Abstract
Multicellular organisms possessing relatively long life spans are subjected to diverse, constant, and often intense intrinsic and extrinsic challenges to their survival. Animal and plant tissues wear out as part of normal physiological functions and can be lost to predators, disease, and injury. Both kingdoms survive this wide variety of insults by strategies that include the maintenance of adult stem cells or the induction of stem cell potential in differentiated cells. Repatterning mechanisms often deploy embryonic genes, but the question remains in both plants and animals whether regeneration invokes embryogenesis, generic patterning mechanisms, or unique circuitry comprised of well-established patterning genes.
Introduction
Developmental studies in plants and animals can be thought of as parallel universes. Each field acknowledges the existence of the other, but rarely do they come in direct contact. Regeneration provided one of those momentous occasions when these parallel universes collided in a stunning discovery that we now take for granted. Over 260 years ago, Abraham Trembley (Trembley, 1744), working under the popular scientific belief that only plants and a few microscopic animals could regenerate, decided to test whether a polyp he had discovered in pond water was or was not a plant (Figure 1A):
Figure 1.
Regeneration in Animals and Plants: A Historical Perspective
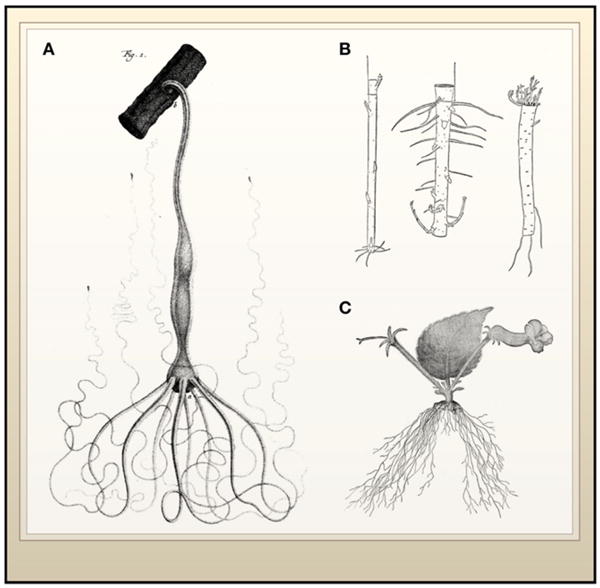
(A) Hydra attached to a small twig with its anterior end pointing down. (B) Regeneration of two pieces of willow suspended in opposite orientations show that polarity is preserved with root and shoot regeneration occurring at the respective bases or apices of the fragments. On the right is a root fragment, also regenerating the corresponding shoots or roots and demonstrating that regenerative capacity is widespread across the anatomy of plants. (C) Regeneration of whole plants from the leaf of the pansy Achimenes haageana. The leaf was removed from a flowering plant and regeneration resulted in roots emerging from the base of the leaf-stalk and flowers emerging near stipules. (Image A from Trembley, 1744; image B from Morgan, 1900 and Vöchting, 1885; image C from Goebel, 1898).
I speculated anew that perhaps these organisms were plants, and fortunately I did not reject this idea. I say fortunately because, although it was the less natural idea, it made me think of cutting up the polyps. I conjectured that if a polyp were cut in two and if each of the severed parts lived and became a complete polyp, it would be clear that these organisms were plants… On November 25, 1740 I sectioned a polyp for the first time…the first polyps I cut were green in color. The two parts extended the same day that I separated them. They were quite easy to distinguish from one another because the first had its anterior end bedecked with those fine threads which serve as the polyp’s arms and legs, whereas the second had none at all… I assumed that the second part was only a kind of tail without the organs vital to the life of the animal… Who would have imagined that it would grow back a head! I was observing this second half to find out how long it would retain the remnants of life; I had not the least expectation of being a spectator to this marvelous kind of reproduction (Lenhoff and Lenhoff, 1986).
The demonstration that simple animals like the polyp (Hydra) described by Trembley were capable of regenerating tissue was soon followed by studies from the likes of Bonnet (Bonnet, 1779) and Spallanzani (Spallanzani, 1769). They unambiguously demonstrated that regeneration was widely dispersed among the metazoans, including earthworms, snails, and salamanders. Similarities between animal and plant regeneration intrigued early investigators. Reviewing the experiments of Vöchting (Vöchting, 1885), T.H. Morgan noted in 1901 that, as in animals, plant regeneration from a twig involves the formation of specialized tissues near the cut ends giving rise to new organs (Figure 1B). At the same time, clear differences in the kingdoms were also apparent, for example, animals typically replace missing tissues, whereas one mode of regeneration in plants leads to entirely new individuals being formed at the wound sites or from severed organs. This was demonstrated by Sachs (Sachs, 1893) and Goebel (Goebel, 1898), who showed that complete individuals can develop de novo from the severed leaves of pansies and begonias (Figure 1C).
Animals and plants have almost certainly evolved multicellularity separately. Thus, the developmental feat of repairing multicellular organs and structures would be expected to have critical differences. However, at a fundamental level, regenerating organisms must turn back the clock on differentiation or freeze developmental youth while at the same time inventing or invoking ways to pattern their new tissue (Figure 2). Even though it is likely that the exact pathways used to activate regeneration in plants and animals may be specific to each kingdom, the mechanistic barriers to totipotency are likely to intersect, and basic principles in regeneration could then be distilled from such comparisons. Do some of the processes called upon during regeneration rely on ancestral functions shared by the two kingdoms, such as nuclear organization and chromatin remodeling? Shared aspects of regeneration, whether ancestral or acquired separately, allow us to focus our attention on those steps that are indispensable during regeneration in multicellular organisms.
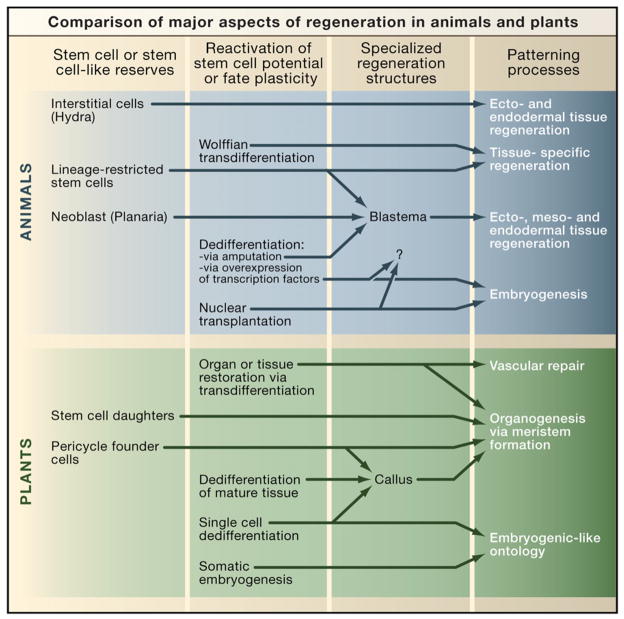
Figure 2. Comparison of Major Aspects of Regeneration in Plants and Animals.
Descriptions of the known cellular origins, regenerative structures, and patterning mechanisms are indicated. We include also an experimental method described in animals (overexpression of transcription factors and nuclear transplantation), which allows differentiated cells to assume stem cell attributes. Whether or not these cells can contribute to regeneration of adult tissues after injury remains untested (Takahashi et al., 2007; Wernig et al., 2007).
This Review aims to compare the knowledge accrued in classic models that show dramatic regenerative capacities in both plants and animals. Particularly, we will focus on two separate phenomena that appear to be basic steps in regeneration in the two kingdoms: (1) acquisition of competence to regenerate through dedifferentiation or by use of pre-existing totipotent cells; and (2) repatterning of regenerating tissues. First, we provide an overview of regeneration in plants and animals, then we canvass the literature preceding the era of molecular biology and use this information as the background against which to examine more recent molecular insights.
Regeneration in Plants and Animals
Plants grow indeterminately, which requires the maintenance of meristems that continually give rise to the major axes of the plant, the root and shoot. Adult plant meristems (Figure 3A) contain stem cell niches (see Essay by J.R. Dinneny and P.N. Benfey, page 553 of this issue) with a group of mitotically less active cells, the quiescent center (QC) in the root and a similar group of cells in the Central Zone (CZ) in the shoot (Mayer et al., 1998; van den Berg et al., 1997). These quiescent cells maintain a group of stem cells (“initials” in botanical terms), mediated by physical contact, that give rise to all the cell types of each respective axis. However, injury frequently removes the stem cell niche completely, and regeneration entails reformation of the stem cell niche in order to resume the continual production of roots and shoots, and thus indeterminate growth. Most plants also maintain other means to either activate or dedifferentiate adult cells to resume meristematic growth, such as pericycle cells that give rise to lateral roots, axillary meristems at the base of leaves, and lateral meristems that contribute to girth (Steeves and Sussex, 1989). In addition, plants can repair damaged tissue by apparent dedifferentiation and respecification of cell types, such as in the repair of severed vascular strands (Sinnott, 1960).
Figure 3. Plant Meristems and Animal Blastemas.
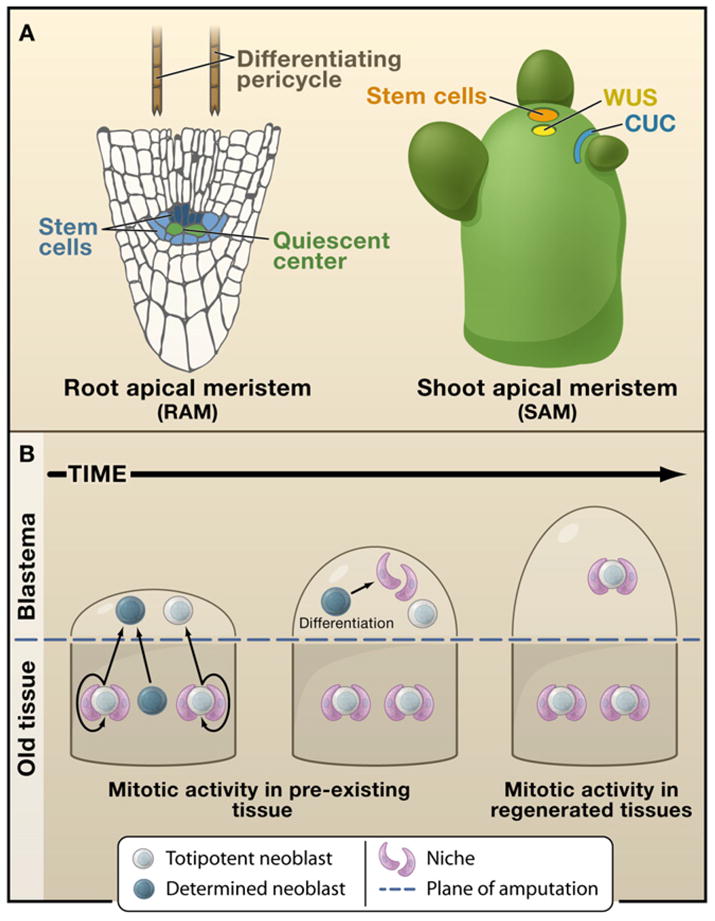
(A) Two of the plant’s indeterminate meristems (root apical meristem, RAM, and shoot apical meristem, SAM, respectively) that can be completely regenerated in adult plants, restoring indeterminate growth. The stem cell niche in the root consists of mitotically less active quiescent center cells (green), which maintain adjacent stem cells (shades of blue) in an undifferentiated state. Cells away from the tip are more differentiated. Differentiating pericycle cells (brown) have been found to give rise to callus, which can in turn recreate the entire root or shoot meristem. The structure of the shoot meristem consists of a group of stem cells in the central zone that lie on top of a group of mitotically active cells that are known to signal and maintain the stem cells, similarly as in root. The largely nonoverlapping domains of CUC2 (blue) and WUS (yellow) are super-imposed on the normal SAM.
(B) A progression series highlighting the cellular processes that take place in the formation of an animal blastema. This structure is assembled by the division progeny of either stem cells residing in the pre-existing tissues (planaria) or a combination of both stem cells and dedifferentiation (salamanders). Once formed, the blastema differentiates, patterns, and functionally integrates itself with the older tissue.
Although most animals do not grow indeterminately, many invertebrate and vertebrate organisms are known to live for very long periods of time (≥100 years) and are referred to as being negligibly senescent (Finch, 1990). Invertebrates such as Hydra and planarian flatworms have unchanging mortality and reproductive rates after many years of culture (Martinez, 1998; Sonneborn, 1930). In mammals, bowhead whales can live more than 200 years (George et al., 1999), and other vertebrates such as the calico rockfish can live for up to 205 years (Cailliet et al., 2001). Although the cellular and molecular mechanisms supporting the longevity of vertebrates remain to be elucidated, the long life span observed in many invertebrates is associated with the lifelong maintenance and regulation of stem cells in somatic tissues. The progeny of these adult stem cells are capable of replacing dying differentiated cells, allowing such organisms to effectively escape death. For instance, animals such as Hydra, planarians, and echinoderms (starfish and crinoids) maintain cells in their body plans that allow for both growth and injury repair. In Hydra and planarians, these stem cells are known as interstitial cells and neoblasts, respectively. Both cell types are capable of replacing all differentiated cells lost to physiological turnover (Holstein et al., 1991; Newmark and Sánchez Alvarado, 2000), and upon injury of the animals these cells undergo robust proliferation to restore the missing tissues (Reddien and Sánchez Alvarado, 2004; Wolpert et al., 1971). Cells with similar functions are found in invertebrates that are phylogenetically more closely related to mammals such as the crinoid echinoderms (Candia Carnevali and Bonasoro, 2001) and the ascidians. Circulating stem cells take part in the regeneration of crinoid arms lost to amputation (Candia Carnevali et al., 1995) or the replacement of complete bodies after physiological turnover in the ascidian Botryllus schlosseri (Lauzon et al., 2002). The lack of cell migration in plants, which is precluded by rigid cell walls, would prevent movement of dispersed stem cells toward injured sites.
In both plants and animals, injury is a stimulus for the formation of specialized wound tissue that initiates regeneration. A regenerative response from these organisms can be elicited by environmental insults, even predatory or pathogenic attacks. Amputation in animals is usually but not always followed by the formation of a specialized structure known as a regeneration blastema (Figures 3B and 4A). This structure consists of an outer epithelial layer that covers mesodermally derived cells and essentially defines a canonical epithelial/mesenchymal interaction, a conserved tissue relationship that is central to the development of complex structures in animals (Sánchez Alvarado and Tsonis, 2006). Other regenerative methods exist in animals, such as the remodeling of pre-existing tissues in planarians to restore both missing body parts and normal scale and proportion (see Figure 5 below) and the regeneration of a whole Hydra by reaggregation of its dissociated cells (Figure 4B) (Gierer et al., 1972). In plants, one frequent but not universal feature of regeneration is the formation of a callus, a mass of growing cells that has lost the differentiated characteristics of the tissue from which it arose. A callus is typically a disorganized growth that arises on wound stumps and in response to certain pathogens (Sinnott, 1960). Similar cell masses can be generated in vitro (Figure 4C), as will be discussed. One common mode of regeneration is the appearance of new meristems within callus tissue. Thus, the plant callus shares with animal regeneration blastemas the property of being a specialized and undifferentiated structure capable of giving rise to new tissues. We emphasize that regeneration in both kingdoms entails a diverse array of phenomena (Figure 2). Furthermore, there are many possible pathways from early to later stages of regeneration. For example, not all cells that have reactivated stem cell potential must pass through either blastema or callus states to regenerate (e.g., Wolffian regeneration, Figure 2). An interesting point is that, despite the diverse array of regeneration mechanisms within kingdoms, almost all phenomena in one kingdom have a counterpart in the other.
Figure 4. Invertebrate and Plant Model Systems for Studying Regeneration.
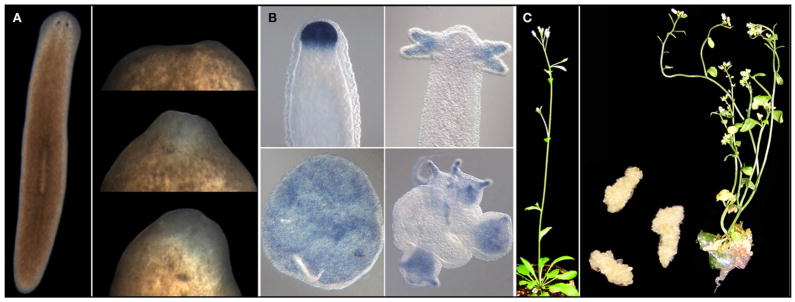
(A) The planarian Schmidtea mediterranea (left) and a regeneration series of the anterior end at ~days 1, 3, and 4 after decapitation.
(B) Head regeneration in Hydra after decapitation (top) and from a cell aggregate of cells obtained after dissociation of a Hydra into individual cells (bottom). The signal corresponds to the expression of a Chordin-like gene in this organism. Images reproduced from Rentzsch et al. (2007), Proc. Natl. Acad. Sci. USA 104, 3249–3254. Copyright (2007) National Academy of Sciences, USA.
(C) Normally developing Arabidopsis with a basal rosette and a reproductive inflorescence that has arisen from the transition of the SAM to a floral meristem (left). Middle, dedifferentiated cell masses of callus that formed from auxin treatment of tissue cuttings from Arabidopsis and the regeneration of a complete shoot from one such callus (right). Images of the callus and regenerating shoot are courtesy of S.P. Gordon and E.M. Meyerowitz.
Figure 5. Remodeling Pre-existing Structures.

A trunk fragment of the land planarian Bipalium is shown undergoing remodeling of pre-existing tissues (see text) and the regeneration of the missing head and tail. The animal is drawn at the same magnification for each of the days illustrated. Note the progressive increase in length and decrease in width of the fragment. These changes result in the regeneration of all the missing structures and ultimately reach the appropriate dimensions relative to the sizes of the newly regenerated head and tail. The table indicates days after amputation and the corresponding change in length to width ratio (L:W) for each specimen pictured. This was accomplished by digitizing the original, drawn-to-scale image after Morgan (1900) and measuring along the midline for length and along the geometric center from lateral edge to lateral edge for width.
Early Regeneration Experiments in Animals
In the late 1800s, the concept of heredity for multicellular organisms was based on the assumption that each differentiated cell in the adult possessed different genetic information, thus restricting their differentiation potential. This theory is better known today as Weismann’s theory of the germ plasm (Weismann, 1893). Weismann postulated that only the cells engaged in producing gametes were totipotent, and that such totipotency was at its highest manifestation in the very early embryo soon after fertilization. As the embryo underwent cleavage, the resulting cells—with the exception of those fated to produce the germline—progressively lost genetic information not needed for the functions of the resulting lineage (that is, muscle cells would eliminate genetic information to make neurons and vice versa). Yet, the regeneration experiments of Trembley, Bonnet, and Spallanzani demonstrated that adult differentiated animal tissues have the capacity to undergo remarkable developmental changes not necessarily associated with their differentiated functions. Weismann noted that the challenge posed to the germ plasm theory by regeneration could be explained by the fact that the tissues being regenerated arose from similar tissues such that limbs only regenerated limbs and tails only regenerated tails (Weismann, 1893). However, in 1895 Wolff demonstrated that a vertebrate cell type believed to be terminally differentiated could undergo a dramatic transformation to produce cell types outside of its normal lineage. The cells in question are the pigmented retinal epithelial cells of the newt eye. After removal of the newt eye lens, these cells can dedifferentiate, proliferate, and then transdifferentiate from their initial epithelial morphology into cells that eventually will regenerate the lens (Wolff, 1895).
Similar plasticity was uncovered later in the studies of other adult animals, particularly in the planarian flatworms in which pre-existing tissue can remodel itself after amputation to restore lost body parts. For instance, if the anterior end of a planarian is cut at any level along the anteroposterior axis, a new head is regenerated. The resulting animal, however, is misproportioned, particularly when the amputation was performed at more posterior regions. What is observed is that the pre-existing tissue undergoes morphological changes such that the normal proportions between the parts are re-established (Reddien and Sánchez Alvarado, 2004). This is clearly illustrated by the work of T.H. Morgan on the land planarian Bipalium kewensi (Morgan, 1900). If a trunk fragment is removed, head and tail structures are regenerated that are nevertheless out of proportion (too small) relative to the size of the trunk fragment (Figure 5). It would be expected that the newly regenerated structures would grow to reach the appropriate size. However, these animals are incapable of feeding until after regeneration of the feeding organ (the pharynx) is completed. Instead, something remarkable happens: the pre-existing trunk actively remodels itself (elongates) as evidenced by a decrease in the width and girth of the fragment and a corresponding increase in length (nearly twice the length of the original fragment). Such remodeling results in the regeneration of both the trunk anatomy and a re-establishment of normal allometry between the trunk and the regenerated cephalic and caudal structures (Figure 5). As such, cellular plasticity is demonstrated with respect to not only regenerating parts but also the tissue remnants from which these parts regenerate.
Regeneration Competency in Animals
Where is the ability to regenerate codified in the adult animal? As early as 1892, it was suspected that a population of undifferentiated cells found in the body plans of adult animals (Randolph, 1892) and triggered to proliferate after injury may be responsible for the regenerative capacities of many invertebrate organisms (Keller, 1894). These specialized cells received the name of neoblasts (Randolph, 1892) and were later shown to be essential for the regeneration of freshwater planarians (Bardeen and Baetjer, 1904). As alluded to earlier, another way for regeneration to manifest itself is through the dedifferentiation of mature, differentiated cells. In the case of appendage (limbs and tail) regeneration in salamanders, the dedifferentiation of mesodermal cells was first observed by Thornton (Thornton, 1938) and later confirmed by the electron microscopy work of Hay (Hay, 1959). Recently, the dedifferentiation process of vertebrate cells has been most exhaustively studied in the muscle fibers of salamanders in culture (Brockes and Kumar, 2002) and in vivo (Echeverri et al., 2001), as well as in the cells of the axolotl spinal cord (Echeverri and Tanaka, 2002). In both cases, the resulting dedifferentiated cells contribute to multiple differentiation lineages (Brockes and Kumar, 2002). In the case of tail regeneration, spinal cord cells switch from an ectodermal lineage and produce mesodermally derived cells such as muscle and cartilage (Echeverri and Tanaka, 2002). That differentiated amphibian cells can maintain access to the inherent totipotentiality codified by their genomes was unambiguously demonstrated by the generation of adult organisms from the transplantation of differentiated cell nuclei into enucleated oocytes in Xenopus (Gurdon, 1962), a finding recently confirmed in mammalian cells (Eggan et al., 2004). As important as dedifferentiation appears to be in salamander regeneration, satellite cells as defined by pax7 expression within the skeletal muscle of these vertebrates can also significantly contribute to the regenerative process (Morrison et al., 2006). This suggests that, like the invertebrates Hydra and planaria, regenerative properties in salamanders correlate with the availability of stem cells residing in the adult body plan.
Another similarity shared by vertebrate and invertebrate animals in their competence to regenerate is the involvement of innervation. In annelid worms, such as Eisenia fetida and Nephtys, removal of the ventral cord or the brain can halt regenerative events (Avel, 1961; Clark, 1965). Conversely, redirecting the ventral cord to the body wall of the annelid Spirographis spallanzanii results in the emergence of an ectopic head at the point of contact (Kiortsis and Moraitou, 1965). In salamanders, similar results are observed in that the denervation of limbs prior to amputation results in an inability to regenerate the limb after amputation (Brockes, 1984), and that deflecting the nerve to the skin at the base of the limb triggers the formation of ectopic limbs (Locatelli, 1924). Moreover, if the nerves innervating the limb are disconnected from the central nervous system by transecting their dorsal roots, normal regeneration is still observed. These combined results suggested that the influence of the nervous system on animal regeneration is due to a soluble factor rather than a direct effect of the innervation proper (Singer, 1947; Singer and Craven, 1948). This has recently been proven by the identification of the anterior gradient protein as being sufficient and necessary to effect the neuroregulation of salamander limb regeneration (Kumar et al., 2007). The newt anterior gradient protein is a ligand secreted by the Schwann cells of the nervous system that binds to the proximodistal specification receptor Prod1/CD59 in salamanders (da Silva et al., 2002). Remarkably, overexpression of the newt anterior gradient protein in rigorously denervated salamander limbs rescued their ability to fully regenerate with high efficiency (Kumar et al., 2007). The existence of trophic factors in promoting regeneration in animals is akin to the central role that phytohormones such as auxins play in the regenerative capacities of plants.
Early Regeneration Experiments in Plants
In 1902, the botanist Gottleib Haberlandt theorized that, under the proper culture conditions, “one could successfully cultivate artificial embryos from vegetative cells” (Haberlandt, 1902; Krikorian and Berquam, 1969). Interestingly, it took decades before effective culture techniques were established in the 1930s and another two decades before single cell totipotency was effectively demonstrated (Gautheret, 2003). Two distinct bodies of work needed to come together. The first was a breakthrough in tissue culture using the critical phytohormone indole-3-acetic acid (auxin). In 1939, several workers finally succeeded in growing perpetual tissue cultures using larger tissue sections, explants, from carrot and other species in media containing auxin (Gautheret, 1985). The auxin treatment and likely endogenous cytokinin appeared to trigger dedifferentiation of root explants, which formed a callus as a first step. Dedifferentiation involves the loss of differentiated cell type characteristics and regression to meristematic states with the ability to grow and divide (Sinnott, 1960). The second critical breakthrough came with the seminal work of Skoog, who found that treating an undifferentiated callus with high ratios of auxin to cytokinin led to a greater frequency of root formation than shoot formation (Skoog and Miller, 1957). In contrast, high cytokinin to auxin ratios led to shoot formation. Thus, Skoog and Miller’s model provided both an experimental tool and conceptual framework for the role of hormones and their interactions in setting up distinct developmental paths during pattern formation and regeneration.
More than 50 years after Haberlandt’s prediction of totipotency, Steward et al. (1958) used secondary phloem cells of carrot root to first form callus cultures from which single cells were isolated to regenerate entire plants. Muir et al. (1958) formed callus directly from single cells of tobacco and used nurse tissue (callus from explants) separated from the regenerating cells by filter paper to apparently condition cells. Ultimately, Vasil and Hildebrandt (1965) used single cells of tobacco to generate reproductive plants without the need for nurse tissue but still with auxin in the conditioning media, providing a firm corroboration that individual cells could be grown into whole plants. The passage of single cells through callus was believed to be the key step that permitted dedifferentiation and the acquisition of competency (Steward et al., 1958). Thus, analogously to the dedifferentiation of cells in salamander regeneration (Brockes and Kumar, 2002) and the regained totipotency of frog and mammalian somatic nuclei transplanted into oocytes (Gurdon, 1962), differentiated cells in plants could be coaxed back into totipotency under a very specific regiment of endogenous hormones that could plausibly mimic an internal environment under regenerative conditions.
The prediction that embryogenesis would be reiterated in isolated cells was borne out in some instances but not others. For example, when Steward cultured single carrot cells into whole plants, the regenerating plantlets clearly resembled embryonic stages after single cells were separated from callus (Steward et al., 1958). However, in other protocols, single cells never passed through embryonic states but developed adult meristems directly from undifferentiated filaments or callus-like growths (Muir et al., 1958; Vasil and Hildebrandt, 1965). Thus, a clear embryonic state, as Haberlandt himself assumed, was not a strict requirement for regeneration from single cells.
Totipotency of Plant Cells
Following the demonstration of totipotency in plants, a body of work established the totipotency of cells from a variety of tissues, such as leaf epidermis and mesophyll, pollen, endosperm, and vascular cells (Halperin, 1986). However, this work led to the potentially misleading dogma that all plant cells are totipotent, a view often propagated by prominent biologists like Steward: “The conclusion is that, in principle, all normally diploid somatic cells are essentially totipotent and that present failures to rear them into plants merely present the challenge to find the right conditions for their development” (Steward et al., 1970). Steward himself used one of the most terminally differentiated cell types, phloem, but his samples started with tissue sections in secondary phloem and individual cells were not isolated until they formed callus (Steward et al., 1958). Thus, the exact starting identity of his cells would not have been known and is arguably obscured by their passage through callus. The totipotency of nonzygotic and at least partially differentiated cells was documented, but the ability of all plant cells to regenerate remains a difficult claim to validate. Binding—whose lab conducted many single cell regeneration studies on different tissues—ultimately questioned whether the origin of many cells that regenerated whole plants from terminally differentiated leaves might arise from meristematic cells in the suspensions of free cells, which were generated by digesting tissue with cell-wall-degrading enzymes (Binding, 1986).
The acceptance of this dogma has important implications for current research. For example, if the reactivation of stem cell potential is associated with specific cellular attributes such as chromatin state or remodeling capability, do we expect all cells to exhibit such attributes? In addition, the dogma obscures an important attribute of plant cell totipotency concerning the developmental age of cells and tissues. For instance, regeneration efficiency was found to be much higher in protoplasts from earlier stages of development as opposed to fully differentiated tissue (Binding, 1974, 1975; Vasil and Vasil, 1974). In carrot, cells of the vasculature, where Steward’s phloem cells originated, readily formed cell lines and somatic embryos whereas, under the same set of conditions, cells from external layers did not (Guzzo et al., 1994). Carrot cotyledons (embryonic leaves) showed exceptional regenerative capacity as they were able to undergo embryogenesis even without addition of hormones to culturing media (Smith and Krikorian, 1989). Thus, it is currently not possible to say whether all plant cells are totipotent, but there do appear to be sharp gradients in the competency of different plant cells to undergo regeneration.
This raises the question of whether plants maintain certain cells outside the stem cell niches in somewhat differentiated states that can still easily access pluripotent states. For example, developmentally young cells, defined here as those cells that have undergone only a few divisions since the asymmetric division of stem cells, can rapidly restore an excised stem cell niche of the root tip (Prantl, 1874). Recent work in animals shows that, in a similar fashion, differentiated animal cells may maintain the ability to exhibit pluripotency during early stages of maturation. In Drosophila, transit-amplifying spermatagonia that have left the stem cell niche can revert back to stem cells with the proper signaling inputs (Brawley and Matunis, 2004). Similar functions for transient amplifying cells have also recently been described in the mammalian testis (Nakagawa et al., 2007). Thus, there appears to be a window in the early developmental stages of a cell in which pluripotency can be readily reconstituted. Together, early regeneration studies established that both kingdoms display a variety of mechanisms to reactivate or re-establish stem cell potential. These cells then participate in patterning steps that resemble embryogenesis in some developmental pathways and bear little resemblance to the stages of embryogenesis at other times.
Regeneration Studies Today
Currently, there is a resurgence in the study of regeneration in animals, which is driven by the re-emergence of classic model systems and the closer examination of established genetic systems. In plants, almost the opposite is true. Classical regeneration experiments developed in a range of species have been adapted for use with powerful new molecular techniques on the workhorse model systems. The result is a burst of new information in both fields that has put classical questions in regeneration back on the front burner and is yielding a new body of work for mechanistic comparisons of regeneration between plants and animals.
In the last few years, the study of animal regeneration has seen the introduction of methodologies aimed at increasing the resolution of molecular analyses such as RNA-mediated genetic interference (RNAi) and the introduction of functional genomics and transgenic methodologies in Hydra, planarians, and salamanders (Sánchez Alvarado and Tsonis, 2006). Moreover, developmental biologists have begun to interrogate the regenerative abilities of genetic model systems such as the fruit fly Drosophila melanogaster (Ohlstein and Spradling, 2007; Singh et al., 2007) and zebrafish (Curado et al., 2007; Lepilina et al., 2006; Stoick-Cooper et al., 2007). In plants, one of the most powerful model systems, Arabidopsis thaliana, has been put to use in regeneration studies recently with the use of green fluorescent protein (GFP) markers and newly developed techniques using timelapse imaging of live tissue (Reddy et al., 2004; Xu et al., 2006). In addition, the substantial knowledge of developmental circuitry in Arabidopsis has provided a starting point for exploring critical genes in regeneration using mutant collections and marker lines. Recent connections between cellular plasticity and chromatin structure in plants have been informed by homology to similar molecular mechanisms that have been well studied in animals (Costa and Shaw, 2007). This work has provided an interesting connection between stem cell states in animals and dedifferentiation in plants.
Chromatin Regulation and Cell Totipotency in Animals
A key characteristic of most metazoan somatic cells is that their nuclei contain all the necessary genetic information to produce whole organisms (Campbell et al., 1996; Eggan et al., 2004; Gurdon, 1962). In animals that regenerate missing body parts, mechanisms to access such inherent totipotentiality must have evolved. Two basic ways in which animals appear to regulate that potential is through cellular dedifferentiation or the maintenance and regulation of adult stem cells in their somatic tissues (Carlson, 2007). Just as there are known differences in chromatin organization between the genomes of embryonic stem cells and differentiated cells (Bernstein et al., 2005), a fundamental difference in the organization of the nucleus and its contents must exist between the adult cells of regenerating and nonregenerating organisms. A hint that such differences may exist is provided by the recently discovered protein nucleostemin (Tsai and McKay, 2002). This protein was found preferentially in both embryonic and adult neuronal stem cells, as well as in many cancer cell lines, but absent from all differentiated adult cells tested (Tsai and McKay, 2002). Nucleostemin derives its name from the fact that in the stem cells in which it was discovered, it resides in the nucleolus—a nuclear organelle traditionally regarded as a ribosome factory but shown in recent years to have many functions (Olson et al., 2000; Pederson, 1998). As embryonic and adult stem cells undergo differentiation, the expression of nucleostemin eventually becomes undetectable. Moreover, elimination of nucleostemin in stem cells prevented their self-renewal and promoted their differentiation (Tsai and McKay, 2002).
In salamanders, a homolog of nucleostemin has been identified (Maki et al., 2007). Although it cannot be detected in differentiated cells, removal of the lens of the salamander eye results in the accumulation of nucleostemin in the nucleoli of dedifferentiating retinal pigmented epithelial cells. Similarly, nucleostemin is also found in the degenerating multinucleate muscle fibers after limb amputation. In both cases, the appearance of nucleostemin in the nucleoli significantly precedes dedifferentiation in the eye and blastema formation in the limb (Maki et al., 2007). These results suggest that nucleostemin is not only necessary to maintain stem cells but is also associated with the dedifferentiation of the cells required to produce the multipotent stem cells of vertebrate regeneration.
Nucleostemin is known to interact with the tumor suppressor p53 both in pulldown experiments and in coimmunoprecipitation experiments of endogenous proteins. Among the many functions ascribed to p53, the local and global modulation of chromatin modifications regulated by this protein is well documented (Allison and Milner, 2004). For example, chromatin immunoprecipitation experiments indicate that p53 recruits the acetyltransferase p300 to the p21 promoter (a direct and natural target of p53), allowing targeted acetylation of chromatin-assembled core histones H2A, H2B, H3, and H4 (Espinosa and Emerson, 2001). p53 also exerts global effects on chromatin through the mediation of histone H3 modifications (Allison and Milner, 2003), which in turn have critical roles in heterochromatin and euchromatin formation (Lachner and Jenuwein, 2002), as well as global chromatin condensation during mitosis (Allison and Milner, 2004; Wei et al., 1999).
Recent studies indicate that the interaction between nucleostemin and p53 inhibits the growth-suppressive activity of p53 (Ma and Pederson, 2007), suggesting that such interactions may play a key role in determining the necessary chromatin organization required by toti/pluripotent cells. Because nucleostemin shuttles between the nucleolus and the nucleoplasm based on its GTP-binding state (Tsai and McKay, 2002), the ability of the nucleoli of stem and dedifferentiation-competent cells to accumulate this protein may reflect cell type-specific functions of this nuclear organelle. Interestingly, a comparison of the nucleolar proteome of animals and plants uncovered a high degree of similarity. Of the 217 proteins identified in Arabidopsis nucleoli, 69% had a direct counterpart (homolog) in the human nucleolar proteome defined for HeLa cells (Leung et al., 2006; Pendle et al., 2005). Given the dynamic nature of the nucleoli in both plants and animals (Meng et al., 2007; Tillemans et al., 2006), it would be interesting to determine to what extent the nucleolar proteome differs between stem and differentiated cells in both plants and animals, and how similar or dissimilar the nucleoli of regeneration-competent plant protoplast and animal stem cells may or may not be to each other.
Regeneration and Tissue Patterning in Animals
Access to totipotentiality and the precise regulation of the identity of the structures being restored appear to be prerequisites for regeneration to occur. Although some progress is being made on the first front, the mechanisms controlling patterning of the resulting regenerate remain poorly understood. Unlike embryogenesis, in which fertilization initiates a self-organizing machinery of stereotypic cell divisions, determination, and differentiation, the starting point for development in regeneration is, if anything, accidental. Depending on the context, the type and extent of the regeneration stimulus (e.g., amputation) depend entirely on experimental design or unplanned, uncontrolled damage in the wild. If a planarian is transected perpendicularly to its midline, the resulting bilaterally symmetric fragments will regenerate the missing tissues. But what if the amputation is oblique, or produces asymmetric animals? How does the animal then regenerate the missing structures and eventually restore its proper form and function?
Rebuilding new tissues from old parts requires robust, specific, and reproducible cell-cell communication. In animals, this is perhaps best illustrated by the regeneration of complete Hydra from aggregates made from dissociated cell suspensions (Figure 4B), and where positional information is presumably completely lost (Gierer et al., 1972). This remarkable process of self-organization is partly mediated by the Wnt/β-catenin pathway (Hobmayer et al., 2000). Initially, uniform expression of Tcf—the transcription factor responsible for mediating Wnt signaling—is detected in the epithelial cells of the entire aggregate. Simultaneously, small domains of about 10–20 Wnt-expressing cells can also be detected inducing surrounding tissues to develop new body axes, which finally separate into intact polyps. Thus, the Hydra orthologs of Wnt and Tcf are likely to be early components of the molecular network responsible for setting up de novo a small number of cells with strong axis-inducing capacity (Hobmayer et al., 2000). The role of β-catenin in regulating anteroposterior polarity has recently been shown in planarians (Figure 6A), where abrogation of this molecule by RNAi results in animals that regenerate heads at posterior ends, whereas abrogation of its inhibitor (the gene Adenomatous Polyposis Coli, or APC) results in the regeneration of tails at anterior ends (Gurley et al., 2008).
Figure 6. Signaling Pathways, Axial Polarity, and Regeneration in Planaria.
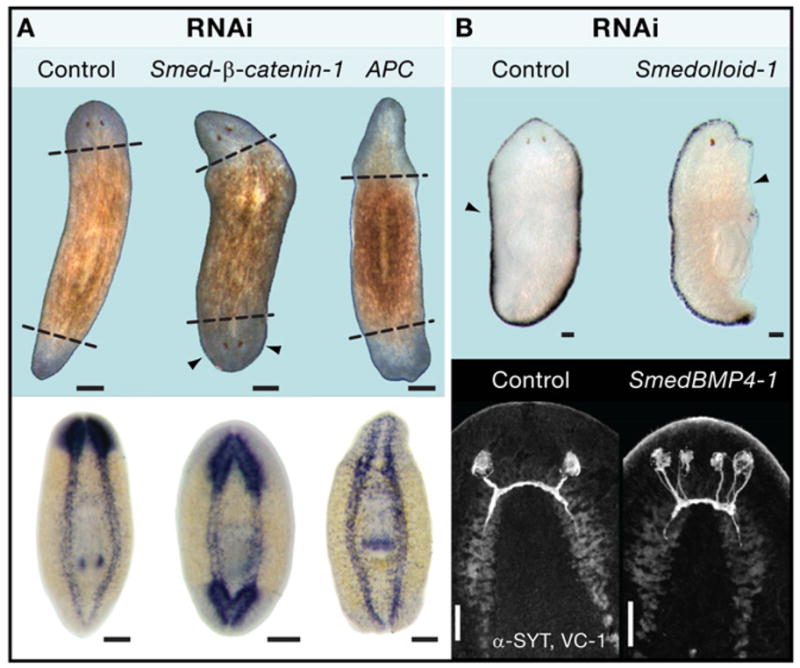
(A) Schmidtea mediterranea planarians subjected to RNAi targeting the key components of Wnt signaling: the Adenomatous polyposis coli (APC) protein and β-catenin (top panel). Abrogation of β-catenin function in amputated animals results in animals that regenerate heads at both ends of the anteroposterior axis. Conversely, accumulation of β-catenin by abrogating its inhibitor (APC) results in animals that regenerate tails and anterior and posterior ends after amputation. In situ panels with a probe against the central nervous system (pro-hormone convertase) demonstrate the presence or absence of brain in RNAi-treated animals. Images from Gurley et al. (2008), Science 319, 323–327. Reprinted with permission from AAAS.
(B) Abrogation of the TGF-β pathway prevents mediolateral regeneration as assayed by a probe (D.21) specific to the animal margin (top panel) and also affects dorso-ventral axis maintenance as evidenced by the generation of an extra pair of photoreceptors in the ventral region of the animal (bottom panel). Images are from Reddien et al. (2007) and are reproduced with the permission of the Company of Biologists. In all cases control refers to unc-22, a gene with a nucleotide sequence not found in planarians. Animals were labeled with antibodies that recognize the photoreceptor neurons (VC-1, anti-Arrestin) and the cephalic ganglia (SYT, anti-Synaptotagmin). Scale bars in (A) are 200 μm and in (B) are 100 μm.
Another signaling pathway involved in the dorsal-ventral patterning of many animal embryos is the transforming growth factor β(TGF-β)/bone morphogenetic protein (BMP) pathway, which has recently been implicated in the re-establishment of bilateral symmetry of irregularly cut adult planarians. Three genes were identified to regulate this process: a BMP1/Tolloid-like gene (smedolloid-1), a SMAD4-like gene (smedsmad4-1), and a BMP2/4/DPP-like gene (smedbmp4-1). BMP signaling was shown to participate in the formation of new tissues at the midline of regeneration, the dorsal-ventral patterning of new tissues, and the maintenance of the dorsal-ventral pattern of existing adult tissue in unamputated animals (Figure 6B). Asymmetric fragments lacking a midline displayed new smedbmp4-1 expression prior to formation of a regeneration blastema, whereas asymmetric fragments containing the midline displayed expanded smedbmp4-1 expression toward the wound. These experiments provide some of the first genetic insights into the topics of blastema specification and restoration of form in regeneration and indicate that injured animals lacking left-right symmetry reset their midline through modulation of BMP activity as an early and necessary event in regeneration (Reddien et al., 2007).
Regeneration of Cellular Morphology in Plants
One testament to the fundamental nature of regeneration is the repair of cellular morphology in the single-celled green alga Acetabularia (also known as the Mermaid’s Wineglass). This 1–10 cm single-celled organism possesses a stalk apex with hairs during vegetative growth and a whorl or cap during reproductive phases. At the other end, it has a rhizoid apex that creates extended projections that root the cell to surfaces. Remarkably, Acetabularia is capable of regenerating its upper hairs or whorls when apical portions are removed (Mandoli, 1998). Even the excised apical stalks, which lack a nucleus, can continue growth and form a reproductive cap if stalks are cut from adult plants. These regeneration experiments in this elegant system have shed light on the respective roles of nuclear and cytoplasmic factors and their influence on phase transitions and regeneration. The regenerative capacity of Acetabularia also demonstrates that the ability to recapitulate development, restore morphology, and re-establish polarity can be an inherent property of the cell independent of cell-cell communication.
Acetabularia also provided an early demonstration of the role of electrical currents in establishing polarity in regeneration. Enucleated stalks of Acetabularia mediterranea grown in the dark can regenerate their whorls at either end when placed in the light (Novak and Sironval, 1975). However, the same stalks failed to regenerate when placed in an apparatus that neutralized internal electrical currents, whereas cells placed in the same apparatus that permitted an electrical potential at opposite ends did regenerate (Novak and Sironval, 1975). Interestingly, in both higher plants and animals, it has been shown that low electrical currents can induce regeneration when applied to callus or to the injured spinal chord of vertebrates (Borgens, 1988; Rathore and Goldsworthy, 1985). In mouse, it has recently been shown that genetic perturbations in inositol-phospholipid signaling disrupted electrically induced migration of epithelium during regeneration (Zhao et al., 2006). Together, these studies show that electrical potential is involved in re-establishing polarity during regeneration in a diverse array of organisms.
Chromatin: Identity Locks and Regeneration Keys?
Soon after the wide potential for single plant cells to regenerate became apparent in the late 1950s, the critical research question shifted from whether cells were totipotent to what limits cell totipotency in normal development. One likely candidate at the moment appears to be chromatin structure, which is regulated by mechanisms with intriguing parallels between plants and animals (Costa and Shaw, 2007). Several different chromatin defects implicate epigenetic changes in controlling totipotency. In Drosophila and Arabidopsis, Polycomb Repressive Groups (PRCs) have been shown to have a role in establishing and maintaining histone H3 methylated on lysine 9 (H3K9) and/or histone H3 trimethylated on lysine 27 modifications. These epigenetic changes are known to maintain genes in a transcriptionally suppressed state at appropriate developmental stages (Cao et al., 2002; Schubert et al., 2006). Only the PRC2 complex has been found in plants so far, and members of this complex have been implicated in gametic imprinting, as is the case for animals, and environmental memory (Pien and Grossniklaus, 2007). Double mutants in CURLY LEAF (CLF) and SWINGER (SWN), partially redundant proteins with homology to E(z) of Drosophila and part of the PRC2 complex, exhibit ectopic callus formation and somatic embryogenesis, implying a role for PRC2 in suppressing totipotency in adult plants (Chanvivattana et al., 2004). A mutant in PICKLE (PKL), a gene that encodes for a putative CHD3 (Chromodomain Helicase DNA-binding protein 3), also showed spontaneous production of callus and persistence of embryonic character (Henderson et al., 2004). CHD3 is part of a complex that contains a histone deacetylase and, like the putative regulatory effects of CLF and SWN, it is expected to suppress transcription. In a screen for mutants with defects in dedifferentiation, a mutant in KRYPTONITE, an H3K9 methyl-transferase, was severely impaired in callus formation (Grafi et al., 2007). Taken together, these studies suggest that multiple types of chromatin modifications may have a role in regulating totipotency and dedifferentiation in plants. Thus, in parallel to connections between chromatin modification and pluri/totipotent states in animals, adjusting epigenetic marks is strongly associated with cellular plasticity in plants.
A more direct link between chromatin remodeling and repatterning came recently from a study of epidermal cells undergoing a fate change. In Arabidopsis, epidermal cells can adopt one of two cell fates, hair and non-hair cells, the latter of which is dependent on the transcription factor GLABRA2 (GL2). Using occasional divisions of epidermal cells that changed cell position and fate cues, Costa and Shaw (2006) analyzed open versus closed chromatin states at the GL2 locus as cells switched their fates. During this transition, it was shown by three-dimensional fluorescent in situ hybridization that the chromatin structure specifically around the GL2 locus changed rapidly from a closed to an open state when GL2 expression was required for the transition to non-hair cell. Accordingly, it changed to a closed state when an extra division landed a cell in the hair cell fate position where GL2 is downregulated (Costa and Shaw, 2006). In addition, the examination of cells that had just undergone cell fate changes suggested that chromatin marks could be removed at mitosis and reset sometime during interphase. Thus, an entire phase of the cell cycle was not required, but the results did imply that cells needed to be competent to divide to switch their fates. A functional link between cell fate plasticity and chromatin state came recently through the gene GL2 expression modulator (GEM) (Caro et al., 2007). Mutations in GEM affected the H3K9 methylation state at the GL2 locus and ultimately disrupted the pattern of fate decisions in the epidermis. In these studies, the chromatin marks on GL2 in cells changing fate were highly labile, but it is unclear if different types of marks are less readily altered. It will be interesting to see if different levels of chromatin remodeling can ultimately explain the gradients of competence exhibited by plant cells and tissues of various age.
Patterning in Plants
A series of new studies has applied modern imaging tools and genomic analysis to regeneration studies, which are now empowered by knowledge of key genes involved in meristem identity. Not surprisingly many of the same genes that play a role in meristem function and embryo patterning appear early during the regeneration process. One set of studies has employed Skoog and Miller’s shoot regeneration system using high cytokinin to auxin ratios to regenerate shoot from callus. Both microarray and imaging analysis showed that the meristem identity genes WUSCHEL and SHOOT MERISTEMLESS were expressed early in the shoot induction phase (Cary et al., 2002; Gordon et al., 2007). Interestingly, both CUP SHAPED COTYLEDON1 and 2 (CUC1/2), which act redundantly in embryonic shoot meristem formation, appeared before shoot formation while root explants were still on the callus-inducing media (Cary et al., 2002; Gordon et al., 2007). This was consistent with the upstream role of CUC genes in triggering the earliest events in the formation of the Shoot Apical Meristem (SAM) (Aida et al., 1999; Daimon et al., 2003; Hibara et al., 2003). Thus, for now, the timing and epistatic relationships of genes during normal shoot formation closely mirror the timing and gene circuitry observed during regeneration.
In plants, as in animals, it is not surprising to see the same genes involved in normal tissue patterning deployed during regeneration as it is unlikely that multicellular organisms would invent different and alternative genetic networks to ultimately produce the same structure. The important difference between normal development and regeneration lies in how these gene networks are activated in both contexts. A critical, defining event may be the derepression of the patterning genes themselves. Che et al. (2007) used the fact that it takes 2–3 days of incubation on high auxin callus-inducing media to make explants competent to regenerate shoots from root explants. This group then exploited microarray technology to ask which genes required treatment in callus-inducing media in order to respond to shoot-inducing media. A total of 57 genes were found to be dependent upon the treatment, showing more than a 20-fold reduction when the auxin-rich callus-inducing media were left out. Among the genes highly dependent on the conditioning phase were WUS and other hormone response regulators. Thus, the work suggested that one role of the competence-inducing phase was to relieve repression of genes that were required for the shoot identity and patterning phase of regeneration.
The work by Che et al. (2007) and another recent paper by Gordon et al. (2007) also addressed the issue of which cells display competency to regenerate under one set of conditions. These studies showed that callus tended to emerge from meristematic tissue and pericycle founder cells, which ultimately dedifferentiate and give rise to lateral roots. In combination with previous studies showing that pericycle cells arrested in the G2 phase of the cell cycle (Dubrovsky et al., 2000), these properties of pericycle cells suggest that they may represent a quasi-stem cell state. This is reminiscent of the neoblasts of planaria. In addition, meristematic cells are partially differentiated but still in a young developmental state. This supports the notion that young cells are readily reverted to stem cell-like states in plants.
Revisiting the Role of Plant Hormones in Regeneration
In normal shoot meristem development, WUS and CUC2 have largely nonoverlapping domains in the central and peripheral zones of the shoot apical meristem, respectively (Figure 3A) (Shani et al., 2006). Using a combination of confocal microscopy and GFP markers, Gordon et al. (2007) observed that the spatial domains of CUC2 and WUS gradually partitioned into separate regions during shoot formation from disorganized callus, reminiscent of the self-partitioning domains of Wnt-expressing cells in Hydra cell aggregates. In plants, hormone activity appeared to underlie the partitioning of CUC2 and WUS as the two genes also showed opposite inductive responses to cytokinin and auxin. CUC2 was responsive to auxin whereas WUS expression was induced in cytokinin-rich media. The cytokinin-rich treatment appeared to lead to a partitioning of CUC2 and WUS accompanied by the apparent spatial partitioning of the hormones themselves. The auxin efflux carrier PIN FORMED1 (PIN1) is expressed early in shoot formation with an apparent role in helping set up auxin gradients, providing evidence that hormone domains are set up early in shoot regeneration. This led to a model in which gradients of auxin and cytokinin partitioned zone identities in the shoot apical meristem at early stages of shoot reformation, a model that is generally consistent with emerging ideas about the instructive roles of differential hormone gradients in normal organogenesis (Benkova et al., 2003; Blilou et al., 2005; Shani et al., 2006). The ability to watch the system self-assemble during regeneration now offers a new tool to dissect the interactions between hormone gradients and genetic circuits that together mediate self-organization in regenerating tissue (Figure 7A).
Figure 7. Regeneration in Plants.
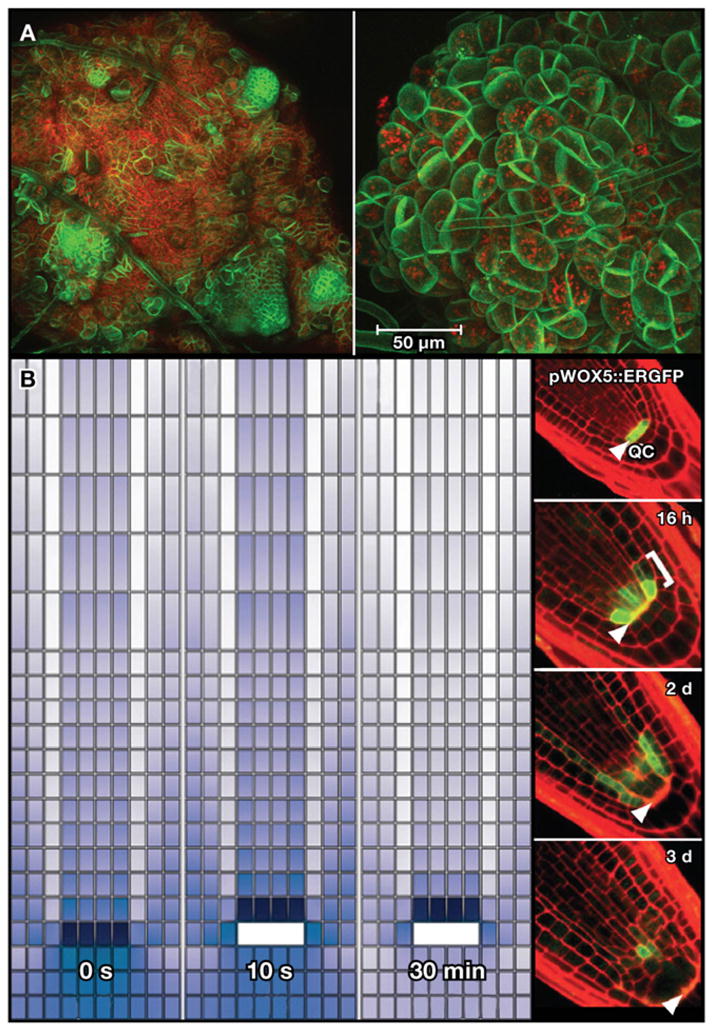
(A) Confocal images of early stage reorganization of SAM in disorganized callus. Red is chlorophyll autofluorescence. Green is a constitutively expressed membrane-bound YFP marker. At left, the membrane-bound YFP marker highlights the small, tightly packed cells that are forming shoot meristems. At right, a close up of the YFP-expressing cells used for imaging analysis and tracking cell division and organ morphology using confocal imaging. Images courtesy of S.P. Gordon and E.M. Meyerowitz.
(B) A graphic display of a simulation of auxin fluxes in the root using auxin transport parameters on the simulated cellular structure of the root (left). The series of panels tracks the simulated ablation of QC and the shift of the maximum concentration (dark purple) away from the tip. Images are from Grieneisen et al. (2007) and are reprinted by permission from Macmillan Publishers Ltd: Nature 449, 1008–1013, copyright 2007. The right panel shows GFP driven by the WOX5 promoter, which specifically marks the QC (panel 1, top small panel, pre-ablation). After ablation, the expression of the WOX5 marker shifts proximally away from the root tip (panel 2, 16 hr post-ablation, panel 3, 2 days post-ablation). At 3 days post-ablation (panel 4), the WOX5 marker re-fines itself to a regenerated QC specified by a newly relocated auxin maxima, in agreement with the simulated shift in auxin concentration. The arrow indicates the position of the original QC. Images are from Xu et al. (2006), Science 311, 385–388. Reprinted with permission from AAAS.
At the other end of the plant, Xu et al. (2006) also used confocal imaging in conjunction with meristem markers to dynamically record regeneration of the stem cell niche after laser ablation. In this case, the mitotically less active QC cells that maintain the stem cell niche were ablated in lines carrying different meristem markers. This group observed a recovery of stem markers several cell tiers above the old stem cell niche within hours after ablation. It has been shown that a high localized concentration of auxin in combination with the overlap of the protein domains of the transcription factors SHORTROOT (SHR) and SCARECROW (SCR) position the stem cell niche (Sabatini et al., 2003). One of the first noticeable events was a shift in the reporter DR5rev::GFP, which marks high auxin concentration, toward the site of the future QC. This showed that, as in the shoot, the early stages of repatterning were characterized by a repositioning of hormone gradients, which are known to localize meristem identity genes and, in turn, to be regulated by them (Blilou et al., 2005; Galinha et al., 2007). Indeed, many of the genes that have been found in forward mutant screens for regeneration mutants are altered in auxin or cytokinin sensing or synthesis (Sugiyama, 1999). One interesting new study modeled auxin transport on the physical root using empirical measures of auxin efflux and influx. This study found that transport alone could largely explain the positioning of the auxin maximum and thus the location of the new stem cell niche (Grieneisen et al., 2007). The model correctly predicted the rapid repositioning of the auxin gradient upon ablation of QC (Figure 7B), suggesting a mechanism by which a totally revised coordinate system could be quickly overlaid on regenerating tissue. This transport system may provide a critical layer of the self-refining and self-organizing mechanism that is needed for regeneration in a system where cells cannot move and the initial morphology, post-damage, is highly unpredictable.
Two Kingdoms, One Common Problem
The study of regeneration can be framed around a single question: How does the disruption of homeostatic mechanisms caused by injury canalize development in plants and animals to regenerate anatomically precise and functionally integrated tissues from such unpredictable starting points? The comparison of regeneration in plants and animals is beginning to provide some answers to this central question.
The first step involves the recruitment of cells competent to undergo the necessary temporal transformations to produce the new tissues. In this respect, plants and animals are quite similar in that they either resort to pre-existing sources of stem or pluripotent cells (for example, pericycle cells and neoblasts) or can dedifferentiate their cells to generate stem cells anew. In both plants and animals, the pool of reactivated stem cells can emerge from stem cell daughters that have partially differentiated, such as in restoration of the root meristem after tip excision in plants and the recent demonstration that spermatagonia can regain totipotency in the Drosophila testis (Brawley and Matunis, 2004; Prantl, 1874). That these strategies may be conserved in multicellular organisms as divergent as plants and animals is unlikely to be a mere coincidence.
At the molecular level, plant and animal genomic DNA is organized into euchromatin and heterochromatin using the astonishingly conserved histone protein family and a similarly conserved suite of histone posttranslational modifications. Moreover, key aspects of animal and plant cell proliferation and differentiation appear to be shared, as reflected by conservation of molecules such as the retinoblastoma protein (Rb) (Wildwater et al., 2005) and the cyclins (Wang et al., 2004). Here is a case where mechanisms used to regulate basic cellular processes, which could be plausibly shared by common ancestry, play a role in multicellular development. Thus, it appears that basic mechanisms involved in the modulation of chromatin structure and the redirection of genomic output may be conserved in both plants and animals, providing fertile ground for comparative and analytical studies aimed at understanding the regulation of cellular totipotentiality.
Patterning, the second step of regeneration, poses more of a challenge in identifying similarities between the two kingdoms. One clear similarity is that both have developed self-organizing systems that can operate on the rough-hewn fragments of injured tissue. In animals and plants, this often involves signals from pre-existing tissues and the specific regeneration structures arising after amputation (blastema and callus). Although the molecular nature of the signaling components is clearly different, basic patterning principles may be shared. BMP signaling plays a role in establishing symmetry in animals whereas the auxin distribution system plays that role in plants. The first signs of polarity in dissociated Hydra cell aggregates involve partitioning ubiquitous Wnt signaling components into smaller domains. Similarly, CUC2 mRNA gradually partitions into specific domains in the disorganized mass of callus cells during plant shoot regeneration. Hence, just as basic developmental principles such as stem cell niches have emerged between plants and animals, a similar vocabulary of mechanistic principles appears to be emerging in regeneration. Interestingly, many of the genes and signaling mechanisms discussed above have a role in embryonic pattern formation. What is not yet clear is how much of the genetic wiring of pattern formation is similar in these developmental scenarios.
Bringing the full power of genetic model organisms into play will be critical to making rapid progress in understanding regeneration. Recent advances in studies of the planarian Schmidtea mediterranea and the model plant Arabidopsis provide a case in point. These model systems and others are now poised to address central questions in regeneration. Is the reactivation of stem cell potential reserved for an elite subset of young or dormant cells or are many fully differentiated cells capable of becoming totipotent? By the same token, how is biological age regulated in those cells capable of producing new progeny in animals like planaria and Hydra? And how is patterning re-established and the new tissue functionally integrated with existing tissues? We expect that vigorous comparisons between the traditionally parallel universes of plants and animals will provide critical insights into unifying principles that could help us unlock the potential of organisms to repair themselves.
Acknowledgments
The authors thank T. Piotrowski and B. Bargmann for their critical reading of the manuscript. A.S.A. is an investigator of the Howard Hughes Medical Institute. NIH-NIGMS RO-1 GM57260 to A.S.A. supported work described in this manuscript. The regeneration research of K.D.B. is supported by NIH-NIGMS RO-1 GM078279.
References
- Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- Allison SJ, Milner J. Loss of p53 has site-specific effects on histone H3 modification, including serine 10 phosphorylation important for maintenance of ploidy. Cancer Res. 2003;63:6674–6679. [PubMed] [Google Scholar]
- Allison SJ, Milner J. Remodelling chromatin on a global scale: a novel protective function of p53. Carcinogenesis. 2004;25:1551–1557. doi: 10.1093/carcin/bgh212. [DOI] [PubMed] [Google Scholar]
- Avel M. L’influence du système nereux sur la régénération chex les urodèles et les oligochètes. Bull Soc Zool Fr. 1961;86:464–483. [Google Scholar]
- Bardeen CR, Baetjer FH. The inhibitive action of the Roentgen rays on regeneration in planarians. J Exp Zool. 1904;1:191–195. [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell . 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Binding H. Regeneration of haplid and diploid plnats from protoplasts of Petunia hybrida L.Z. Plant Mol Biol Newsl. 1974;1:77–95. [Google Scholar]
- Binding H. Reproducibly high planting efficiencies of isolated proplasts from shoot cultures of tobacco. Plant Physiol. 1975;35:325–327. [Google Scholar]
- Binding H. Regeneration from Protoplasts. In: Vasil IK, editor. Cell Culture and Somatic Cell Genetics of Plants 2. Amsterdam: Elsevier Science & Technology Books; 1986. pp. 259–269. [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Bonnet C. Oeuvres d’Histoire Naturelle et de Philosophie de Charles Bonnet. Neuchatel, Switzerland: Samuel Fauche; 1779. [Google Scholar]
- Borgens RB. Stimulation of neuronal regeneration and development by steady electrical fields. Adv Neurol. 1988;47:547–564. [PubMed] [Google Scholar]
- Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- Brockes JP. Mitogenic growth factors and nerve dependence of limb regeneration. Science. 1984;225:1280–1287. doi: 10.1126/science.6474177. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat Rev Mol Cell Biol. 2002;3:566–574. doi: 10.1038/nrm881. [DOI] [PubMed] [Google Scholar]
- Cailliet GM, Andrews AH, Burton EJ, Watters DL, Kline DE, Ferry-Graham LA. Age determination and validation studies of marine fishes: do deep-dwellers live longer? Exp Gerontol. 2001;36:739–764. doi: 10.1016/s0531-5565(00)00239-4. [DOI] [PubMed] [Google Scholar]
- Campbell K, McWhir J, Ritchie W, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Candia Carnevali MD, Bonasoro F. Microscopic overview of crinoid regeneration. Microsc Res Tech. 2001;55:403–426. doi: 10.1002/jemt.1187. [DOI] [PubMed] [Google Scholar]
- Candia Carnevali MD, Bonasoro F, Lucca E, Thorndyke MC. Pattern of cell proliferation in the early stages of arm regeneration in the feather star Antedon mediterranea. J Exp Zool. 1995;272:464–474. [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carlson BM. Principles of Regenerative Biology. London: Elsevier Inc; 2007. [Google Scholar]
- Caro E, Castellano MM, Gutierrez C. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature. 2007;447:213–217. doi: 10.1038/nature05763. [DOI] [PubMed] [Google Scholar]
- Cary AJ, Che P, Howell SH. Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J. 2002;32:867–877. doi: 10.1046/j.1365-313x.2002.01479.x. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- Che P, Lall S, Howell SH. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta. 2007;226:1183–1194. doi: 10.1007/s00425-007-0565-4. [DOI] [PubMed] [Google Scholar]
- Clark ME. Cellular aspects of regeneration in the polychaete Nephtys. In: Kiortsis V, Trampusch HAL, editors. Regeneration in Animals and Related Problems. Amsterdam: North-Holland Publishing Company; 1965. pp. 240–248. [Google Scholar]
- Costa S, Shaw P. Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature. 2006;439:493–496. doi: 10.1038/nature04269. [DOI] [PubMed] [Google Scholar]
- Costa S, Shaw P. ‘Open minded’ cells: how cells can change fate. Trends Cell Biol. 2007;17:101–106. doi: 10.1016/j.tcb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- da Silva SM, Gates PB, Brockes JP. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell. 2002;3:547–555. doi: 10.1016/s1534-5807(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Daimon Y, Takabe K, Tasaka M. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 2003;44:113–121. doi: 10.1093/pcp/pcg038. [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colon-Carmona A, Rost TL. Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 2000;124:1648–1657. doi: 10.1104/pp.124.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science. 2002;298:1993–1996. doi: 10.1126/science.1077804. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Clarke JD, Tanaka EM. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev Biol. 2001;236:151–164. doi: 10.1006/dbio.2001.0312. [DOI] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Finch CE. Longevity, Senescence and the Genome. Chicago: University of Chicago Press; 1990. [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gautheret RJ. History of plant tissue and cell culture: A personal account. In: Vasil IK, editor. Cell Culture and Somatic Cell Genetics of Plants: Cell Growth, Nutrition, Cytodifferentiation and Cryopreservation. Amsterdam: Elsevier Science & Technology Books; 1985. [Google Scholar]
- Gautheret RJ. Plant Tissue Culture: 100 Years since Gottlieb Haberlandt. New York: SpringWien; 2003. Plant tissue culture: the history; pp. 205–214. [Google Scholar]
- George JC, Bada J, Zeh J, Brown SE, O’Hara T, Suydam R. Age and growth estimates of bowhead whales (Balaena mysticetus)via aspartic acid racemization. Can J Zool. 1999;77:571–580. [Google Scholar]
- Gierer A, Berking S, Bode H, David C, Flick K, Hansmann G, Schaller H, Trenkner E. Regeneration of hydra from reaggregated cells. Nat New Biol. 1972;239:98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- Goebel K. Organographie der Pflanzen. Jena: I. Allgemeine Organographie; 1898. [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007;134:3539–3548. doi: 10.1242/dev.010298. [DOI] [PubMed] [Google Scholar]
- Grafi G, Ben-Meir H, Avivi Y, Moshe M, Dahan Y, Zemach A. Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Dev Biol. 2007;306:838–846. doi: 10.1016/j.ydbio.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelial cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Alvarado AS. {beta}-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo F, Baldan B, Mariani P, Lo Schiavo F, Terzi M. Studies on the origin of totipotent cells in explants of Daucus carota L. J Exp Bot. 1994;45:1427–1432. [Google Scholar]
- Haberlandt G. Culturversuche mit isolierten Pflanzenzellen. Sitzungsber Akad Wiss Wien Math Nat. 1902;111:69–91. [Google Scholar]
- Halperin W. Plant Regeneration and Genetic Variability. Vol. 3. Orlando, FL: Academic Press, Inc; 1986. Attainment and retention of morphogenetic capacity in vitro. In Cell Culture and Somatic Cell Genetics of Plants; pp. 3–47. [Google Scholar]
- Hay ED. Electron microscopic observations of muscle dedifferentiation in regenerating Amblystoma limbs. Dev Biol. 1959;1:555–585. [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 2004;134:995–1005. doi: 10.1104/pp.103.030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K, Takada S, Tasaka M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 2003;36:687–696. doi: 10.1046/j.1365-313x.2003.01911.x. [DOI] [PubMed] [Google Scholar]
- Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 2000;407:186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- Holstein TW, Hobmayer E, David CN. Pattern of epithelial cell cycling in hydra. Dev Biol. 1991;148:602–611. doi: 10.1016/0012-1606(91)90277-a. [DOI] [PubMed] [Google Scholar]
- Keller J. Die ungeschlechtliche Fortpflanzung der Süsswasserturbellarien. Jen Zeit Naturw. 1894:3823–3827. [Google Scholar]
- Kiortsis V, Moraitou M. Factors of regeneraiton in Spirographis spallanzanii. In: Kiortsis V, Trampusch HAL, editors. Regeneration in Animals and Related Problems. Amsterdam: North-Holland Publishing Company; 1965. pp. 250–261. [Google Scholar]
- Krikorian AD, Berquam DL. Plant cell and tissue cultures: The role of haberlandt. Bot Rev. 1969;35:59–88. [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lauzon RJ, Ishizuka KJ, Weissman IL. Cyclical generation and degeneration of organs in a colonial urochordate involves crosstalk between old and new: a model for development and regeneration. Dev Biol. 2002;249:333–348. doi: 10.1006/dbio.2002.0772. [DOI] [PubMed] [Google Scholar]
- Lenhoff HM, Lenhoff SG. A Translation from the French of Memoires, pour servir a L’histoire d’ un genre de polypes d’eau douce, a bras en forme de cornes. Pacific Grove, CA: Boxwood Press; 1986. [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI. NOPdb: Nucleolar proteome database. Nucleic Acids Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli P. L’influenza del sistema nervoso sui processi dirigenerazione. Arch Sci Biol (Bologna) 1924;5:362–376. [Google Scholar]
- Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;7:2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki N, Takechi K, Sano S, Tarui H, Sasai Y, Agata K. Rapid accumulation of nucleostemin in nucleolus during newt regeneration. Dev Dyn. 2007;236:941–950. doi: 10.1002/dvdy.21027. [DOI] [PubMed] [Google Scholar]
- Mandoli DF. ELABORATION OF BODY PLAN AND PHASE CHANGE DURING DEVELOPMENT OF ACETABULARIA: How is the complex architecture of a giant unicell built? Annu Rev Plant Physiol Plant Mol Biol. 1998;49:173–198. doi: 10.1146/annurev.arplant.49.1.173. [DOI] [PubMed] [Google Scholar]
- Martinez DE. Mortality patterns suggest lack of senescence in hydra. Exp Gerontol. 1998;33:217–225. doi: 10.1016/s0531-5565(97)00113-7. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Meng L, Zhu Q, Tsai RY. Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol Cell Biol. 2007;27:8670–8682. doi: 10.1128/MCB.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH. Regeneration in Bipalium. Arch Entwick Mech. 1900;9:563–586. [Google Scholar]
- Morrison JI, Loof S, He P, Simon A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J Cell Biol. 2006;172:433–440. doi: 10.1083/jcb.200509011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir WH, Hildebrandt AC, Riker AJ. The preparation, isolation, and growth in cutlure of single cells from higher plants. Am J Bot. 1958;45:589–597. [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Novak B, Sironval C. Inhibition of regeneration of acetabularia mediterranea enculated posterior stalk segments by electrica isolation. Plant Sci Lett. 1975;5:183–188. [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JWS, Shaw PJ. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell. 2005;16:260–269. doi: 10.1091/mbc.E04-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim Biophys Acta. 2007;1769:375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Prantl K. Untersuchungen uber die Regeneration des Vegetations-punktes an Agiospermenwurzeln. Arb Bot Inst Wurzburg. 1874;4:546–562. [Google Scholar]
- Randolph H. The regeneration of the tail in lumbriculus. J Morphol. 1892;7:317–344. [Google Scholar]
- Rathore KS, Goldsworthy A. Electrical control of shoot regeneration in plant tissue cultures. Biotechnology. 1985;3:1107–1109. [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Guder C, Vocke D, Hobmayer B, Holstein TW. Proc. Natl Acad Sci USA. 2007;104:3249–3254. doi: 10.1073/pnas.0604501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs J. Physiologische Notizen, I. Ueber einige Beziehungen der specifischen Grosse der Pflanzen zu ihrer Organisation. Flora. 1893;77:49–81. [Google Scholar]
- Sánchez Alvarado A, Tsonis PA. Bridging the regeneration gap : Genetic insights from diverse animal models. Nat Rev. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Yanai O, Ori N. The role of hormones in shoot apical meristem function. Curr Opin Plant Biol. 2006;9:484–489. doi: 10.1016/j.pbi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Singer M. The nervous system and regeneration of the forelimb of adult Triturus. VII The relation between number of nerve fibers and surface area of amputation. J Exp Zool. 1947;104:251–265. doi: 10.1002/jez.1401040205. [DOI] [PubMed] [Google Scholar]
- Singer M, Craven M. The growth and morphogenesis of the regenerating forelimb of adult Triturus following denervation at various stages of development. J Exp Zool. 1948;108:279–308. doi: 10.1002/jez.1401080207. [DOI] [PubMed] [Google Scholar]
- Singh SR, Liu W, Hou SX. The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem Cell. 2007;1:1191–1203. doi: 10.1016/j.stem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott EW. Plant Morphogenesis. New York: McGraw-ill; 1960. [Google Scholar]
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;54:118–130. [PubMed] [Google Scholar]
- Smith DL, Krikorian AD. Release of somatic embryogenic potential from excised zygotic embryos of carrot and maintenance of proembryonic cultures in hormone-free medium. Am J Bot. 1989;76:1832–1843. [PubMed] [Google Scholar]
- Sonneborn TM. Genetic studies on stenostomum incaudatum (nov. spec.) I The nature and origin of diferences among individuals formed during vegetative reproduction. J Exp Zool. 1930;57:57–108. [Google Scholar]
- Spallanzani L. Prodromo di un opera da imprimersi sopra la riproduzioni animali (An Essay on Animal Reproduction) London: T. Becket & de Hondt; 1769. [Google Scholar]
- Steeves TA, Sussex IM. Patterns in plant development. 2. Cambridge: Cambridge University Press; 1989. [Google Scholar]
- Steward FC, Mapes MO, Mears K. Growth and organized development of cultured cells. II Organization in cultures growtn from freely suspended cells. Am J Bot. 1958;45:705–708. [Google Scholar]
- Steward FC, Ammirato PV, Mapes MO. Growth and development of totiptent cells: Some problems, procedures, and perspectives. Ann Bot (Lond) 1970;34:761–787. [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Sugiyama M. Organogenesis in vitro. Curr Opin Plant Biol. 1999;2:61–64. doi: 10.1016/s1369-5266(99)80012-0. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thornton CS. The histogenesis of the regenerating forelimb of larval Amblystoma after exarticulation of the humerus. J Morphol. 1938;62:219–241. [Google Scholar]
- Tillemans V, Leponce I, Rausin G, Dispa L, Motte P. Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell. 2006;18:3218–3234. doi: 10.1105/tpc.106.044529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley A. Mémoires pour Servir a L’histoire d’un Genre de Polypes d’eau Douce, a Bras en Forme de Cornes. Leiden: Jean and Herman Verbeek; 1744. [Google Scholar]
- Tsai RYL, McKay RDG. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- Vasil V, Hildebrandt AC. Differentiation of tobacco plants from single, isolated cells in microcultures. Science. 1965;150:889–892. doi: 10.1126/science.150.3698.889. [DOI] [PubMed] [Google Scholar]
- Vasil V, Vasil IK. Regeneration of tobacco and Petunia plnats from protoplasts and culture of corn protoplasts. In Vitro. 1974;10:83–96. doi: 10.1007/BF02615342. [DOI] [PubMed] [Google Scholar]
- Vöchting H. Über die regeneration der marchantieen. Jahrb f wiss Botanik . 1885;XVI [Google Scholar]
- Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, dePamphilis CW, Ma H. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 2004;135:1084–1099. doi: 10.1104/pp.104.040436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Weismann A. A Theory of Heredity. New York: Charles Scribner’s Sons; 1893. The Germ-Plasm. Vol. First English edition. [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Wolff G. Entwickelungsphysiologische Studien. I Die Regeneration der Urodelenlinse. Arch Entw-Mecho Org. 1895;1:380–390. [Google Scholar]
- Wolpert L, Hicklin J, Hornbruch A. Positional information and pattern regulation in regeneration of hydra. Symp Soc Exp Biol. 1971;25:391–415. [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. A molecular framework for plant regeneration. Science. 2006;311:385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, et al. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442:457–460. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]