Abstract
The human brain cortex is a highly convoluted sheet. Mapping of the cortical surface into a canonical coordinate space is an important tool for the study of the structure and function of the brain. Here, we present a technique based on least-square conformal mapping with spring energy for the mapping of the cortical surface. This method aims to reduce the metric and area distortion while maintaining the conformal map and computation efficiency. We demonstrate through numerical results that this method effectively controls metric and area distortion, and is computational efficient. This technique is particularly useful for fast visualization of the brain cortex.
Keywords: conformal mapping, brain cortex, spring energy, brain mapping, angle distortion, area distortion
1. Introduction
The cortical surface of the human brain is extremely convoluted, with a plethora of gyri and sulci. Because of the significant differences in the cortical folding among different individuals, the study of the cortical structure across the population is a challenging task. The cortical surface can be considered as a two-dimensional sheet bending in the three-dimensional space. One promising approach to studying and visualizing the cortex is based on mapping the cortical surface to a canonical space, while retaining the best possible geometric information regarding the original structure [8, 10].
Several methods that substantially minimize the metric and area distortion have been proposed in the literature [8, 10]. The Riemann Mapping theorem [2] indicates that continuously differentiable surfaces with the same topology topologies can be mapped onto each other without angular distortion. Based on this a variety of approaches have been proposed to flatten the cortical surface while minimizing the angular distortion. For example, Gu and Yau [1] presented a method for conformally mapping surface structures by minimizing the harmonic energy iteratively on a sphere. Hurdal et al. [6] proposed a technique for quasi-conformal flattening or unfolding of cortical surfaces using circle packing methods. To reduce the computation complexity in conformal mapping, Levy et al. [3] proposed a least-square conformal map method (LSCM) by representing the conformal energy as the square sense of the Cauchy-Riemann equation. This method produces reasonable results in terms of both angular distortion and computation. Ju et al. [4] extended this method to spherical conformal maps.
In this work, we design and add a spring energy to the least-squares conformal maps in order to reduce the metric and area distortion while maintaining the conformal map and computation efficiency. The spring energy was chosen because of its ability to controls edge distortion efficiently. Moreover, the spring energy can be implemented by a least-square formulation and solved efficiently. We extend our method to spherical maps using a technique similar to that of [4]. A brief description of our method is given in [15].
2. Method
2.1 Conformal Mapping
The conformal mapping, or conformal equivalence [11], defines a one-to-one mapping between two surfaces that preserves the local angle and the local similarity. Mathematically, the conformal mapping is defined as: when the mapping U maps a domain (u, v) to a surface U (u, v), each (u, v) satisfies:
| (1) |
In (1) N(u, v) is the unit norm vector on the surface U (u, v). It should be noted that the angle preserving in the conformal mapping is defined on the Riemann surface. When considering a triangular representation of the surface, there would be errors in calculation of Riemann angles around each vertex. An approximate method called the “market share” method has been proposed to obtain the angle on Riemann surfaces [6]. As shown in Figure 1, supposing that there are n angles around vertex v, each angle on Riemann surface is defined as:
| (2) |
where θi is the angle on Euclidean plane.
Figure 1.
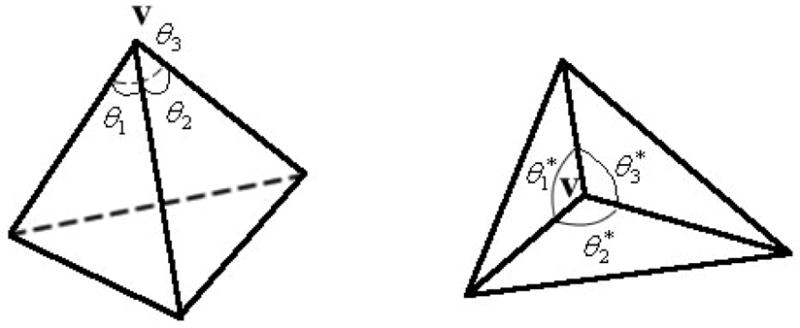
Illustration of “market share” angle.
2.2 Least-square Conformal Mapping
As introduced in [3], a new quasi-conformal parameterization method is proposed based on a least-square approximation of the Cauchy-Riemann equations. In this section we present a brief introduction of least-square conformal maps.
Consider a triangulation mesh K = {V,T}, where V = {v1, v2,···, vn}, vi ∈ R3 is a set of vertex positions, and T = {t1,t2, ···,tm}, ti = {vi1,vi2,vi3} is a set of triangles consisting of triples of vertices, with i1, i2, and i3 denoting the vertical index in V. Since each triangle ti has a uniquely defined normal, ti can be imposed on a local orthonormal basis (x, y) with the normal direction along the z axis.
Based on the Riemann equation [3], a mapping U: (x, y) ↦ (u,v) is said to be conformal on a triangle ti if and only if the following equation holds true:
| (3) |
As (3) cannot be strictly enforced on the whole surface, the violation of the equation can be defined as the conformal energy in a square sense:
| (4) |
where A(ti) is the area of the triangle ti.
For each triangle ti = {vi1, vi2, vi3}, its three vertices can be imposed on a local orthonormal basis {(xi1, yi1),(xi2, yi2),(xi3, yi3)} with the normal direction along the z axis. When ti is mapped to {ui1,ui2,ui3}, where ui = (ui + ivi), we obtain:
| (5) |
Then the Cauchy-Riemann equation on ti can be written as:
| (6) |
where
| (7) |
Based on Eq. (6), the conformal energy can be written as:
| (8) |
Denote u = (u1,u2, ···,un), where ui = (ui + ivi), the Ec can be re-written in a quadratic form with the complex vector u as:
| (9) |
where Mc = {mij} is a sparse m × n complex matrix whose components are:
| (10) |
Suppose p (p ≥ 2) of the ui is pre-fixed, then the minimization of Ec has a unique and non-trivial solution. We rearrange u as , where uf is the vector of n − p free coordinates of u, and u p is the vector of p pre-fixed coordinates of u. We can then decompose Mc in block matrices as Mc = (M cf, M cp), where M cf is a m × (n − p) matrix and M cp is a m × p matrix. Then Eq. (9) can be re-written as:
| (11) |
where ||u||2 denotes the inner product <u, ū>, and ū is the conjugate of u.
To solve this problem, we rewrite Ec in the form of real matrices and vectors as:
| (12) |
with
where , and the superscripts 1 and 2 denote the real and imaginary parts of matrix or vector respectively.
This minimization problem has a unique solution u′ = (AT A)−1 AT b, when p ≥ 2. And it can be efficiently minimized using the conjugate gradient method as in [9]. In order to obtain a planar mapping with better conformality, the variable p should set to 2. As suggested in [3], the two vertices maximizing the length of the shorted path between them were pinned in our experiments.
2.3 Least-squares Conformal Maps with Spring Energy
To maintain the efficiency of the algorithm and control the metric distortion at the same time, the spring energy Espring is added into the LSCM model. In the literature, similar techniques have been introduced, e.g., Hoppe [5] used the spring energy into mesh optimization as a regularizing term.
Supposing that all edges in the mesh are denoted as an edge set E = {e1,e2,···,el}, ei = {vi1,vi2}, the Espring can be placed on each edge of the mesh, represented as the parameter κ:
| (13) |
This can be written as a quadratic form with the complex vector u as:
| (14) |
where M spring = {mij} is a sparse l × n complex sparse matrix whose coefficient is
| (15) |
Let the matrix , M is a (m + l) × n complex sparse matrix. Then the model becomes:
| (16) |
where λ is a weight parameter which balances conformal and spring energy. The conjugate gradient method used in Section 2.1 can be used to solve this equation.
2.4 Spherical Mapping
The method described in above sections can map the surface of topological disk to a planar region. A method for mapping a genus zero surface to a topological sphere was introduced in [4]. First, an arbitrary vertex v* is chosen from V. Then v* and all its connected edges and triangles are removed from the triangulation K. Hence, the remaining surface becomes a topological disk and thus can be mapped to a planar region. Finally, the planar region is stereographically projected to a unit sphere while keeping v* at the “north pole” of the sphere. The spherical mapping is not unique since there is a group of conformal transforms on the sphere that are so-called Mobius transforms:
| (17) |
As this method might produce a highly distorted spherical map, a Mobius transform is selected from Ψ which minimizes the metric distortion [4]. When v* is fixed to the “north pole”, the Ψ becomes:
| (18) |
Since it is complicated to compute the derivation information, the Powell Method [9] is used for this minimization problem instead.
2.5 Hemispherical Mapping
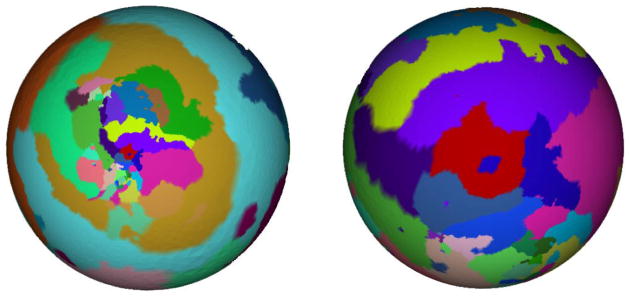
It is noted that different choices of vertex v* generate different mapping results. For example, though local angles are well preserved over the surface with different v*, the mapped surfaces that shrink or expand in local regions appear different, as illustrated in Figure 2. Since it is difficult to choose a best v* that minimizes the distortion, it is challenging to generate a standard mapping result with an automatic choice of v*. Moreover, since the cortical surface is approximately bilateral symmetric, bilateral symmetric mapping results are expected when mapping the whole cortical to a sphere. However, the spherical mapping method introduced above usually generates unbalanced spheres, that is, one of the left or right hemispherical cortical surfaces is shrunk and the other one is enlarged.
Figure 2.
Two spherical mapping results using different cutting vertex v*
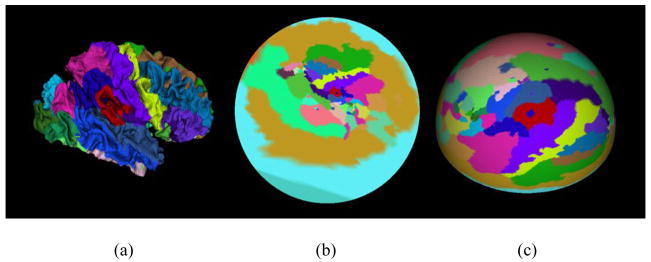
To avoid choosing cut vertex v* that maps the cortical surface symmetrically to a sphere, a “hemispherical” mapping method is proposed here. First, the whole cortex is automatically cut into two approximately symmetric parts: left hemisphere and right hemisphere. Then, these two parts are mapped to planar regions respectively with all boundary vertices pinned on a circle. These two circles can be easily projected to hemispheres, as illustrated in Figure 3. Since only one parameter, namely, the radius of the mapping sphere, needs to be decided in this procedure, it is convenient to determine the radius to minimize the metric distortion [4] using the Powell Method [9].
Figure 3.
The procedure of hemisphere mapping. (a) Right cortical surface. (b) Mapping to planar circle region. (c) Projection to hemisphere.
2.6 Selection of Parameter κi
As noted in Section 2.2, κi is the spring coefficient of edge ei. Larger κi tends to map ei to a smaller length. In [5], κi is set to a constant for each edge. This method makes the mapping result more regular, that is, each edge tends to have the same length. This is important in texture mapping and mesh simplification. However in order to reduce the metric distortion, we desire that each edge could keep the length unchanged after mapping. Here, we set each parameter κi as a function of the length of ei:
| (19) |
where d(vi1, vi2) is the geodesic distance between the two vertices of edge ei.
When applying spherical mapping, the surface is first mapped to a planar region and is then stereographically projected into a sphere. As the stereographical projection is nonlinear, the parameter κi defined above can only control the metric distortion on planar and might cause more distortion on sphere. To control the metric distortion on the sphere, we proposed an approximate method to determine κi. First, the surface is mapped to a sphere without the spring energy. From this result, the metric distortion on each edge can be estimated. Subsequently, this distortion is multiplied by κi as a weight. The updated κi becomes:
| (20) |
where d(ui1 − ui2) is the geodesic distance on the first time mapping sphere.
3. Result
3.1 Data and Preprocessing
The method described above is applied to inner cortical surfaces (GM/WM interface) that are reconstructed from T1-weighted SPGR brain images for ten subjects. For the T1-weighted volumetric MR image, we utilize the Oxford FSL tools for preprocessing and brain segmentation. First, non-cerebral tissues including bone, skin, fat, and etc. are removed by Oxford BET tool [17]. Since only the cerebral cortex will be tested in our algorithm, we also remove the cerebellum and brain stem using the HAMMER package [16]. We then use the automated segmentation tool FAST [17] to segment the brain into GM, WM and CSF compartments, where the spatial bias is corrected simultaneously.
Reconstructing topologically correct and geometrically accurate inner surfaces is a challenging problem. An automated graph-based algorithm for topology correction in a binary volume image was proposed in [12]. The graphs are constructed to represent the connectivity of both the foreground and background voxels respectively. Assuming that no cycle exists in the connectivity graphs if the expected surface is homeomorphic to a sphere, the handle removing problem can be considered as removing the cycles in the connectivity graphs. The graph-based method operates on the voxel membership set of the volume, and makes minimal changes to force it to have the topology of a sphere [12]. After topology correction, a voxel-based method such as marching cubes algorithm [14] can be used to reconstruct an isosurface of the WM volume.
In our experiment, the BrainSuite [13] tools that implement the algorithm in [12] were used to remove the topology errors and also to generate the inner cortical surface. Using these tools, the entire cortex can automatically be cut into two approximately symmetric parts. Then the whole inner cortical surfaces are parcellated into 76 anatomic regions with different colors marked for identification using the method in [7, 19].
3.2 Distortion Measurements
To measure the distortion between the original surface and the flat map, some measurements that are invariant to the similarity transformation are defined. We used a method similar to that used for measuring angular and metric distortions in [4]. The angular distortion is defined as:
| (21) |
where θ*i1i2i3 and denote the “market share” [6] angles ∠vi1 vi2 vi3 and ∠U(vi1)U(vi2)U(vi3) respectively.
The metric distortion is defined as:
| (22) |
where N(i) denotes the set of predefined neighbors of vi, dij. The symbol denotes the normalized geodesic distance between vi and vj on the mesh and on its resulted map respectively, and s ∈ R+ is a scale parameter that is used to avoid the difference of similarity transformations. Only the directly connected neighbors (1-neighborhood) of vi are used and illustrated in our experiments. The 2- and 3- neighborhoods are tested, but there is no significant difference (less than 1%), compared to the 1-neighborhood.
The area distortion is defined as:
| (23) |
where A(ti) and AU (ti) are normalized geodesic areas of triangle ti on original mesh and on its corresponding flat map respectively.
The three measurements defined above can well evaluate the local distortions over the surface. To measure the global distortion, we define the region-based area distortion as:
| (24) |
where Re is the set of 76 anatomic regions defined in section 3.1, ri indicates each region in Re, and A(ri) and AU (ri) are normalized geodesic areas of region ri on the original mesh and on its corresponding flat map respectively.
3.3 Spherical Mapping
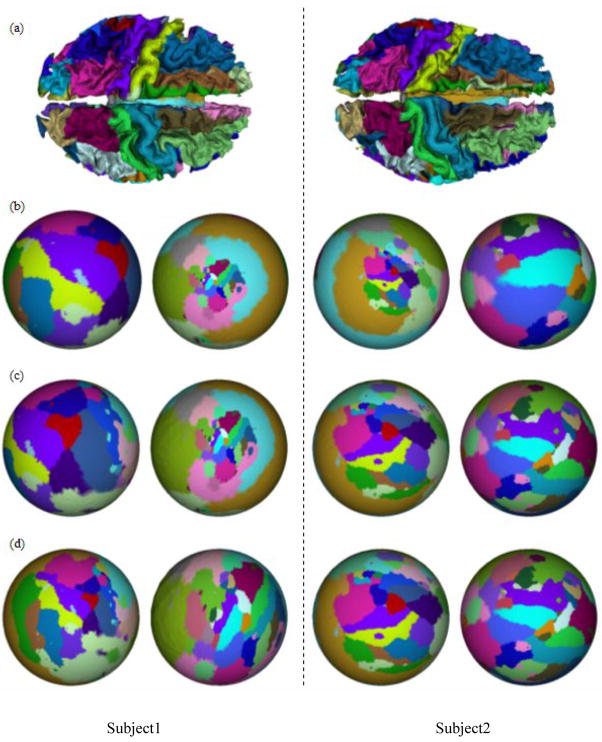
We have applied the spherical conformal brain mapping method in section 2.4 to 10 cases of inner cortical surfaces, which are obtained using the method in section 3.1. As mentioned in section 3.1, each inner cortical surface is parcellated into 76 anatomic regions with different colors marked for identification using the method in [7]. We map each of the cortical surface into spheres with different values of λ. For visual evaluation, Figure 4 shows two examples of spherical mapping results. The results of this mapping for λ = 0, λ = 0.5, and λ = 1.0 are shown in Figures 4a, 4b, and 4c respectively. It is noted that the method of LSCM with spring energy is equivalent to the LSCM method when parameter λ = 0. It is apparent that larger values of λ produce less area distortions, as shown by the relative area ratios of labeled neuroanatomic regions.
Figure 4.
Spherical mapping results of two subjects. (a) Cortical Surfaces. (b) Mapping results with λ= 0. (c) Mapping results with λ=0.5. (d) Mapping results with λ=1.0.
We have demonstrated a quantitative evaluation of the method of LSCM with spring energy in Tables 1–4. The values of angle distortion in the spherical mapping for 10 cases are presented in Table 1, where we have shown that the value of angle distortion increases as the value of parameter λ becomes larger. The results in Table 2–4 show that other three measurements of metric distortion, area distortion and region distortion decrease consistently as the value of parameter λ increases. Specifically, the average metric distortion over 10 cases decreases from 0.59 to 0.49, the average area distortion decreases from 0.82 to 0.76, and the average region distortion decreases from 0.79 to 0.76. This result indicates that the method of LSCM with spring energy reduces the metric and area distortions than LSCM method as expected. Based on the results in Table 1–4, we can see that the method of LSCM with spring energy reduces metric and area distortion, while increasing the angle distortion. Note that in some applications, such as for visualization purposes, the problem of angle distortion is less important than the problem of metric and area distortion. Thus the method of LSCM with spring energy provides the opportunity of sacrificing angle distortion while prioritizing metric and area distortion.
Table 1.
Spherical mapping results of Dangle (U) (degree).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 | λ=10 |
|---|---|---|---|---|---|---|
| 1 | 6.65 | 6.34 | 7.97 | 10.31 | 17.59 | 20.21 |
| 2 | 10.30 | 9.84 | 9.65 | 10.62 | 16.67 | 19.59 |
| 3 | 6.54 | 6.78 | 8.66 | 11.69 | 18.77 | 21.49 |
| 4 | 8.10 | 7.54 | 7.97 | 9.99 | 17.78 | 20.65 |
| 5 | 8.71 | 8.38 | 8.13 | 8.83 | 13.40 | 15.55 |
| 6 | 6.38 | 6.54 | 8.27 | 10.28 | 16.35 | 18.56 |
| 7 | 6.45 | 6.56 | 7.83 | 9.34 | 14.83 | 17.08 |
| 8 | 8.81 | 7.90 | 10.40 | 14.04 | 24.62 | 28.38 |
| 9 | 10.66 | 10.65 | 10.72 | 11.20 | 14.30 | 15.95 |
| 10 | 7.10 | 7.09 | 8.53 | 10.30 | 15.92 | 18.08 |
| Average | 7.97 | 7.76 | 8.81 | 10.66 | 17.02 | 19.56 |
Table 4.
Spherical mapping results of Dregion (U).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 | λ=10 |
|---|---|---|---|---|---|---|
| 1 | 0.773 | 0.770 | 0.764 | 0.815 | 0.746 | 0.734 |
| 2 | 0.779 | 0.781 | 0.800 | 0.787 | 0.750 | 0.748 |
| 3 | 0.792 | 0.792 | 0.803 | 0.794 | 0.756 | 0.748 |
| 4 | 0.790 | 0.783 | 0.764 | 0.749 | 0.731 | 0.733 |
| 5 | 0.753 | 0.751 | 0.764 | 0.745 | 0.689 | 0.667 |
| 6 | 0.784 | 0.783 | 0.785 | 0.800 | 0.825 | 0.817 |
| 7 | 0.771 | 0.764 | 0.747 | 0.744 | 0.742 | 0.731 |
| 8 | 0.838 | 0.818 | 0.805 | 0.802 | 0.819 | 0.837 |
| 9 | 0.774 | 0.772 | 0.771 | 0.769 | 0.803 | 0.788 |
| 10 | 0.843 | 0.837 | 0.813 | 0.803 | 0.790 | 0.787 |
| Average | 0.790 | 0.785 | 0.781 | 0.781 | 0.765 | 0.759 |
Table 2.
Spherical mapping results of Dmetric (U).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 | λ=10 |
|---|---|---|---|---|---|---|
| 1 | 0.593 | 0.588 | 0.573 | 0.524 | 0.488 | 0.476 |
| 2 | 0.620 | 0.620 | 0.554 | 0.538 | 0.519 | 0.515 |
| 3 | 0.603 | 0.595 | 0.539 | 0.532 | 0.507 | 0.499 |
| 4 | 0.593 | 0.583 | 0.570 | 0.561 | 0.514 | 0.513 |
| 5 | 0.574 | 0.562 | 0.511 | 0.491 | 0.441 | 0.429 |
| 6 | 0.590 | 0.586 | 0.573 | 0.560 | 0.492 | 0.480 |
| 7 | 0.588 | 0.582 | 0.564 | 0.544 | 0.471 | 0.459 |
| 8 | 0.576 | 0.530 | 0.514 | 0.506 | 0.497 | 0.499 |
| 9 | 0.628 | 0.623 | 0.609 | 0.599 | 0.553 | 0.545 |
| 10 | 0.531 | 0.529 | 0.529 | 0.525 | 0.518 | 0.519 |
| Average | 0.590 | 0.580 | 0.554 | 0.538 | 0.500 | 0.493 |
It is noted that the cortical mapping process is very fast. For a typical cortical surface with about 50,000 vertices and 100,000 triangles, the average computation time on a P4 2.0GHz CPU is around 15 minutes. In terms of computation efficiency, this method is comparable to the LSCM method in [4], as the method of LSCM with spring energy also employs a linear system similar to that of Eq. (16). Thus, comparing to other time-consuming methods [18], this method is well-suited for fast visualization purposes.
3.4 Hemispheric Mapping
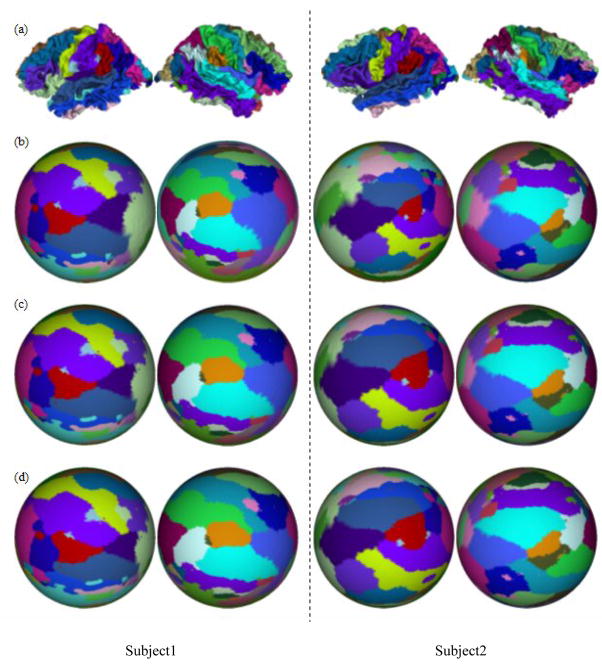
We have also applied the hemispheric conformal brain mapping method in section 2.5 to the same 10 cases of inner cortical surfaces presented in section 3.3. Each the hemispherical cortical surface is mapped to hemispheres with different values of λ. Figure 5 presents the hemispherical mapping results for two subjects. The mapping results with λ = 0, λ = 0.5, and λ = 1.0 are shown in Figure 5a, 5b, and 5c respectively. Similarly, the method of LSCM with spring energy is equivalent to the LSCM method when the parameter λ is set to 0. As expected, larger λ produces less area distortions, as shown by the relative area ratios of labeled neuroanatomic regions.
Figure 5.
Hemispherical mapping results of two subjects. (a) Cortical Surfaces. (b) Mapping results with λ=0. (c) Mapping results with λ=0.5. (d) Mapping results with λ=1.0.
Table 5–8 are the quantitative evaluations of the method of LSCM with spring energy. Table 5 shows the angle distortion in the hemispherical mapping for 10 cases. The average angle distortion decreases slightly from 13.84 to 13.12, when the parameter λ changes from 0 to 0.1. But when λ is above 0.1, the angle distortion increases when the parameter λ becomes larger. The results in Table 6–8 demonstrate that the measurements of metric distortion, area distortion and region distortion decreases consistently with the increasing of parameter λ. Specifically, the average metric distortion over 10 cases decreases from 0.47 to 0.41, the average area distortion decreases from 0.71 to 0.56, and the average region distortion decreases from 0.70 to 0.63. This result further supports that the method of LSCM with spring energy reduces the metric and area distortions than LSCM method. Again, based on the results in Table 5–8, it is apparent that the method of LSCM with spring energy reduces metric and area distortion, while at the same increases the angle distortion. This method of LSCM with spring energy provides the opportunity of being able to sacrifice angle distortion while prioritizing metric and area distortion. This is specially significant when angle distortion is not important in applications such as visualization of cortex.
Table 5.
Hemispherical mapping results of Dangle (U) (degree).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 |
|---|---|---|---|---|---|
| 1 | 15.47 | 15.72 | 14.89 | 16.09 | 21.39 |
| 2 | 13.99 | 14.13 | 14.97 | 15.93 | 21.31 |
| 3 | 14.56 | 14.56 | 15.09 | 14.07 | 19.75 |
| 4 | 13.59 | 13.62 | 12.08 | 14.27 | 20.48 |
| 5 | 9.94 | 11.36 | 13.89 | 14.56 | 19.35 |
| 6 | 13.02 | 8.64 | 16.67 | 13.18 | 20.09 |
| 7 | 14.03 | 13.96 | 14.68 | 15.85 | 18.94 |
| 8 | 16.45 | 14.51 | 15.08 | 15.88 | 19.41 |
| 9 | 14.92 | 14.94 | 15.55 | 17.97 | 20.09 |
| 10 | 12.44 | 9.74 | 11.03 | 14.89 | 20.59 |
| Average | 13.84 | 13.12 | 14.39 | 15.27 | 20.14 |
Table 8.
Hemispherical mapping results of Dregion (U).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 |
|---|---|---|---|---|---|
| 1 | 0.665 | 0.655 | 0.626 | 0.604 | 0.558 |
| 2 | 0.629 | 0.624 | 0.609 | 0.599 | 0.622 |
| 3 | 0.693 | 0.687 | 0.673 | 0.716 | 0.716 |
| 4 | 0.668 | 0.661 | 0.698 | 0.702 | 0.621 |
| 5 | 0.709 | 0.689 | 0.635 | 0.631 | 0.628 |
| 6 | 0.814 | 0.865 | 0.717 | 0.730 | 0.712 |
| 7 | 0.712 | 0.699 | 0.663 | 0.642 | 0.663 |
| 8 | 0.646 | 0.643 | 0.627 | 0.612 | 0.580 |
| 9 | 0.693 | 0.684 | 0.660 | 0.642 | 0.600 |
| 10 | 0.735 | 0.729 | 0.704 | 0.659 | 0.631 |
| Average | 0.696 | 0.693 | 0.661 | 0.654 | 0.633 |
Table 6.
Hemispherical mapping results of Dmetric (U).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 |
|---|---|---|---|---|---|
| 1 | 0.450 | 0.443 | 0.413 | 0.400 | 0.395 |
| 2 | 0.450 | 0.444 | 0.425 | 0.411 | 0.405 |
| 3 | 0.457 | 0.450 | 0.432 | 0.445 | 0.437 |
| 4 | 0.463 | 0.456 | 0.460 | 0.460 | 0.405 |
| 5 | 0.477 | 0.468 | 0.436 | 0.425 | 0.401 |
| 6 | 0.480 | 0.491 | 0.421 | 0.433 | 0.394 |
| 7 | 0.456 | 0.447 | 0.422 | 0.406 | 0.400 |
| 8 | 0.456 | 0.440 | 0.422 | 0.409 | 0.390 |
| 9 | 0.505 | 0.498 | 0.478 | 0.473 | 0.437 |
| 10 | 0.501 | 0.506 | 0.476 | 0.434 | 0.406 |
| Average | 0.470 | 0.464 | 0.438 | 0.429 | 0.407 |
In the hemispherical brain mapping, the procedure of automatically cutting the whole cortical surface into two hemispheres and their mappings to planar regions are fully automatic, and very fast. This step does not add much computational cost to the least-square conformal mapping with spring energy step. Thus the method of hemispherical brain mapping using LSCM with spring energy is desirable for visualization of human brain cortex.
4. Conclusion
It has been shown that the method of LSCM with spring energy can reduce the metric and area distortion while preserving the advantage of fast computation of LSCM in conformal brain mapping. The spring energy weighting parameter λ can be fine-tuned as a user adjusts different types of distortion such as angle distortion, metric distortion, area distortion and region distortion according to different brain mapping applications. Our future work will focus on development of more suitable methods for reducing these distortions simultaneously.
Table 3.
Spherical mapping results of Darea (U).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 | λ=10 |
|---|---|---|---|---|---|---|
| 1 | 0.799 | 0.798 | 0.798 | 0.840 | 0.779 | 0.757 |
| 2 | 0.826 | 0.851 | 0.849 | 0.847 | 0.812 | 0.798 |
| 3 | 0.823 | 0.838 | 0.854 | 0.836 | 0.790 | 0.776 |
| 4 | 0.829 | 0.836 | 0.818 | 0.809 | 0.805 | 0.795 |
| 5 | 0.814 | 0.826 | 0.833 | 0.806 | 0.715 | 0.681 |
| 6 | 0.789 | 0.788 | 0.788 | 0.799 | 0.789 | 0.763 |
| 7 | 0.798 | 0.795 | 0.788 | 0.800 | 0.772 | 0.743 |
| 8 | 0.823 | 0.861 | 0.831 | 0.806 | 0.756 | 0.751 |
| 9 | 0.826 | 0.822 | 0.815 | 0.809 | 0.831 | 0.807 |
| 10 | 0.863 | 0.859 | 0.837 | 0.819 | 0.781 | 0.772 |
| Average | 0.819 | 0.827 | 0.821 | 0.817 | 0.783 | 0.764 |
Table 7.
Hemispherical mapping results of Darea (U).
| Subject index | λ=0 | λ=0.1 | λ=0.5 | λ=1 | λ=5 |
|---|---|---|---|---|---|
| 1 | 0.655 | 0.643 | 0.604 | 0.575 | 0.527 |
| 2 | 0.669 | 0.659 | 0.626 | 0.599 | 0.552 |
| 3 | 0.693 | 0.682 | 0.647 | 0.685 | 0.642 |
| 4 | 0.693 | 0.682 | 0.710 | 0.695 | 0.546 |
| 5 | 0.754 | 0.726 | 0.654 | 0.633 | 0.563 |
| 6 | 0.762 | 0.811 | 0.633 | 0.682 | 0.555 |
| 7 | 0.696 | 0.680 | 0.632 | 0.596 | 0.578 |
| 8 | 0.666 | 0.654 | 0.623 | 0.598 | 0.541 |
| 9 | 0.717 | 0.707 | 0.675 | 0.650 | 0.581 |
| 10 | 0.766 | 0.786 | 0.740 | 0.640 | 0.553 |
| Average | 0.707 | 0.703 | 0.654 | 0.635 | 0.564 |
Acknowledgments
Parts of the datasets and source code were provided by Dr. Lili Ju of the University of South Carolina.
This research was funded by a HCNR research grant awarded to Prof. Stephen T.C. Wong.
Biographies
Jingxin Nie: Mr. Nie is a PhD student at the Department of Automation, Northwestern Polytechnic University, Xi’an, China. He is working with Prof. Guo, Dr. Liu, and Prof. Wong on human brain mapping, cortical surface reconstruction and mapping, deformable surface model, hybrid volume and surface registration, and statistical inference.
Tianming Liu: Dr. Liu is an Instructor of Radiology at Harvard Medical School and Brigham and Women’s Hospital. Dr. Liu received his PhD in computer science from Shanghai Jiaotong University in 2002. Dr. Liu was a Microsoft Fellow, and conducted his PhD dissertation work in Microsoft Research Asia from 2000–2002. Dr. Liu did two years postdoc research at the Department of Radiology at the University of Pennsylvania. His research interests include biomedical imaging and image informatics.
Gang Li: Mr. Li is a PhD student at the Department of Automation, Northwestern Polytechnic University, Xi’an, China. He is working as a research assistant at the Center for Bioinformatics, Harvard Center for Neurodegeneration and Repair, Harvard Medical School. His research interests include deformable registration and warping, neuroimaging, and human brain mapping.
Ashley Tarokh: Dr. Tarokh is a postdoctoral research fellow at the Harvard Medical School and Brigham and Women’s Hospital. She received her PhD in Electrical Engineering from Northeastern University in 2005. Her research interests include medical imaging and reconstruction.
Lei Guo: Dr. Guo is a Professor at the Department of Automation, Northwestern Polytechnic University, Xi’an, China. He has been working in the area of computer vision, neural networks, machine learning, and pattern recognition for over 20 years.
Geoffrey Young: Dr. Young is a clinical instructor at the Brigham and Women’s Hospital and Harvard Medical School. Dr. Young has over 10 years of experience in clinical neuroradiology.
Stephen Wong: Dr. Wong is the Director of HCNR Center for Bioinformatics, Harvard Medical School, and the Executive Director of Functional and Molecular Imaging Center, Department of Radiology, Brigham and Women’s Hospital, and Associate Professor of Radiology, Harvard University. Dr. Wong has 20 years of experience in image analysis, bioinformatics, software development, and scientific management.
References
- 1.Gu X, Wang Y, et al. Genus zero surface conformal mapping and its application to brain surface mapping. IEEE Transactions on Medical Imaging. 2004 Aug;23(8):949–957. doi: 10.1109/TMI.2004.831226. [DOI] [PubMed] [Google Scholar]
- 2.Ahfors LV. Complex Analysis. McGraw-Hill Book company; New York: 1996. [Google Scholar]
- 3.Levy B, Petitjean S, Ray N, Malliot J. Least squares conformal maps for automatic texture atlas generation. Proceedings of ACM SIGGRAPH’02; Addison Wesley. 2002. [Google Scholar]
- 4.Ju L, Stern Josh, et al. Cortical surface flattening using least square conformal mapping with minimal metric distortion. IEEE ISBI; 2004; 2004. pp. 77–80. [Google Scholar]
- 5.Hoppe H, DeRose T, et al. Mesh optimization. SIGGRAPH ’93 Proceedings; 1993. pp. 19–26. [Google Scholar]
- 6.Hurdal MK, Bowers PL, et al. Quasi-conformally flat mapping the human cerebellum. Lecture Notes in Computer Science. 1999;1679:279–286. [Google Scholar]
- 7.Liu T, Shen D, Davatzikos C. Deformable registration of cortical structures via hybrid volumetric and surface warping. NeuroImage. 2004;22(4) doi: 10.1016/j.neuroimage.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Van Essen H, Drury A, Joshi S, Miller MI. Functional and structural mapping of human cerebral cortex: solutions are in the surfaces. PNAS. 1998;95(3):788–95. doi: 10.1073/pnas.95.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipe. C. Cambridge Univ. Press; 1994. [Google Scholar]
- 10.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 11.Steven H, Sigurd A, et al. Conformal Surface Parameterization for Texture Mapping. IEEE Transactions on Visualization and Computer Graphics. 2004 April;6(2):1–9. [Google Scholar]
- 12.Shattuck DW, Leahy RM. Automated Graph-Based Analysis and Correction of Cortical Volume Topology. IEEE Trans Med Imag. 2001;20(11):1167–1177. doi: 10.1109/42.963819. [DOI] [PubMed] [Google Scholar]
- 13.Shattuck DW, Leahy RM. BrainSuit: An automated cortical surface identification tool. Medical Image Analysis. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 14.Lorensen WE, Cline HE. Marching cubes: A high-resolution 3-D surface construction algorithm. ACM Comput Graph. 1987;21(4):163–170. [Google Scholar]
- 15.Nie Jingxin, Liu Tianming, Young Geoffrey, Guo Lei, Wong Stephen TC. Leaset square conformal mapping with spring energy. International Symposium on Biomedical Imaging; 2006. [Google Scholar]
- 16.Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 17.http://www.fmrib.ox.ac.uk/fsl/
- 18.Ju Lili, Hurdal Monica K, Stern Josh, Rehm Kelly, Schaper Kirt, Rottenberg David. NeuroImage. Quantitative evaluation of three cortical surface flattening methods. 2005 December;28(4):869–880. doi: 10.1016/j.neuroimage.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Young G, Huang L, Chen NK, Wong ST. 76-Space analysis of grey matter diffusivity: Methods and applications. NeuroImage. 2006 May 15;31(1):51–65. doi: 10.1016/j.neuroimage.2005.11.041. [DOI] [PubMed] [Google Scholar]