Abstract
Ultrasound-guided brachytherapy using titanium-shelled radioactive seeds is a popular, effective means of treating prostate cancer. Unfortunately, implantation using needles inserted transperitoneally causes gland movement and distortion, which often results in seed misplacement and dosimetry errors. If actual seed locations could be determined in the operating room, then corrections to dosimetry errors could be made immediately. However, seed specularity, shadowing, and tissue clutter make imaging seeds difficult using conventional ultrasound. Singular spectrum analysis (SSA) shows promise for reliably imaging radioactive seeds implanted in the prostate and enabling additional corrective implantations to be made in the operating room. SSA utilizes eigenvalues derived from the diagonalized correlation matrix of envelope-detected radiofrequency echo signals to yield a P-value indicative of the likelihood of a seed-specific repetitive signal. We demonstrated the potential of SSA for seed detection and imaging and illustrated the trade-off considerations for optimization of SSA in clinical applications using simulations assessing performance as a function of different levels of noise and the presence of repetitive signals with various repetition periods; experiments in an ideal scattering environment; and experiments using seeds implanted in beef.
I. INTRODUCTION
Implantation of seeds that contain a radioactive material (e.g., iodine or palladium) into the prostate is becoming a commonly used means of treating gland-confined cancer.1,2 Seed implantation also is gaining acceptance as a means of treating other cancers as well, e. g., breast cancer.3–5 The standard imaging modality for treatment-planning dosimetry and for guiding and monitoring seed implantation is transrectal ultrasound (TRUS).6,7 However, movement of the prostate during seed insertion, shadowing caused by hemorrhage or by other seeds, clutter from calcifications and other hyperechogenic scattering objects, and the specularity of seeds themselves make detecting implanted seeds, determining their actual location, and verifying proper dosimetry difficult in an intraoperative time frame. As a consequence, real-time, intra-operative corrections cannot be made for insufficient treatment doses that result from seed misplacements. Currently, information regarding actual seed location typically is obtained from X-ray computed tomography (CT) or magnetic resonance imaging (MRI) performed three hours to four weeks after the implantation procedure.8–10 At that time, dose corrections can be made only through subsequent external-beam radiation therapy. Radiation oncologists would find a real-time imaging system that is capable of providing accurate, post-insertion, seed-location information to be very valuable because the information would enable timely feedback for intraoperatively correcting deficiencies in the radiation dose delivered to the gland. Implanting dose-correcting seeds in the operating room would permit an optimal treatment effect to be achieved by brachytherapy alone. Therefore, a remaining challenge for TRUS-guided brachytherapy of prostate cancer is accurate detection and localization of the seeds upon their insertion. This article describes initial investigations of a method that may offer a means of meeting this challenge.
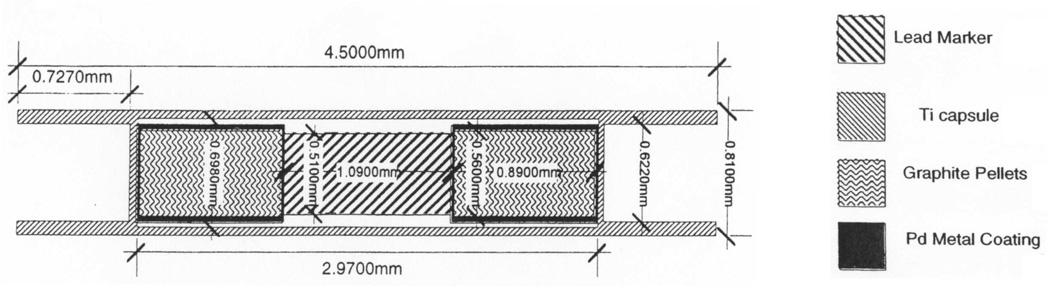
Figure 1 shows a cross-section of a commonly used seed that contains radioactive palladium (Pd125). Seeds used in the prostate nominally are 5 mm long by 0.8 mm in diameter.(The actual length of the Pd seed is 4.5 mm.) Like the Pd seed shown in Fig. 1, most types of prostate-brachytherapy seeds consist of a thin, rigid, cylindrical, titanium shell with enclosed radioactive material; the smooth metal shell causes strong specular reflections of ultrasound waves. However, other commonly occurring strongly reflecting entities, such as calcifications, also often exist in the prostate and may produce false-positive indications if seed detection were to be based solely on backscattered echo-signal strength. Furthermore, the cylindrical shape of most seeds makes their echo strength dependent on the angle of beam incidence. During brachytherapy, seeds are inserted into the prostate using needles that are orthogonal to the scan plane, i.e., to the ultrasound-beam axis. However, because of gland distortion and movement during insertion, an orthogonal orientation with respect to the beam axis can be difficult to maintain after needle withdrawal even when stranded seeds are used.
FIG. 1.
Structure of the brachytherapy seeds used in our study. (Printed with the permission of American Association of Physicists in Medicine.)
The difficulties in achieving optimal seed placement and dose distribution in prostate brachytherapy led us to formulate this study with the following general goal: to improve detection and imaging of brachytherapy seeds using ultrasound signals. This paper describes our initial efforts to identify a seed “signature” and to develop a straightforward, robust, signal-processing method for reliably detecting that signature and depicting the corresponding seed location. The article is organized into the following sections: Sec. II establishes that a distinctive seed signature exists; Sec. III details the signal-processing approach we have developed for signature detection; Sec. IV presents our results from simulations and in vitro experiments; and Sec. V discusses the study and draws relevant conclusions.
II. SEED SIGNATURE
Our initial aim in this study was to determine whether a seed signature exists that would be amenable to detection and localization. To determine whether we could detect a seed ultrasonically, we conducted a simple experiment. We inserted a seed into an acoustically transparent gel standoff pad (Aquaflex, Parker Laboratories, Fairfield, NJ). Following seed insertion, we scanned the gel pad using three, different, single-element, focused ultrasound transducers. The nominal center-frequencies of the imaging transducers were 3.5, 5.0, and 10.0 MHz. These frequencies were chosen in the range of those used to clinically image prostate (near 6 MHz). An Acqiris DP-110 (Acqiris, Monroe, NY) A/D board digitized 8-bit samples of radio-frequency (RF) echo-signals at a sampling rate of 50 MS/s, and a desktop computer stored the digitized signals for subsequent processing, as described below. Figure 2 shows the B-mode image reconstructed from RF echo-signal data obtained with a nominally 5-MHz transducer (V326, Panametrics, Inc., Waltham, MA). The dynamic range of the image is approximately 50 dB. The transducer has an F-number of 5 with a focal distance of 5 cm and an aperture of 1 cm. The shadow below the seed on Figure 2 suggests that the backscattered signals from the seed have a very long “tail” of repeating echoes. This phenomenon also was consistently present for all three transducers.
FIG. 2.
B-mode image of a seed in an acoustically transparent gel pad obtained with a 5-MHz center-frequency transducer.
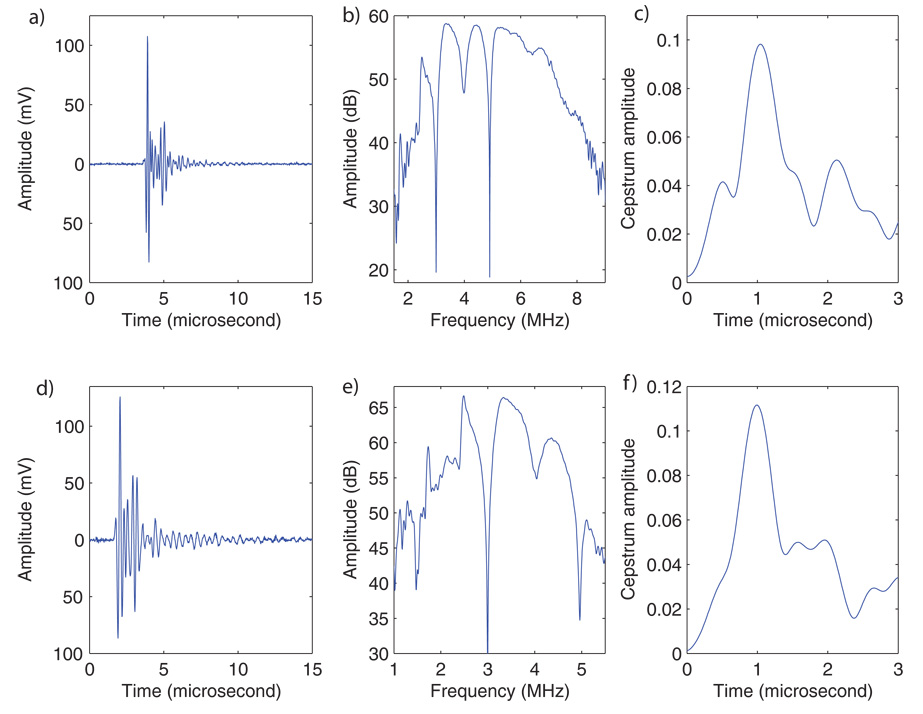
Figures 3a and 3d show typical backscattered RF signals from the seed acquired with the 5- and 3.5-MHz (V383, Panametrics Inc., Waltham, MA) transducers respectively. These signals seem to show a first echo followed about 1 µs later by a second echo that is very similar in shape to the first echo. Figures 3b and 3e show the log power spectra of the backscattered signals shown in Figs 3a and 3d. The spectra contain periodic ripples that are about 1 MHz apart. Scallops in spectra indicate repetitions in the corresponding time signal; they typically are caused by interference between echoes from closely spaced parallel reflecting surfaces. A standard method to test for repetitions in time signals is to compute the cepstrum of the time signal.11 (The cepstrum is the inverse Fourier transform of the log power spectrum.) Peaks in the cepstrum indicate repetitions in the time-signals. The locations of the cepstral peaks in time denote the period of the repetitions. Figures 3c and 3f show the computed cepstra derived from the RF signals of Figs. 3a and 3d. These cepstra contain a single, very clear peak near 1 µs, which indicates that repetitions exist in the original signal with a repetition period (i.e., time separation) of approximately 1 µs. In particular, both RF signals seem to have the same repetition period even though they were acquired using two different transducers. Thus, a repetition frequency near 1 µs may be typical of (and possibly specific for) these titanium-shelled, Pd, brachytherapy seeds. This determination achieved our initial aim by establishing that a signature in fact exists in the form of the distinctive pattern of echo-signal repetitions that we observed. Accordingly, the second aim of this study was to determine whether seeds can be detected and located by using a signal-processing technique that is able to recognize specific repetitions in the echo signals from seeds.
FIG. 3.
RF signals (left), log spectrum (middle), and cepstrum (right) obtained from a seed with a 5-MHz center-frequency transducer (top) and a 3.5-MHz center-frequency transducer (bottom).
III. METHODS
A. Approaches considered
We considered four approaches for sensing characteristic signal repetitions and specifying their locations: cepstral, autocorrelation, spectral autocorrelation, and singular-spectral methods.
The use of cepstra to detect and characterize repetitions in signals that arise from repetitive spacings in tissue architecture is not new to the medical ultrasound community. Cepstra have been used for tissue characterization for more than two decades. In particular, cepstra were used for estimation of mean-scatterer spacing, which is a diagnostically significant parameter for diffuse liver diseases.12 Several methods subsequent to the original cepstral approach developed by Lizzi et al11 were applied successfully. Wear also showed that the use of autoregressive analysis to derive the spectrum for cepstrum analysis improved the accuracy of the cepstral estimate of the mean-scatterer spacing (i.e., provided a lower bias and smaller variance) with a smaller time-gate (i.e., improved spatial resolution).13 The cepstrum is a valuable tool for detecting repetitions in signals (as illustrated in Fig. 3), but cepstra have several drawbacks that make them unsuitable for seed imaging. First, cepstrum computation is sensitive to the bandwidth of the log spectrum used prior to Fourier inversion. For example, to compute the cepstra displayed in Figs. 3c and 3f, only the 2–7.5 MHz and 1–6 MHz bandwidths of the log spectra displayed on Figs. 3b and 3e were used, respectively. Frequency-dependent attenuation decreases the usable bandwidth for computing the cepstrum as a function of depth into tissue, and thus obtaining reliable cepstra may necessitate schemes to correct for bandwidth reduction as a function of depth. Second, specific to our imaging problem, detecting a maximum near 1 µs and then applying a decision criterion for the presence or absence of a seed would be difficult. Third, the presence of an unambiguous cepstral peak near 1 µs, would establish only that a repetition exists, but it would not establish the location of the seed within an A-line, as this study seeks to provide. Using a sliding window to compute short-time cepstrum would be computer intensive. Furthermore, cepstrum quality degrades quickly with decreasing signal length.13
Autocorrelation functions are a second candidate method for detecting and characterizing periodicity in tissue. Wear recently compared cepstral and autocorrelation approaches for evaluating the cortical thickness of the tibia.14 The thickness of the cortical layer of bone is an indicator of fracture risk, and ultrasound offers a less-expensive, non-ionizing alternative to X-ray methods. In his study, Wear demonstrated that both techniques performed comparably for this application.
Varghese and Donohue refined the autocorrelation technique by introducing the spectral autocorrelation function.15,16 Spectral autocorrelation is a third candidate for assessing signal periodicity and Varghese and Donohue showed to be clearly superior to basic autocorrelation methods when strong, diffuse (i.e., unresolved) scatterers were present. In particular, they were able to obtain reliable estimates using shorter signals which provided improved spatial resolution.
Singular spectrum analysis (SSA) is a fourth candidate method for detecting and characterizing signal repetitions. Vautard et al introduced the SSA approach and successfully used it for atmospheric sciences.17,18 The SSA method was used to extract trends in periodicity from 130 years of temperature data.19 Pereira et al applied the SSA approach in ultrasound,20 and it proved to be effective for characterizing properties of trabecular bones.21 The SSA method provided information about bone microstructures that could not be appreciated in B-mode images. Furthermore, Pereira established that his SSA algorithm performed at least as well as the advanced autocorrelation methods.
When used alone, none of these methods (cepstrum, autocorrelation, spectral autocorrelation, and SSA) provides localization information, which is needed for clinically useful imaging of brachytherapy seeds. All of these methods return an estimate of the signal-repetition period over a given region-of-interest (ROI), but all are devoid of information regarding the location of structures of interest within the ROI. In estimating the mean-scatterer spacing of tissue, specific localization information is not expected or needed because the scatterers are smaller than the wavelength and are not resolved. In such applications, the mean-scatterer estimates serve as useful statistical descriptors of the periodic organization of the underlying stochastic tissue architecture. However, to image seeds, we require location information and therefore we are developing a theoretical framework that is based on SSA but that is augmented by a method to retrieve localization information.
B. Singular Spectrum Analysis
The SSA framework is very well-suited to our problem because it allows detecting repetitions with very little sensitivity degradation due to noise, and a heuristic criterion easily can be formulated to detect specific repetition periods (e.g., in our case near 1 µs.). A good sensitivity to a specific periodic echo-signal pattern is necessary because structures other than seeds may exist in tissue (e.g., liver, prostate, etc.) that generate repetitions in the backscattered signals that need to be distinguished from the repetition signals associated with seeds.
The SSA framework assumes that the time-signal is a linear combination of the orthonormal basis that is deduced from the correlation matrix of the time signal. In particular, this orthonormal basis is different for each RF time-signal. The diagonalization of the correlation matrix allows for optimal denoising and easy detection of signal repetitions. Theoretical proofs of these mathematical statements can be found in Vautard and Ghil,17 and they are outside the scope of this paper.
Assume that our time-signal, s, is composed of N samples and that, a sliding window of M (< N) samples is used to estimate the correlation matrix. In our SSA framework for seed detection, s is the zero-mean envelope-detected backscattered RF signal. The correlation matrix, A,of s is of size M by M and element (i,j) is estimated by:
| (1) |
Eq. (1) shows that values of the matrix elements depend only on the absolute value of i−j (for a given signal s), i.e., the matrix A is Toeplitz and symmetric. The diagonalization of A is the first step of our SSA method. After diagonalization, the eigenvalues are ordered from the largest to the smallest, λ1 > … > λM, (the eigenvalues are real and positive because A is symmetric). The eigenvectors, E1,…,EM, are called the empirical orthonormal functions (EOF), and the projections of the original time-signal onto the EOFs yields the principal components (PCs), a1,…,aM.17,19
Each eigenvalue is equal to the variance of the signal in the corresponding PC-direction. Therefore, only the largest eigenvalues are really signal components, and the smallest eigenvalues can be interpreted as noise. In order to filter our time-signal, we selected a heuristic criterion to reject the eigenvalues assumed to correspond to PCs of the noise. We decided to discard all the PCs that corresponded to eigenvalues beyond 90% of the total variance of the signal (i.e., the sum of all eigenvalues: ). Eigenvalue λk was rejected if:
| (2) |
A particularly relevant property of SSA is that periodic components (i.e., repetitions) generate eigenvalue pairs.17,19 Therefore, from eigenvalues that remained after rejection of noise-associated eigenvalues, we retained only the eigenvalues that existed in pairs. However, problems with automated implementation are that 1) two eigenvalues are not likely to be exactly equal, and 2) noise in the system can create spurious pairs. Therefore, we adopted a heuristic criterion for detecting “actual” eigenvalue pairs: λ, and λ, were selected as a non-noisy “actual” periodic pair if the following conditions are simultaneously true:
i < k and j < k, where k is defined in Eq. (2).
The frequencies, fi and fj, at which the spectra (i.e., the magnitude of the Fourier transform) of eigenvectors Ei and Ej reach their maxima, respectively, are such that
Condition 1 discards all eigenvalues assumed to be associated with noise. Condition 2 allows for 4% tolerance to detect “equal” eigenvalues. Finally, condition 3 is added because eigenvalue pairs corresponding to periodic behavior should have EOFs with a very similar spectral content. The set of selected eigenvalues is denoted by Γ.
Following eigenvalue selection, a new time-signal, sr, is constructed from the selected pairs by summing the remaining PCs17:
| (3) |
where the subscripts i and j represent the ith and jth component of the PC and EOF, respectively. As stated above, the PCs are obtained by projection of s over the EOF17:
| (4) |
C. Seed Detection
The signal, sr, constructed from the selected eigenvalue pairs is used first to determine seed presence and location. According to the selection criteria applied to the eigenvalues, the possibility exists that no eigenvalue pair can be selected. In that case, sr vanishes, and the algorithm determines that no seed is present in this time-signal.
If eigenvalues are selected, the Fourier transform, Sr(f), of sr is computed. The spectral power, P, in the bandwidth of interest (i.e., that of the repetition frequency) is computed:
| (5) |
P represents the power of the periodic part of the signal after optimal denoising in the frequency range close to that expected to be generated if a seed were present. Therefore, P can be interpreted as a score for the likelihood of a seed being present. However, P is specific to the entire portion of the original signal, s, within the analysis window, and information on the location of the seed within the window does not exist; that is, the seed signal can be located anywhere in the analysis window. We observed experimentally that the first part of a repetition signal has a greater magnitude than the second part, and we determined that the seed actually is located at the first echo. This is illustrated in Fig. 3a. Therefore, we assume that the location of the seed is where sr achieves its maximum amplitude.
To obtain a color-coded image that depicts seed location based on our detection criteria, each backscattered signal is evaluated using a window size of N samples over which the SSA processing is conducted and leads to the assignment of a P value. This P is displayed at the location where sr achieves its maximum amplitude. The window is then shifted to the next location and the computation is repeated. Windows are designed to provide an 87.5% overlap between two consecutive windows. After the SSA processing has been conducted over the entire A-line, the algorithm moves to the next A-line. Processing ends when all the A-lines are analyzed. The resulting P-map is encoded in false color and imposed over the conventional B-mode image. This color-coded image is called a P-mode image. Higher values of P indicate a higher likelihood of the presence of a seed. A discussion of how to chose and interpret the values of N, M, and the overlap percentage is provided in Sec. V.
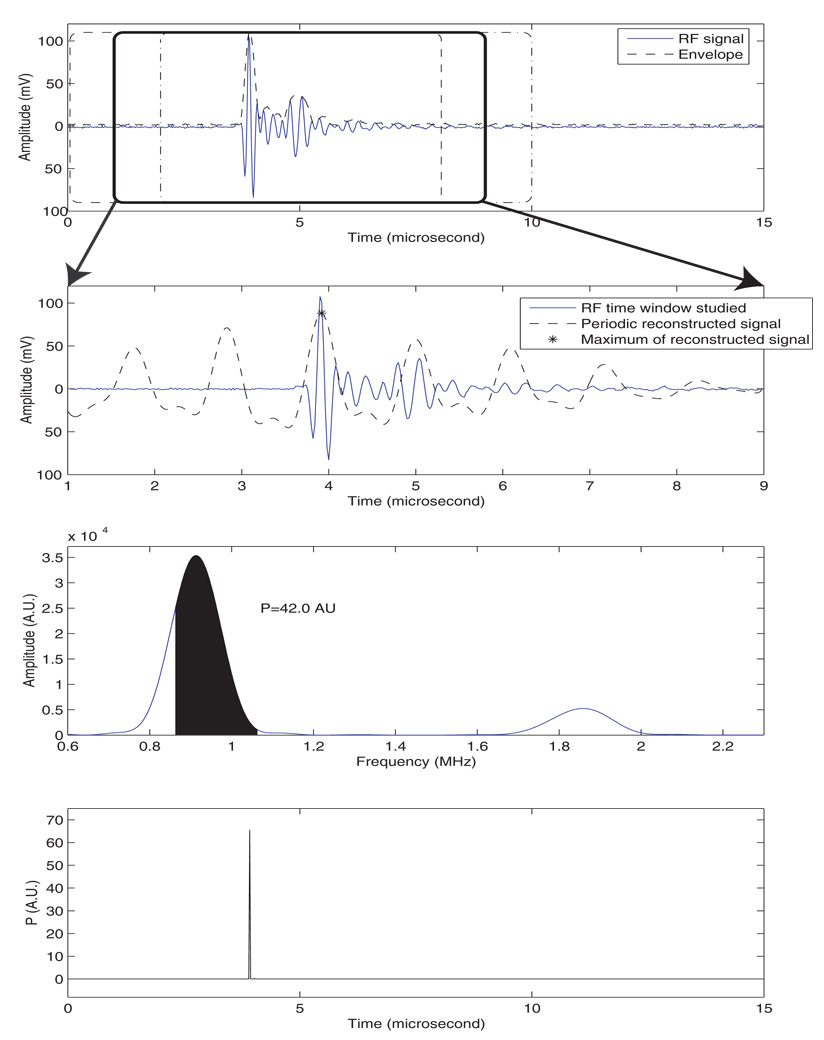
To illustrate the method, we applied it to the center A-line of the B-mode image of Fig. 2. The corresponding RF signal is shown in Fig. 3a. The first N samples were analyzed within the window indicated by the dashed rectangle in Fig. 4a; in this case, N is 400 and the sampling frequency was 50 MHz, so that 400 samples span 8 µs. M was chosen to be the half of N (i.e., 200 samples and 4 µs). The method is illustrated on the second 400-sample window with 87.5 % overlap (i.e., spanning samples 51–450) as indicated by the bold rectangle in Figure 4a). The 200-by-200 correlation matrix of this signal was estimated using Eq. (1), diagonalized, and eigenvalues were ordered from the largest to the smallest. Then, we detected the cut-off eigenvalue by finding the lowest value of index, k, such that Eq. (2) is true. For this example, k was 17. Among these 17 eigenvalues, 2 pairs of eigenvalues met the three requirements of Sec. III.B. The reconstructed signal using these 2 pairs of eigenvalues is shown on Figure 4b. The symbol, *, indicates where the reconstructed signal achieved its maximum amplitude.
FIG. 4.
Illustration of the proposed SSA method. a) RF signal, envelope, and three 8-µs windows with 7-µs overlap (i.e., 87.5 %). b) Reconstructed signal from the SSA method. c) Spectrum of the reconstructed signal, and illustration of the evaluation of Eq. (5). d) P-line.
The spectrum (i.e., magnitude of the Fourier transform) of the reconstructed signal is shown on Fig. 4c. For this signal of a seed in a gel pad, the power integration of Eq. (5) was conducted over a frequency range of 0.86 to 1.06 MHz, because we experimentally noticed a repetition period of approximately 1.04 µs (which corresponds to a repetition frequency of 0.96 MHz) as illustrated by the location of the cepstral peak on Fig. 3c. The dark area under the curve in Fig. 4c depicts this integration, and the value of P for this segment was 42.0 in Arbitrary Units (AU). This value was added to the P-plot at time sample 3.92 µs because it was the time sample at which sr reached its maximum, as denoted by the symbol, *, in Fig. 4b.
The algorithm continued by analyzing the next N-point window with 87.5% overlap (i.e., samples 101–500) and repeated itself until the entire signal was processed. The output of the algorithm was a P-line, i.e., a signal with a length equal to the length of the original signal and with values indicating the relative likelihood of a seed being present. This P-line is shown in Fig. 4d. It contained a very sharp peak near 3.94 µs. The amplitude of the peak was 65.5 AUs, which is greater than 42.0 AUs because the other N-sample windows also contributed to the P-value at this time point. Every A-line was analyzed; the resulting P-lines were merged together to form a 2D matrix of P-values. Then, the P-values were normalized, log-compressed, color-encoded and superimposed on the B-mode image to form the P-mode image. (The corresponding P-mode image will be presented in greater detail in the Results section.)
IV. RESULTS
A. Simulations
To demonstrate and validate the proposed detection method, we conducted several simulations with increasing complexity and levels of noise. All the simulations were performed using a single, computer-generated RF signal (i.e., one A-line) over which our SSA algorithm was conducted. The output of the simulation was a P-line, i.e., the plot of P as a function of time. For all these simulations, the P integration was conducted over the frequency range of 0.9 to 1.1 MHz because we were looking for repetition period near 1 µs.
1. One repetition and white noise
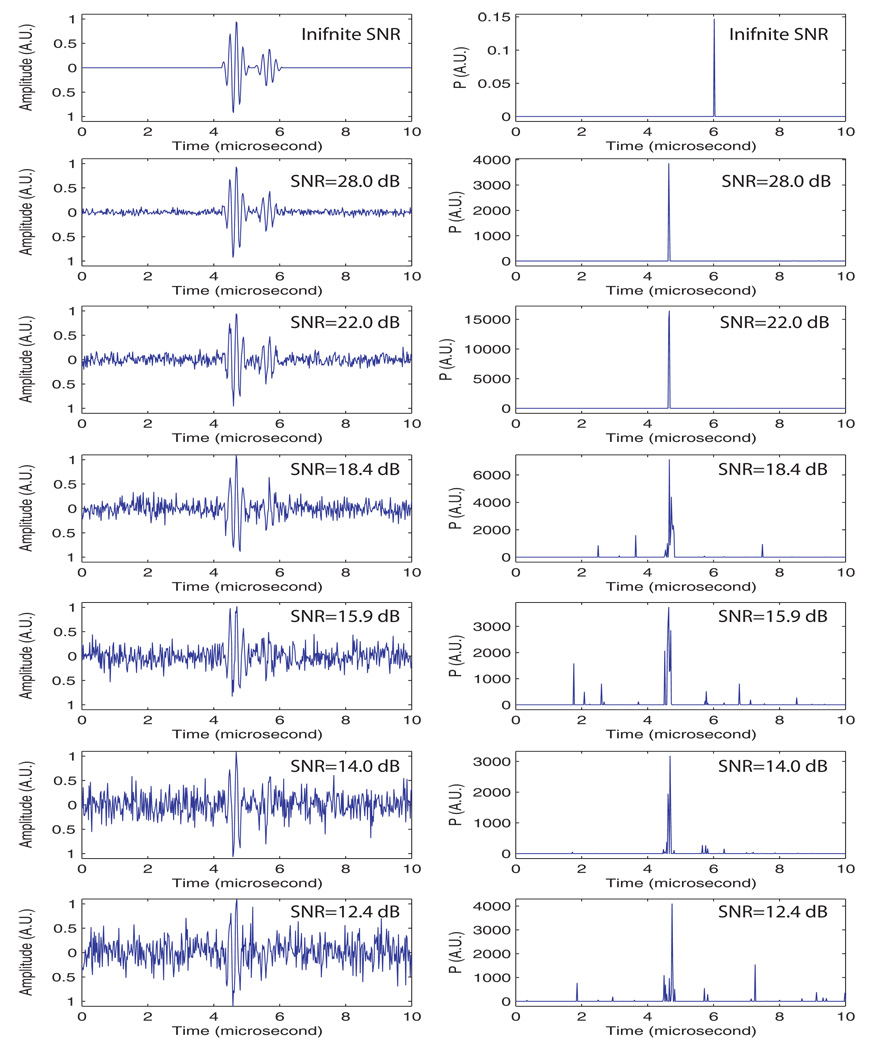
The purpose of this first set of simulations was to assess the robustness of the method in the presence of white noise. An RF-signal that contained an obvious repetition was generated. It contained two, 5-cycle, Hanning-windowed pulses at a center frequency of 5 MHz. The first 5-cycle pulse had an amplitude of 1 while the second had an amplitude of 0.4. These numbers were consistent with what is observed in Figure 3a. The two pulses were separated by 1 µs, and the first 5-cycle pulse always started at 4.25 µs; and its peak was at 4.5 µs. Seven simulations were conducted. The first simulated signal was free of noise, and increasing amounts of white noise were added to the simulated signal in subsequent simulations. The SNR in noise-containing signals ranged from 28 dB to 12.4 dB. For each case, 10 signals were generated and the resulting 10 P-lines were averaged. The left side of Fig. 5 shows examples of signal realizations with a prescribed SNR, and the right side shows the corresponding, averaged P-line.
FIG. 5.
First set of simulations: one repetition in white noise. Left displays one realization at a specific SNR, right displays resulting averaged P-line from 10 realizations at the same SNR.
The first striking observation that can be made from these results is that the worst case was actually the noise-free case for which the P-curve virtually vanished at every sample; in this case, the peak at 6 µs was several orders of magnitude smaller than the other peaks in the other cases. The reason for this virtually-zero P-curve was our criterion for the selection of eigenvalues in which we discarded eigenvalues that were beyond 90% of the total variance of the signal (selection criterion 1), but for a noise-free signal, all eigenvalues are meaningful and important descriptors. Consequently, important information about the noise-free signal was lost and sr did not contain a significant amount of spectral energy near the repetition frequency of 1 MHz.
The next two P-lines (SNRs of 28.0 and 22.0 dB) showed excellent results; a single and sharp peak existed near 4.5 µs, which is where the maximum amplitude of the original signal is located. These two cases show that the algorithm is robust in the presence of noise, and it easily achieves excellent results even with only a moderate SNR.
The last four P-curves showed less-satisfactory results. In particular, two noise effects degraded the results. First, the presence of noise led to the appearance of several side peaks that did not correspond to the actual repetition. Second, the peaks that did correspond to the repetition were broader, i.e., P values spread to the sides and widened the peak. This leakage had the detrimental effects of decreasing the effective SNR (i.e., the peak magnitude was lower, i.e., closer to the noise plateau), and degrading the resolution of the reconstructed P-mode image (i.e., it introduced uncertainty in the seed location depicted by the P-mode image). Nevertheless, the presence of a peak near the actual repetition location demonstrates that the algorithm continues to perform at these low SNRs. However, the possibility exists that the side peaks presented by signals with very low SNRs could cause false-positive seed determinations in some P-mode images.
This set of simulations established that the threshold used for discarding the eigenvalues needs to be a function of noise. The lower the noise level is, the closer the threshold should be to 100% so that the greatest amount of information is retained. However, a number of eigenvalues express the noise, and therefore the threshold needs to be reduced as the SNR becomes lower so that the noise can be separated effectively from the useful information. The threshold value of 90% adopted here was selected simply by trial and error on several seed-imaging cases. Nevertheless, this set of simulations demonstrates that the algorithm behaves very well at reasonable noise levels (e.g., for an SNR > 22 dB), and we expect that the algorithm will perform satisfactorily with the SNRs of signals acquired under clinical conditions.
2. Two different repetitions and white noise
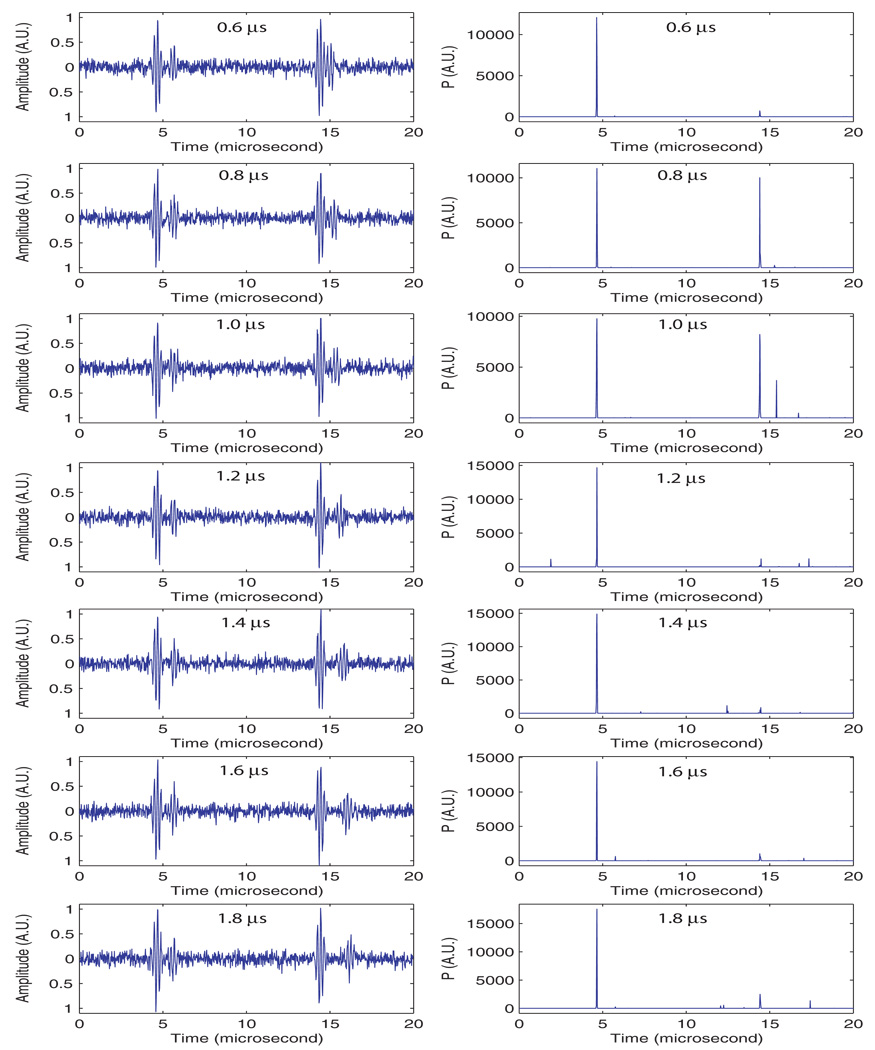
The purpose of this second set of simulations was to evaluate how the algorithm performs when a repetitive signal from a structure other than the seed is present in addition to the signal from the seed. For this set of simulations, the noise was kept at a constant SNR of 22.0 dB. This noise level was chosen based on the results of the previous simulations. The signal consisted of two repetitions: the first one was the same as in the seed-based signal used in the previous simulation, i.e., two 5-cycle pulse with 1 µs time difference. The second, non-seed repetition also was composed of two 5-cycle pulses; for the non-seed signal, the first 5-cycle pulse was always located at 14 µs, the second pulse was located from 14.6 µs to 16.4 µs in 0.2-µs increments giving time differences that ranged from 0.6 to 2.4 µs (i.e., the repetition frequency ranged from approximately 1.67 MHz to 0.42 MHz).
Figure 6 presents the results in the same format that was used for the previous set of simulations. The left side of Fig. 6 shows one realization of each different case, and the right shows the averaged, reconstructed P-line. This figure demonstrates that the selectivity of the algorithm seems excellent because it shows no significant P values near 14 µs except for the two cases with delays of 0.8 and 1 µs for the second repetition signal. For the 1 µs, these relatively large P values occurred because the interval between pulses was within the repetition band (0.90 to 1.10 MHz) to which the algorithm was sensitive. Just as in the previous set of simulations, P leakage existed into the location of the second cycle (at 15 µs) of the repetition. This probably was an effect of noise. The presence of a strong peak for the case in which the delay was 0.8 µs was surprising because the algorithm should not be sensitive to this repetition frequency (i.e., is outside the integration range of 0.9 to 1.1 MHz). The existence of this peak means that the algorithm found significant energy near 1 MHz; it also suggests that the combination of two, 5-cycle bursts led to the creation of a spurious energy component near 1 MHz.
FIG. 6.
Second set of simulations: two repetitions in white noise. Left displays one realization for specific time difference for the second repetition, right displays resulting averaged P-line from 10 realizations. SNR was kept at 22 dB.
This set of simulations demonstrates that the SSA algorithm generally is very selective and capable of differentiating among repetitions having different frequencies. These capabilities are important for many clinical, imaging situations where periodic scattering structures may exist that are regularly organized and capable of creating systematic repetitions in the RF signals.
3. One repetition and speckle noise
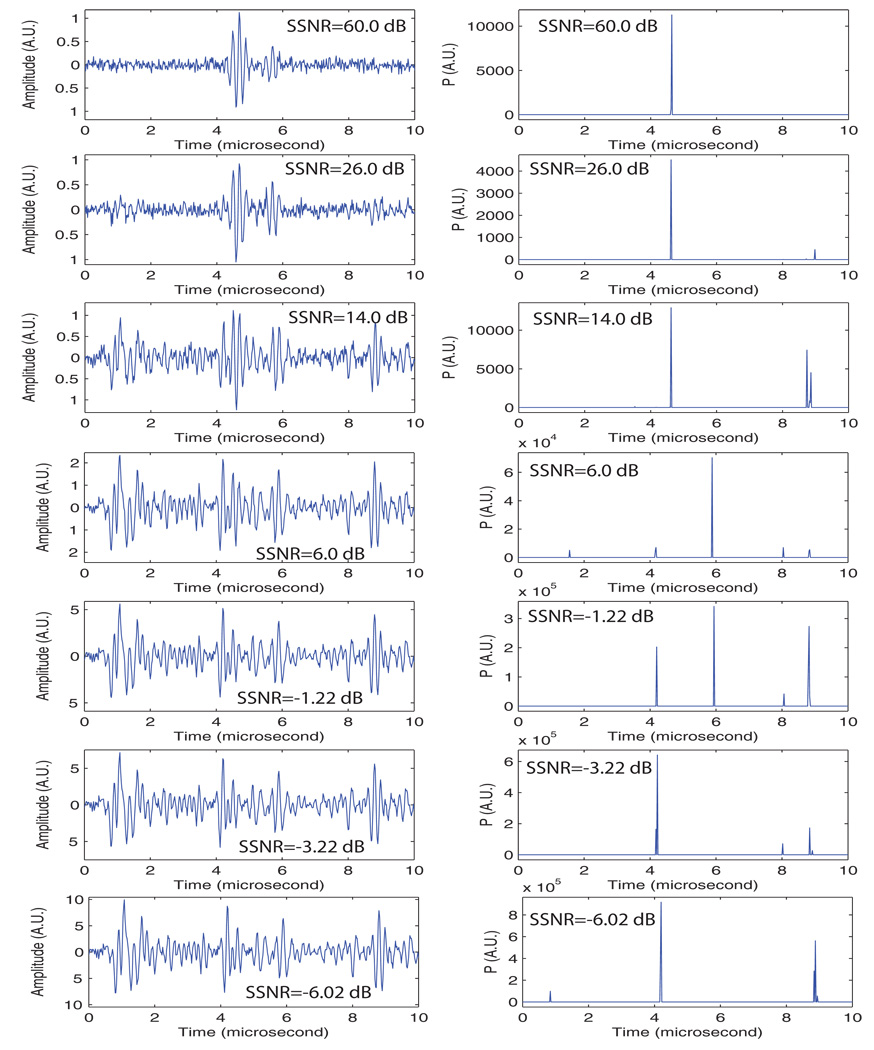
The purpose of this third set of simulations was to test the SSA approach in a setting that would occur when imaging biological tissues. For this set of simulations, speckle noise was added to the simulated signal containing the original repetition. Speckle was obtained by acquiring signals from degassed beef. We used the 5-MHz transducer and a sampling frequency of 50 MHz; these parameters are the same as those used for generating the simulated repetition. A signal with a single repetition was generated, and the acquired “speckle noise signal” was added. We also added white noise so that the SNR was kept at 22 dB. We varied the signal-to-speckle-noise ratio (SSNR) from 60 dB to −6 dB. We defined the SSNR as the ratio of the maximum of the simulated signal to the standard deviation of the speckle noise signal.
Figure 7 shows the results. The 60-dB SSNR case was essentially equivalent to the case of Fig. 5 for which the SNR was 22 dB. The seed is clearly detected and no artifacts or false-positive are found on the P-line. The next two cases (i.e., SSNR of 26.0 and 14.0 dB) showed satisfactory results with a large peak at the actual seed location; however a smaller, but significant, peak also was present at a location far away from the actual seed (near 9 µs). The 6-dB case shows an important peak at the location of the second part of the repetition (near 6 µs). This phenomenon also is apparent on certain P-lines in Fig. 5. The −1.22-dB case showed several peaks near 4, 6, 8, and 9 µs. The algorithm apparently broke down at this SSNR. Surprisingly, fairly satisfactory results were obtained for the last two cases with even lower SSNRs.
FIG. 7.
Third set of simulations: one repetition in speckle noise. Left displays one realization at a specific SSNR, right displays resulting averaged P-line from 10 realizations at the same SSNR. Speckle noise signal was acquired with 5-MHz center frequency transducer from a degassed beef steak. SNR was kept at 22 dB.
This set of simulations demonstrates that the algorithm is quite robust even in the presence of speckle noise acquired from biological tissues. However, it also shows that, when a repetition is present, it may assign a high P-value to the second pulse of the repetition, which in turn may result in a P-mode image showing a seed very slightly deeper than its actual location. (Fortunately, the resulting error would be less than a millimeter, and would not affect the delivered dose in a clinically significant manner.)
B. Seed in gel pad
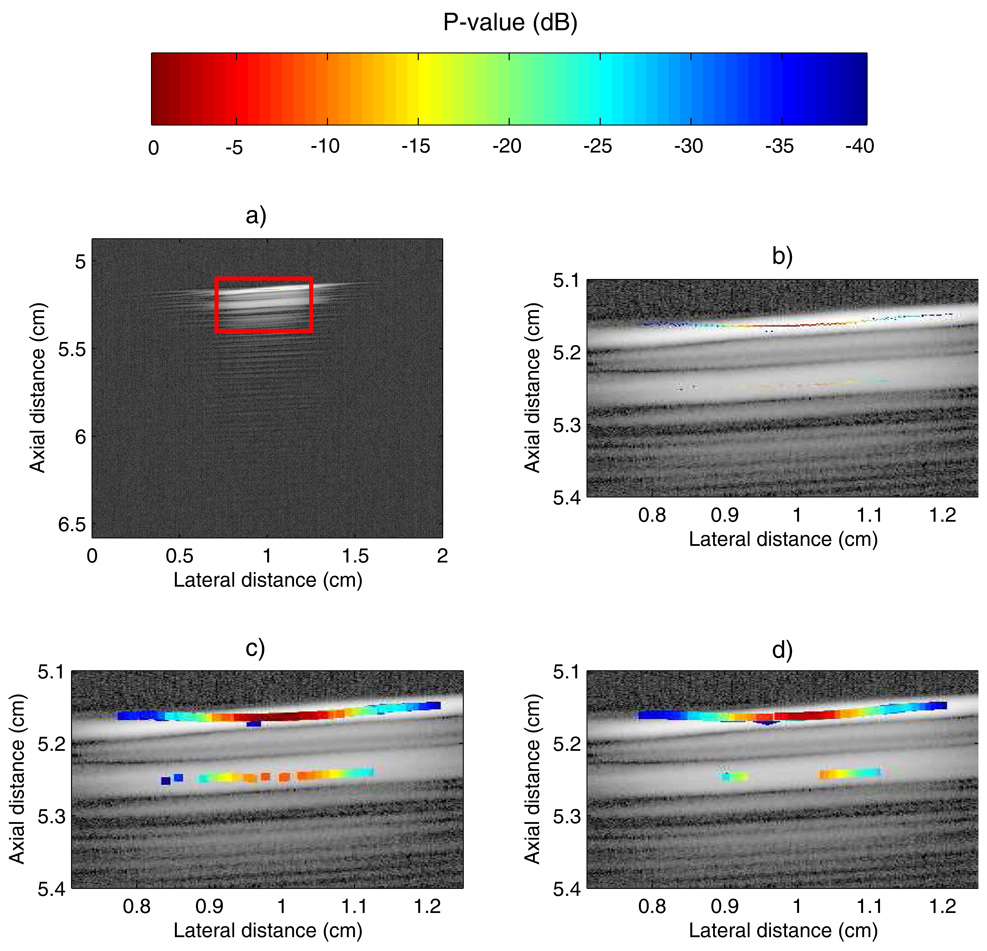
We evaluated the SSA method in the ideal case of a 5-MHz scan of a seed in an acoustically transparent gel pad (as shown in Fig. 2). The reconstructed P-mode image is displayed in Figure 8a. This image was reconstructed by retaining the P-values that fall within a 40-dB dynamic range. Specifically, if a value of P was outside that dynamic range, then it was replaced by the B-mode data. As shown on Figure 8a, all of the P-values in the 40-dB dynamic range are very well localized in the red rectangle. For this case, basically no leakage of P-values occurred outside the actual seed location. Unfortunately, the color-encoded information shown in Fig. 8a is difficult to appreciate; Fig. 8b shows an expanded version of the red rectangle. However, even in the expanded version, the color-coded information remains somewhat difficult to assess.
FIG. 8.
Images of a single seed in an acoustically transparent gel stand-off pad. a) P-mode image of a seed in gel pad. b) Zoomed-in version of red rectangle of a). c) Dilation of b). d) Median-filtered version of c).
To improve the reconstructed P-mode image, we consecutively applied two non-linear image-processing steps. First, we used a morpho-mathematical operation called dilation with a square mask of side length 100 µm. This operation essentially expands the color-coded region. A more-mathematical description of dilation is outside the scope of this paper, but can be found in Serra.22 The resulting dilated image is shown in Fig. 8c. Second, we applied a median filter with a square mask of side length 200 µm. This operation reduced the noise and also allowed for the “shadow” of the seed (i.e., the lower intensity P-values contained below the main line of large P-values in the red rectangle) to be somewhat mitigated. In some cases, the seed shadow appears because the reconstructed signal has its maximum at the second repetition echo complex instead of the first one. However, the “shadow” P-values are less connected so that the median filter is able to reduce their effect as shown in Fig. 8d. In the remainder of this paper, the displayed P-mode images always include these two image-processing operations.
The final P-mode image (Fig. 8d) shows excellent results, the seed is clearly visible and the SSA algorithm did not produce any false-positive indications. The “shadow” persists, but with P-values approximately 20 dB lower than those at the true seed location. This example shows that the SSA method performs very well for this controlled case.
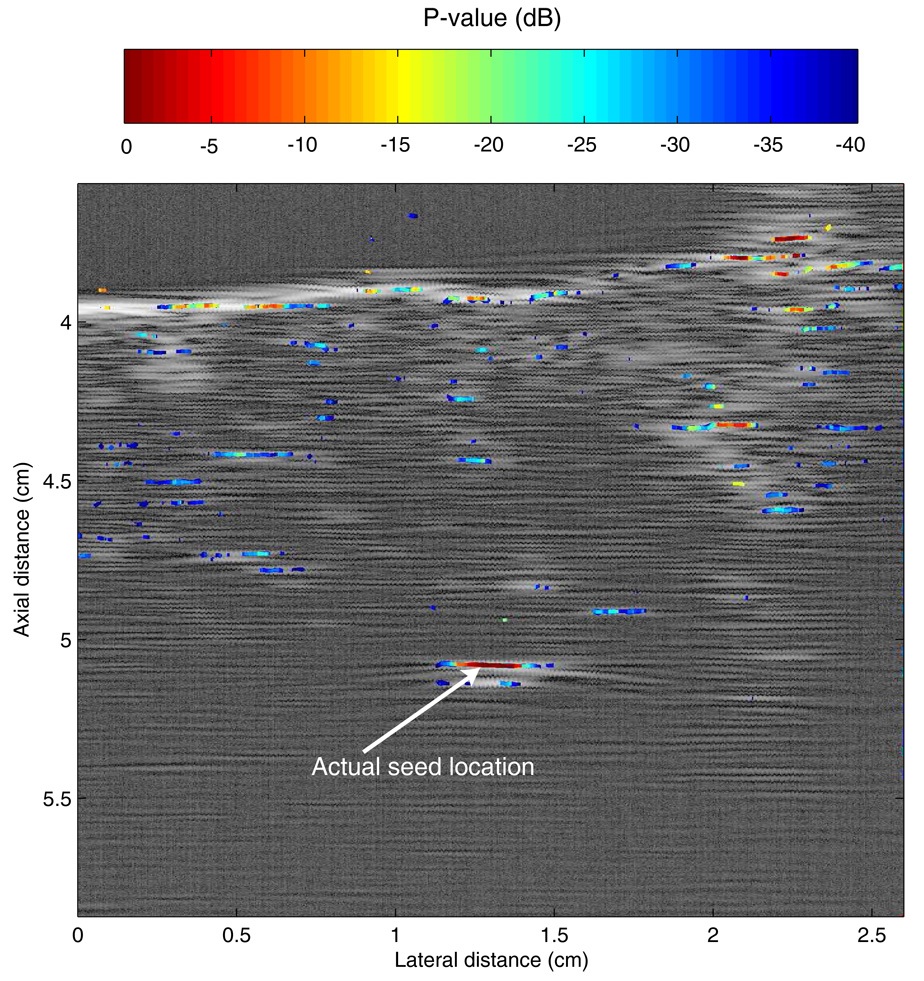
C. Ex vivo experiment
The last step of this preliminary study was to evaluate the SSA algorithm ex vivo. A seed was implanted in a piece of fresh, degassed beef, and several planes were scanned with the 3.5-MHz transducer and processed using our SSA method. In this setting, the true location of the seed was known because it was implanted into the piece of beef with a needle after the beef was degassed. This allowed for direct assessment of the method. The experiment also is interesting because biological tissues are strong generators of speckle and also the small, spatial variations of acoustical properties that exist in tissue create scattering and phase aberrations. This experiment was meant to evaluate the performance of the method in a more-challenging setting that is more presentative of conditions to be encountered in clinical applications.
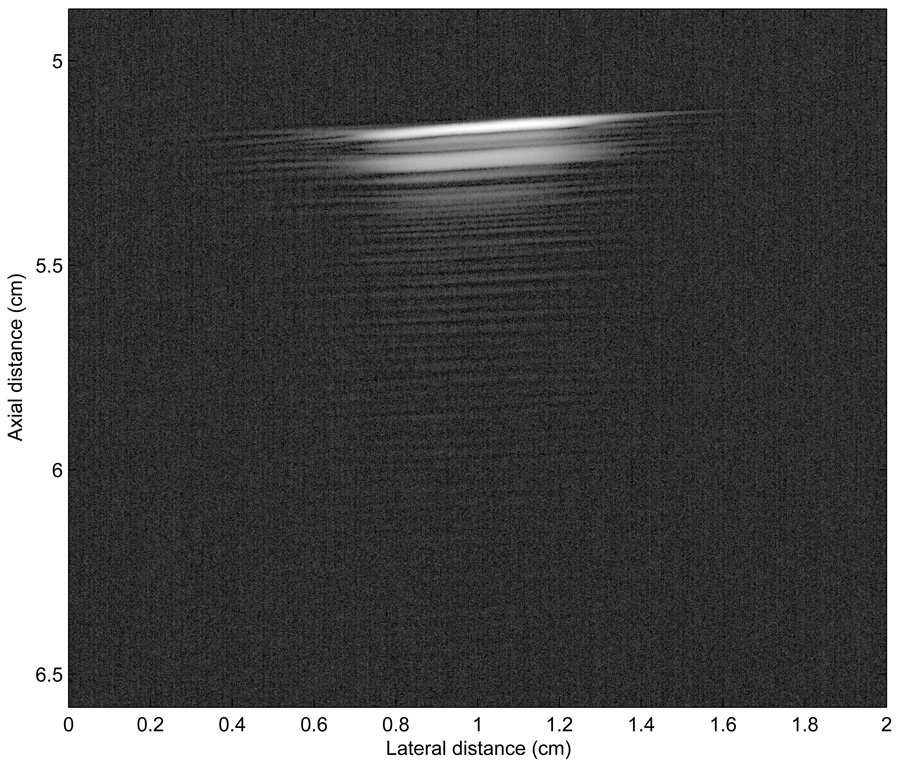
Figure 9 shows a P-mode image obtained ex vivo. The seed location is indicated by an arrow. The image shows that for this more-challenging case, P-value leakage clearly exists as indicated by the scattered light to dark blue squares present on the image. However, the leakage P-values remain low (< −30 dB); therefore, the leakage is recognized clearly as noise. In Fig. 9, the largest P-value is at the actual seed location, and we can state confidently that, in this image, the seed is localized using our SSA method. However, in the same image, significant P-values are scattered along the water-meat interface. In particular, a strong (about −3 dB) P-value is displayed at an axial distance of approximately 3.7 cm and a lateral distance of 2.25 cm. In the absence of prior knowledge, this component might be interpreted as a seed, and if so, would represent a false-positive. However, in the present case, prior knowledge of the water-meat boundary allows for easy dismissal of this potential false-positive. The only other possible false positive is located at an axial distance of 4.4 cm and a lateral distance of 2.1 cm. The P-values at that location are about −10 dB so that even though significantly lower than those of the actual seed, this connected component possibly might be interpreted incorrectly as a seed.
FIG. 9.
P-mode image obtained with a 3.5-MHz center-frequency transducer of a seed implanted in a piece of degassed beef.
In the clinic, a priori knowledge would be applied clinically on a 3-D basis using adaptations of current clinical dosimetry-planning software to depict apparent seed locations in 3-D. E.g., if 40 seeds were implanted and 43 seeds were depicted, but 40 were shown to be within the gland and 3 were shown to be outside the gland, then a reasonable initial determination would be that all seeds were within the gland and their locations would be used to compute the actual dose distribution; a dose correction could be made by adding seeds if necessary. Similarly, if 40 seeds were implanted and 43 seeds were depicted, but only 37 were within the gland while 6 were shown to be 3 outside it, then an inadequate dose to the gland would be established and a correction could be made immediately.
V. DISCUSSION
The proposed algorithm appears to be an attractive choice for seed-localization and imaging because it allows seed detection and depiction to be performed independently of transducer technology or beam-forming algorithms. These advantages obviously are attractive for real-time clinical imaging of the prostate. However, the top-priority remaining task in developing this algorithm is optimization of the design parameters. These parameters include eigenvalue-pair selection (conditions 1 to 3 in Sec. III.B), integration frequencies (f0 and f1) for the computation of P in Eq. (5), and windowing parameters (N, M, and the overlap percentage, OP).
When making the choice of N, M, and OP, the first obvious engineering consideration is processing time. Smaller values of N and M, and larger values of OP require longer times for the algorithm to complete. In this study, computation time was not considered critical, and the algorithm implementations in Matlab (Mathworks Inc., Natick, MA) took several seconds for treating a single A-line with the initially used window parameters (N = 400, M = 200, and P = 87.5%). Using the current Matlab implementation, processing a complete, conventional, clinical, B-mode image to generate the color-coded P-mode image would require several minutes. We are confident that the algorithm can be implemented in a more-rapid manner (e.g., using C++), and that available computer technology will allow for clinically practical, real-time computation of P-mode images.
More important is making the optimal choice of N and M with respect to the repetition frequencies to which the algorithm is sensitive. N must be sufficiently large to span the anticipated range in repetition period within the N-sample signal; M must be sufficiently large to encompass a single cycle of the repetition. Thus, to properly design the SSA implementation, the nature of repetitions to be detected must be determined with care. A statistical interest also exists in having N and M as large as possible so that the accuracy of the correlation estimation described in Eq. (1) is maximized.
The choice of OP can be related to the SNR of the resulting P-line. In particular, a larger OP will lead to a higher SNR. Specifically, if a sufficiently large overlap exists, then a repetition will be contained in several consecutive windows; this effect will produce a high combined P-value at the true seed location. If noise is dominant, a very broad noise spectrum would lead to the production of several eigenvalue pairs with spectral peaks near the repetition frequency. However, because of the random nature of noise, the spurious P-values arising from noise are unlikely to combine at the same time-sample. Thus, the spurious P-values are most likely spread randomly and simply lead to an increased SNR of the P-line. The tradeoff in the choice of OP is in effect a tradeoff between computational time and SNR in the reconstructed P-line.
As demonstrated in the simulations, the eigenvalue-selection threshold is related to noise and numerical analysis. The eigenvalue cut-off threshold can be related to the SNR in the system, and it easily can be tuned to optimize sensitivity and specificity. Furthermore, according to the first set of simulation results, threshold selection is fairly robust on a large range of SNR. (We used a 90% threshold value for all the presented analyses and results.) In the absence of noise, the eigenvalues of a periodic process theoretically are exactly equal if the correlation matrix is estimated accurately. Noise has two effects: it can separate actual pairs, and it can create spurious pairs. We did not experience difficulty in tuning these threshold parameters, but the problem is “discontinuity” in the processing. A discontinuity occurs when an infinitesimal change in the data causes a large change in the results; this occurs if a pair of eigenvalues near the threshold either is selected or discarded thus resulting in a different reconstructed signal and hence a different P-value. Therefore, P is not a continuous function of the data, which potentially could cause robustness issues for the algorithm in certain “close” cases.
Finally, the integration window (i.e., f0 and f1 in Eq. (5) must be specified by studying signals from seeds in a controlled environment, as we did in our laboratory experiment. A reasonable possibility is that look-up tables of values for f0 and f1 could be designed in vitro and then input to medical scanners for in vivo imaging.
Frequency-dependent attenuation may affect the robustness of the algorithm because the seed signature may change as a function depth. The effect of frequency-dependent attenuation currently is under investigation. Nevertheless, we anticipate that the algorithm can be implemented with depth-dependent parameters to correct for possible changes in seed-signature as a function of depth.
A remaining step in refining the method is to evaluate its dependence on seed orientation. While the implanted seed axis ideally is orthogonal to the transducer beam axis and scan plane, orthogonality rarely is maintained, and variations exist in seed orientation with respect to the scan plane. The cylindrical geometry and highly specular reflective symmetry of the seed may make the efficacy of the SSA method depend upon the angle between the seed and the beam axis. We have begun to examine at cases where the seed orientation was varied, and have found that the repetition period may be a function of the angle between the seed and beam axes. Thus, evaluation of angle-dependence of the method is being strongly pursued, and results will be reported in subsequent papers.
The physical origin of the repetition signal backscattered from a seed also is under investigation. We have not yet determined conclusively what the origin of this signal is. However, experiments with a range of frequencies extending from 3 MHz to 25 MHz showed that the repetition signal occurred independent of frequency. These preliminary measurements did not show any statistically significant differences among the seed repetition-signal periods at different insonification frequencies. Previous unpublished studies by Feleppa et al. using 5, 10, and 15 MHz suggested that the repetition period is not frequency-dependent. Because all our measurements led to repetition periods in the narrow range of 0.8 to 1.2 µs, we conclude that if the repetition period is a function of frequency, then the dependence is not strong. A physical understanding of the cause of the signal periodicity may permit improvement of the SSA algorithm. Therefore, we are pursuing this understanding actively.
A necessary extension of the completed study is assessing errors made by the algorithm, specifically, an evaluation of the false-positive and false-negative determinations. Certainly, error management can be improved by adding a priori knowledge to the detection algorithm. The clinician knows how many seeds are injected. This number can be input into the algorithm to limit the algorithm to detecting a fixed number of seeds within or near the gland. Even more important, although misplacement of seeds occurs because of movement and distortion of the gland, the clinician always knows approximately where the seed should be located. In fact, clinical dosimetry software currently utilizes 3D depictions of the gland shape to compute optimal needle loadings and seed placements. The software-prescribed locations can be exploited to engineer a penalty for finding a seed very far from the prescribed locations.
After algorithm development is complete, the algorithm will be tested over a large number of actual clinical cases to obtain ROC (receiver operator characteristic) curves that will provide an objective assessment of algorithm performance under actual clinical conditions.
ACKNOWLEDGEMENTS
The authors thank Dr. W. C. A. Pereira for many stimulating and informative discussions regarding SSA and Dr. C-S Wuu for his insights and perspectives regarding current methods of seed placement and dosimetry in prostate brachytherapy. We also thank E. Girard for her support in the experimental aspects of these studies.
This research is supported in part by NIH grant CA098465.
Footnotes
PACS numbers: 43.60.Lq, 43.80.Vj, 43.35.Yb
REFERENCES
- 1.Holm HH, Juul N, Pedersen JF, Hansen H, Stroyer I. Transperineal 125iodine seed implantation in prostatic cancer guided by transrectal ultrasonography. J. Urol. 1983;130:283–286. doi: 10.1016/s0022-5347(17)51108-8. [DOI] [PubMed] [Google Scholar]
- 2.Blasko JC, Ragde H, Schumacher D. Transperineal percutaneous iodine-125 implantation for prostatic carcinoma using transrectal ultrasound and template guidance. Endocurie./Hypertherm. Oncol. 1987;3:131–139. [Google Scholar]
- 3.Niehoff P, Ballardini B, Polgar C, Major T, Hammer J, Richetti A, Kovacs G. Early european experience with the mammosite radiation therapy system for partial breast brachytherapy following breast conservation operation in low-risk breast cancer. Breast. 2006;15:319–325. doi: 10.1016/j.breast.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Niehoff P, Polgar C, Ostertag H, Major T, Sulyok Z, Kimmig B, Kovacs G. Clinical experience with the mammosite((r)) radiation therapy system for brachytherapy of breast cancer: Results from an international phase ii trial. Radiother Oncol. 2006;79:316–320. doi: 10.1016/j.radonc.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Patel RR, Das RK. Image-guided breast brachytherapy: an alternative to whole-breast radiotherapy. Lancet Oncol. 2006;7:407–415. doi: 10.1016/S1470-2045(06)70692-1. [DOI] [PubMed] [Google Scholar]
- 6.Crook J, McLean M, Catton C, Yeung I, Tsihlias J, Pintilie M. Factors influencing risk of acute urinary retention after trus-guided permanent prostate seed implantation. Int J Radiat Oncol Biol Phys. 2002;52:453–460. doi: 10.1016/s0360-3016(01)02658-x. [DOI] [PubMed] [Google Scholar]
- 7.Wei Z, Ding M, Downey D, Fenster A. 3d trus guided robot assisted prostate brachytherapy. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2005;8:17–24. doi: 10.1007/11566489_3. [DOI] [PubMed] [Google Scholar]
- 8.Brinkmann DH, Kline RW. Automated seed localization from ct datasets of the prostate. Med Phys. 1998;25:1667–1672. doi: 10.1118/1.598346. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, Cheng G, Yu Y, Brasacchio R, Rubens D, Strang J, Liao L, Messing E. Automatic localization of implanted seeds from post-implant ct images. Phys Med Biol. 2003;48:1191–1203. doi: 10.1088/0031-9155/48/9/308. [DOI] [PubMed] [Google Scholar]
- 10.Miquel ME, Rhode KS, Acher PL, Macdougall ND, Blackall J, Gaston RP, Hegde S, Morris SL, Beaney R, Deehan C, Popert R, Keevil SF. Using combined x-ray and mr imaging for prostate i-125 post-implant dosimetry: phantom validation and preliminary patient work. Phys Med Biol. 2006;51:1129–1137. doi: 10.1088/0031-9155/51/5/005. [DOI] [PubMed] [Google Scholar]
- 11.Lizzi FL, Feleppa EJ, N Y. Differentiation of tumors types in vivo by scatterer property estimates and parametric images using ultrasound backscatter. Proceedings of the 2003 IEEE Ultrasonics Symposium. 2003:1014–1017. [Google Scholar]
- 12.King DL, Lizzi FL, Feleppa EJ, Wai PM, Yaremko MM, Rorke MC, Herbst J. Focal and diffuse liver disease studied by quantitative microstructural sonography. Radiology. 1985;155:457–462. doi: 10.1148/radiology.155.2.2984720. [DOI] [PubMed] [Google Scholar]
- 13.Wear K, Wagner IMF, Hall TJ. Application of autoregressive spectral analysis to cepstral estimation of mean scatterer spacing. IEEE Trans Ultrason Ferroelectr Freq Control. 1993;40:50–58. doi: 10.1109/58.184998. [DOI] [PubMed] [Google Scholar]
- 14.Wear KA. Autocorrelation and cepstral methods for measurement of tibial cortical thickness. IEEE Trans Ultrason Ferroelectr Freq Control. 2003;50:655–660. doi: 10.1109/tuffc.2003.1209552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varghese T, Donohue KD. Mean-scatterer spacing estimates with spectral correlation. J Acoust Soc Am. 1994;96:3504–3515. doi: 10.1121/1.410611. [DOI] [PubMed] [Google Scholar]
- 16.Varghese T, Donohue KD. Estimating mean scatterer spacing with the frequency-smoothed spectral autocorrelation function. IEEE Trans Ultrason Ferroelectr Freq Control. 1995;42:451–463. [Google Scholar]
- 17.Vautard R, Ghil M. Singular spectrum analysis in nonlinear dynamics, with applications to paleoclimatic time series. Physica D. 1989;35:395–424. [Google Scholar]
- 18.Broomhead D, King G. Extracting qualitative dynamics from experimental data. Physica D. 1986;20D:217–236. [Google Scholar]
- 19.Vautard R, Yiou P, Ghil M. Singular-spectrum analysis: a toolkit for short, noisy chaotic signals. Physica D. 1992;58:95–126. [Google Scholar]
- 20.Pereira WC, Maciel CD. Performance of ultrasound echo decomposition using singular spectrum analysis. Ultrasound Med Biol. 2001;27:1231–1238. doi: 10.1016/s0301-5629(01)00423-9. [DOI] [PubMed] [Google Scholar]
- 21.Pereira WC, Bridal SL, Coron A, Laugier P. Singular spectrum analysis applied to backscattered ultrasound signals from in vitro human cancellous bone specimens. IEEE Trans Ultrason Ferroelectr Freq Control. 2004;51:302–312. [PubMed] [Google Scholar]
- 22.Serra J. Image analysis and mathematical morphology. New York, NY: Academic Press; 1982. [Google Scholar]