S, Grubbs C. Chemoprevention by grape seed extract and genistein in carcinogen-induced mammary cancer in rats is diet dependent. J Nutr. 2004;134:3445S–3452S. doi: 10.1093/jn/134.12.3445S. [DOI] [PubMed] [Google Scholar]
- 27.Shu XO, Jin F, Dai Q, Wen W, Potter JD, Kushi LH, Ruan Z, Gao YT, Zheng W. Soyfood intake during adolescence and subsequent risk of breast Cancer among Chinese women. Cancer Epidemiol Biomarkers Prev. 2001;10:483–488. [PubMed] [Google Scholar]
- 28.Wu AH, Wan P, Hankin J, Tseng CC, Yu MC, Pike MC. Adolescent and adult soy intake and risk of breast cancer in Asian-Americans. Carcinogenesis. 2002;23:1491–1496. doi: 10.1093/carcin/23.9.1491. [DOI] [PubMed] [Google Scholar]
- 29.Whitsett T, Carpenter M, Lamartiniere CA. Resveratrol, but not EGCG, in the diet suppresses DMBA-induced mammary cancer in rats. J Carcinog. 2006;5:15. doi: 10.1186/1477-3163-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SJ, Ahn JK, Kim SH, Kim JT, Han SJ, Jung MY, Chung IM. Variation in isoflavone of soybean cultivars with location and storage duration. J Agric Food Chem. 2003;51:3382–3389. doi: 10.1021/jf0261405. [DOI] [PubMed] [Google Scholar]
- 31.Caldwell CR, Britz SJ, Mirecki RM. Effect of temperature, elevated carbon dioxide, and drought during seed development on the isoflavone content of dwarf soybean [Glycine max (L.) Merrill] grown in controlled environments. J Agric Food Chem. 2005;53:1125–1129. doi: 10.1021/jf0355351. [DOI] [PubMed] [Google Scholar]
- 32.Simonne AH, Smith M, Weaver DB, Vail T, Barnes S, Wei CI. Retention and changes of soy isoflavones and carotenoids in immature soybean seeds (Edamame) during processing. J Agric Food Chem. 2000;48:6061–6069. doi: 10.1021/jf000247f. [DOI] [PubMed] [Google Scholar]
- 33.Horn-Ross PL, Barnes S, Lee M, Coward L, Mandel E, Koo J, John EM, Smith M. Assessing phytoestrogen exposure in epidemiologic studies: development of a database (United States) Cancer Causes and Control. 2000;11:289–298. doi: 10.1023/a:1008995606699. [DOI] [PubMed] [Google Scholar]
- 34.Coward L, Barnes NC, Setchell KDR, Barnes S. The antitumor isoflavones, genistein and daidzein, in soybean foods of American and Asian diets. J Agric Food Chem. 1993;41:1961–1967. [Google Scholar]
- 35.Sfakianos J, Coward L, Kirk M, Barnes S. Intestinal uptake and biliary excretion of the isoflavone genistein in rats. J Nutr. 1997;127:1260–1268. doi: 10.1093/jn/127.7.1260. [DOI] [PubMed] [Google Scholar]
- 36.Barnes S, Kirk M, Coward L. Isoflavones and their conjugates in soy foods: extraction conditions and analysis by HPLC-mass spectrometry. J Agric Food Chem. 1994;42:2466–2474. [Google Scholar]
- 37.Coward L, Smith M, Kirk M, Barnes S. Chemical Modification of Isoflavones in Soy Foods During Cooking and Processing. Am J Clin Nutr. 1998;68:1486S–1491S. doi: 10.1093/ajcn/68.6.1486S. [DOI] [PubMed] [Google Scholar]
- 38.Setchell KD, Brown NM, Lydeking-Olsen E. The clinical importance of the metabolite equol-a clue to the effectiveness of soy and its isoflavones. J Nutr. 2002;132:3577–3584. doi: 10.1093/jn/132.12.3577. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Oka J, Higuchi M, Tabata I, Toda T, Fujioka M, Fuku N, Teramoto T, Okuhira T, Ueno T, Uchiyama S, Urata K, Yamada K, Ishimi Y. Cooperative effects of isoflavones and exercise on bone and lipid metabolism in postmenopausal Japanese women: a randomized placebo-controlled trial. Metabolism. 2006;55:423–433. doi: 10.1016/j.metabol.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Wu J, Oka J, Ezaki J, Ohtomo T, Ueno T, Uchiyama S, Toda T, Uehara M, Ishimi Y. Possible role of equol status in the effects of isoflavone on bone and fat mass in postmenopausal Japanese women: a double-blind, randomized, controlled trial. Menopause. 2007;14:866–874. doi: 10.1097/gme.0b013e3180305299. [DOI] [PubMed] [Google Scholar]
- 41.Clerici C, Setchell KD, Battezzati PM, Pirro M, Giuliano V, Asciutti S, Castellani D, Nardi E, Sabatino G, Orlandi S, Baldoni M, Morelli O, Mannarino E, Morelli A. Pasta Naturally Enriched with Isoflavone Aglycons from Soy Germ Reduces Serum Lipids and Improves Markers of Cardiovascular Risk. J Nutr. 2007;137:2270–2278. doi: 10.1093/jn/137.10.2270. [DOI] [PubMed] [Google Scholar]
- 42.Setchell KD, Clerici C, Lephart ED, Cole SJ, Heenan C, Castellani D, Wolfe BE, Nechemias-Zimmer L, Brown NM, Lund TD, Handa RJ, Heubi JE. S-equol, a potent ligand for estrogen receptor beta, is the exclusive enantiomeric form of the soy isoflavone metabolite produced by human intestinal bacterial flora. Am J Clin Nutr. 2005;81:1072–1079. doi: 10.1093/ajcn/81.5.1072. [DOI] [PubMed] [Google Scholar]
- 43.Clerici C, Setchell KD, Battezzati PM, Pirro M, Giuliano V, Asciutti S, Castellani D, Nardi E, Sabatino G, Orlandi S, Baldoni M, Morelli O, Mannarino E, Morelli A. Pasta naturally enriched with isoflavone aglycons from soy germ reduces serum lipids and improves markers of cardiovascular risk. J Nutr. 2007;137:2270–2278. doi: 10.1093/jn/137.10.2270. [DOI] [PubMed] [Google Scholar]
- 44. [Accessed 10/21/07];National Fruit and Vegetable Program under the direction of the Centers for Disease control. Available at http://www.fruitsandveggiesmatter.gov.
- 45. [Accessed 10/21/07];National Heart, Lung and Blood Institute recommendation on intake of fruits and vegetables. Available at http://www.nhlbi.nih.gov/hbp/prevent/h_eating/h_e_dash.htm.
- 46.Koushik A, Kraft P, Fuchs CS, Hankinson SE, Willett WC, Giovannucci EL, Hunter DJ. Nonsynonymous polymorphisms in genes in the one-carbon metabolism pathway and associations with colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2408–2417. doi: 10.1158/1055-9965.EPI-06-0624. [DOI] [PubMed] [Google Scholar]
- 47.Kaput J, Ordovas JM, Ferguson L, van Ommen B, Rodriguez RL, Allen L, et al. The case for strategic international alliances to harness nutritional genomics for public and personal health. Br J Nutr. 2005;94:623–632. doi: 10.1079/bjn20051585. [DOI] [PubMed] [Google Scholar]
- 48.European Nutritional Genomics Organization (NuGO) [Accessed 10/21/07]; See http://www.nugo.org/everyone.
- 49.Nutritional Genomics Society. [Accessed 10/21/07]; See http://www.nugo.org/nip/24015.
- 50.Symposium in 2007 at the University of North Carolina. [Accessed 10/21/07];Who We Are and What We Eat: The Role of Metabolomics and Nutritional Genomics in Creating Healthful Foods and Healthy Lives. Available at http://www.uncnri.org/news/conference/proceedings.html.