Abstract
Non-enzymatic glycation of tissue proteins has important implications in the development of complications of diabetes mellitus. While electron transfer dissociation (ETD) has been shown to outperform collision-induced dissociation (CID) in sequencing glycated peptides by tandem mass spectrometry, ETD instrumentation is not yet widely available and often suffers from significantly lower sensitivity than CID. In this study, we evaluated different advanced CID techniques (i.e., neutral-loss-triggered MS3 and multi-stage activation) during liquid chromatography/multi-stage mass spectrometric (LC/MSn) analyses of Amadori-modified peptides enriched from human serum glycated in vitro. During neutral-loss-triggered MS3 experiments, MS3 scans triggered by neutral losses of 3 H2O or 3 H2O + HCHO produced similar results in terms of glycated peptide identifications. However, neutral losses of 3 H2O resulted in significantly more glycated peptide identifications during multi-stage activation experiments. Overall, the multi-stage activation approach produced more glycated peptide identifications, while the neutral-loss-triggered MS3 approach resulted in much higher specificity. Both techniques are viable alternatives to ETD for identifying glycated peptides.
Glycation of proteins is a non-enzymatic process. It begins with reaction between an aldehyde group of a reducing sugar (glucose, fructose, etc.) and a primary amine of a protein to form a reversible Schiff base intermediate, which then rearranges to form a relatively stable ketoamine or Amadori adduct.1 Under oxidative conditions, the Amadori adduct decomposes into more reactive carbonyl compounds that can further modify proteins. These advanced glycation end products (AGEs) have been implicated in the development of nephropathy, neuropathy, retinopathy, and cardiovascular disease during the progression of diabetes mellitus.2-4 In addition, protein glycation is relevant to the pharmaceutical industry, as reducing sugars are used as stabilizers in the formulation of therapeutic proteins.5 Inevitably, glycated proteins will be generated as impurities in these processes, negatively impacting the quality of the final products.
Liquid chromatography (LC) coupled to mass spectrometry (MS) via electrospray ionization (ESI) has long been used for the study of protein glycation, from analyzing fructoselysine (lysine containing the Amadori adduct) produced by total protein hydrolysis to sequencing glycated peptides generated by proteolytic digestion.6-8 However, there are significant challenges associated with sequencing glycated peptides using collision-induced dissociation (CID), the most broadly available and applied ion dissociation method in mass spectrometry. During CID of glycated peptide ions, the labile Amadori adduct tends to dissociate preferentially, resulting in poor production of sequence specific ions from the peptide backbone. As a result, ions corresponding to various neutral losses dominate the mass spectrum, which hampers the identification of peptide sequences and sites of glycation.8,9 To overcome this limitation, a precursor-ion scanning method based on the Amadori-modified lysine immonium ion (192.1 Da) was recently used to map glycation sites on glycated bovine serum albumin.10 In addition, a prescreening and sequencing method based on a neutral-loss scan of the Amadori moiety (162 Da) was developed to identify glycation sites in human serum albumin.11 However, both methods relied on quadrupole-time-of-flight instruments employing features unavailable on ion-trap instruments, which are more commonly utilized for LC/MS-based proteomics experiments. An additional limitation of these methods is that both were evaluated only for a single protein. It is unclear how these approaches would perform with a complex peptide mixture produced from proteolytic digestion of biofluid or cellular proteins.
The recent incorporation of electron transfer dissociation (ETD) in ion-trap instruments has enabled the high-throughput identification of peptides containing labile post-translational modifications (PTMs).12-14 Labile PTMs remain intact during peptide fragmentation by ETD, which greatly improves backbone fragmentation and peptide identification. A similar technology (electron capture dissociation (ECD)) is available on Fourier transform ion cyclotron resonance mass spectrometers and is amenable to the identification of polypeptides, particularly large peptides and peptides carrying labile PTMs;15 however, the lower efficiency of ECD limits its application in high-throughput LC/MS-based proteomics. We recently used ETD tandem mass spectrometry to sequence glycated peptides in complex proteomic samples.9,16,17 Our data showed that ETD outperformed CID in sequencing glycated peptides.9,16 However, ETD is not without its own limitations. For example, it is generally less sensitive than CID and also is less effective for dications and for ions with m/z over 850.18 In addition, ETD is not yet widely available.
Alternative methods are in development to more fully utilize the MSn capability of ion-trap instruments for sequencing peptides containing labile modifications when ETD is unavailable. For example, data-dependent neutral-loss-triggered MS3 (NLMS3) and multi-stage activation (MSA) methods have been developed for the analysis of phosphopeptides,19-22 due to the intense neutral loss of phosphoric acid generated by this PTM upon CID fragmentation. Data-dependent NLMS3 functions by first isolating a product ion produced during the MS2 scan and corresponding to a specified neutral loss, which is then activated and dissociated in the following MS3 stage. Similarly, MSA, or pseudo-MS3, was developed to avoid the loss of sequence informative ions from the backbone of peptides containing labile PTMs in the MS2 stage. In this approach, a precursor ion is first fragmented in the MS2 stage, and product ions corresponding to specified neutral losses are further activated and fragmented in the MS3 stage. The product ions (MS2) from the initial precursor ion and the activated neutral-loss product ions (MS3) are then simultaneously stored, and a composite spectrum is generated containing ions from all of the activated precursors.
In this work, we report the first use of NLMS3 and MSA as fragmentation techniques for sequencing D-glucose glycated peptides from a complex proteomic sample – human serum glycated in vitro. The efficacy of these two fragmentation methods in identifying glycated peptides is compared.
EXPERIMENTAL
Chemicals and materials
All chemicals, in vitro glycated human serum, and peptide desalting solid-phase extraction (SPE) cartridges (Supelco Discovery DSC-18) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Micro-BCA protein assay kits and ICON 9K concentrators were purchased from Pierce (Rockford, IL, USA). Sequencing-grade trypsin was obtained from Promega (Madison, WI, USA), and Glycogel II boronate affinity gel (Pierce) was a gift from Dr. Bart Haigh of the Institute for Bioanalytics (Branford, CT, USA).
Protein digestion and enrichment of glycated peptides by boronate affinity chromatography
The lyophilized powder of human serum glycated in vitro was reconstituted in 100 mM NH4HCO3 (pH 8.0). Glucose and other salts were removed with an ICON 9K concentrator per the manufacturer's instructions. Glycated human serum (5 mg) was dissolved in 8 M urea/100 mM NH4HCO3 (pH 8.0) at a concentration of 10 mg/mL. Disulfide bonds were reduced with 5 mM dithiothreitol for 1 h at 37°C, followed by alkylation of free sulfhydryl groups with 20 mM iodoacetamide at 37°C for 1 h in the dark. Samples were then diluted with 50 mM NH4HCO3 (pH 8.0) to reduce the urea concentration to below 1 M, and CaCl2 was added to a final concentration of 1.5 mM. Sequencing-grade trypsin was then added at a ratio of 1:40 (w/w, enzyme/protein), and the samples were digested at 37°C overnight. The final digestion mixtures were passed through C18 SPE cartridges for desalting, and eluted peptide solutions were dried by speed-vac before being subjected to boronate affinity chromatography.
The enrichment of glycated peptides was carried out as described previously.16,17 Briefly, the dried peptides were reconstituted in mobile phase A (see below) at a concentration of 1 μg/μL and injected onto the home-packed Glycogel II boronate affinity column (Tricorn, 5 mm × 100 mm; GE Healthcare, Piscataway, NJ, USA) using an Agilent 1100 series LC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a fraction collector. The LC mobile phases were (A) 50 mM MgCl2 and 250 mM NH4OAc (pH 8.1) in water and (B) 0.1 M HOAc in water. A gradient (0% B for 10 min; 0–100% B in 0.1 min, 100% B for 10 min; 100–0% B in 0.1 min; 0% B for 10 min) was used to separate glycated peptides from non-glycated peptides at a flow rate of 1.0 mL/min. The LC effluent was monitored at 280 nm with a UV detector. Fractions corresponding to glycated peptides were collected,16,17 concentrated by speed-vac, and subsequently desalted with C18 SPE cartridge.
LC/MS/MS analyses of glycated peptides
Glycated peptides were analyzed using an automated four-column capillary LC system coupled on-line with an LTQ ion-trap mass spectrometer (ThermoElectron, San Jose, CA, USA) via an ESI interface.23 The reversed-phase capillary columns (75 μm i.d. × 65 cm) were slurry packed with 3 μm Jupiter C18 particles (Phenomenex, Torrence, CA, USA). The mobile phase solvents consisted of (A) 0.2% HOAc and 0.05% trifluoroacetic acid (TFA) in water and (B) 0.1% TFA in 90:10 CH3CN/H2O. An exponential gradient was used for the separation, which started at 100% A and gradually increased to 60% B over 100 min. The instrument was operated in data-dependent mode with an m/z range of 350–2000, and dynamic exclusion of 60 s was applied to avoid repeated analyses of the same abundant precursor ion. The top seven most intense ions from each MS scan were selected for further dynamic MSn analyses using a normalized collision energy of 40%. The following neutral losses from Amadori-modified lysine were considered in either NLMS3 or MSA experiments as shown in Table 1: (1) formation of pyrylium ion (loss of 3 H2O): 18.01 (3+), 27.02 (2+) and 54.03 (1+); (2) formation of furylium ion (loss of 3 H2O and HCHO): 28.01 (3+), 42.02 (2+), 84.04 (1+); (3) loss of the Amadori moiety (C6H10O5): 54.02 (3+), 81.03 (2+) and 162.05 (1+); and (4) a combination of all neutral losses listed in (1) to (3). If these neutral losses were present in the top ten most intense ions from the MS2 spectra, then either a MS3 fragmentation or multi-stage activation would be triggered for NLMS3 or MSA experiments, respectively. For each analytical method, the sample was analyzed on the same capillary LC column in duplicate.
Table 1.
Acquisition and database search parameters for various NLMS3 or MSA experiments
| Acquisition parameters |
Database search parameters |
||
|---|---|---|---|
| Neutral loss category | Neutral losses | NLMS3 | MSA |
| 3 H2O | 18.01 (3+), 27.02 (2+), 54.03 (1+) | K+108.02 | K+162.05, with neutral loss of 54.03 |
| 3 H2O+HCHO | 28.01 (3+), 42.02 (2+), 84.04 (1+) | K+78.01 | K+162.05, with neutral loss of 84.04 |
| C6H10O5 | 54.02 (3+), 81.03 (2+), 162.05 (1+) | none | K+162.05, with neutral loss of 162.05 |
| All neutral losses | All of the above | K+78.01 or K+108.02 | K+162.05, with neutral loss of 54.03, 84.04 or 162.05 |
Data analyses
Instrument .raw files were converted into .dta files with extract_msn.exe software (ThermoElectron). For .raw files acquired during NLMS3 experiments, a Python script was further employed to subset the created .dta files to only contain MS3 scans. The resultant .dta or subset-.dta files were concatenated with a Python script to create XML encapsulated .dta files as input for the Open Mass Spectrometry Search Algorithm (OMSSA) search engine (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD, freely downloadable from the internet24). OMSSA was then used to match the tandem MS data to the protein sequences in the human Swiss-Prot protein library. Carbamidomethylation of cysteine (57.02 Da) was specified as a static modification for all searches, and up to two missed tryptic cleavages were allowed. Depending on the data acquisition method, various dynamic modifications of lysine were specified, as outlined in Table 1. Other search parameters were kept as the default values. An E-value of <0.01 was used as a cut-off for peptide identifications, which typically results in less than 1% of false peptide identifications.18,25
RESULTS AND DISCUSSION
Due to the nature of the Amadori adduct, tandem MS analyses of glycated peptides under CID conditions tend to generate abundant neutral losses (i.e., neutral losses of H2O, 2 H2O, 3 H2O, 4 H2O, 3 H2O + HCHO, as well as neutral loss of the Amadori adduct, C6H10O5 in the MS2 spectrum (Fig. 1). These neutral losses are characteristic for Amadori-modified peptides and are independent of any additional modifications that may be present on the N- and C-termini.8-10,26 While neutral losses of H2O and 2 H2O can also occur on other peptides (e.g., neutral loss of H2O from peptides containing hydroxyl or carboxylic acid residues), neutral losses of 3 H2O, 4 H2O, 3 H2O + HCHO, and C6H10O5 are less common and may be exploited in NLMS3- and MSA-based methods to sequence glycated peptides. In the following experiments, we have compared the efficacy of utilizing these characteristic neutral losses individually, as well as in combination, for sequencing glycated peptides using a linear ion-trap mass spectrometer.
Figure 1.
Product-ion spectrum produced from fragmentation of the [M + 3H]3+ ion of peptide RHPYFYAPELLFFAK*R, where * represents the Amadori adduct modification site. Different neutral losses are shown in the zoomed inset.
Evaluation of NLMS3-based approaches
As described above, data-dependent NLMS3 functions by first isolating a product ion produced by the MS2 scan and corresponding to a specified neutral loss, which is then activated and dissociated in the following MS3 stage. Those neutral losses used to trigger MS3 scans in our NLMS3 experiments are listed in Table 1, and the results of these experiments are summarized in Table 2. Approximately 1500 spectra were confidently identified by each NLMS3 approach, with the exception of neutral loss of Amadori-adduct-triggered fragmentation, which resulted in only 614 spectra identified. The latter approach resulted in ∼60% fewer identifications due to the low intensity of the ion corresponding to the neutral loss of the Amadori adduct (Fig. 1). The low intensity of this ion often results in its exclusion for fragmentation during subsequent MS3 scans. Regardless of the NLMS3-based approach employed, an average of 99% of the spectra identified corresponded to peptides containing Amadori modification at lysine, which highlights the high specificity of NLMS3-based approaches to identifying glycated peptides.
Table 2.
Summary of peptides identified by different NLMS3- and MSA-based approaches
| Category | Neutral losses | Total # of identified spectra |
Total # of spectra containing glycated peptides |
Percentage* of spectra containing glycated peptides |
# of unique glycated peptides |
|---|---|---|---|---|---|
| NLMS3 | 3 H2O | 1578 | 1558 | 98.7% | 168 |
| 3 H2O+HCHO | 1564 | 1558 | 99.6% | 186 | |
| C6H10O5 | 614 | 614 | 100.0% | 115 | |
| All neutral losses | 1434 | 1424 | 99.3% | 165 | |
| MSA | 3 H2O | 5246 | 2474 | 47.2% | 253 |
| 3 H2O+HCHO | 3982 | 1222 | 30.7% | 158 | |
| C6H10O5 | 3326 | 632 | 19.0% | 54 | |
| All neutral losses | 4979 | 2790 | 56.0% | 175 |
Calculated as the ratio of the number of spectra containing the Amadori adduct at lysine to the total number of spectra identified by the respective database search parameters.
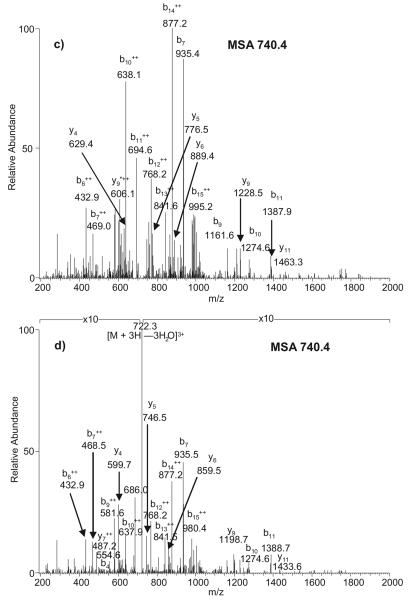
When all potential neutral losses were considered in the same LC/MSn analysis the majority (88.6%) of glycated peptide identifications were from MS3 scans triggered by neutral loss of 3 H2O (Table 3). In contrast, neutral loss of 3 H2O + HCHO produced only 11.4% of glycated peptide identifications, whereas no glycated peptides were identified in MS3 scans triggered by neutral loss of the Amadori adduct. This again is in line with the intensity of the ion corresponding to the neutral loss in the MS2 stage. Lower ion intensity may result in exclusion from subsequent MS3 scans, and, as illustrated in Fig. 1, the intensity of the ion corresponding to neutral loss of 3 H2O is typically more intense than the intensity of the ion corresponding to neutral loss of 3 H2O + HCHO. However, the neutral loss of 3 H2O + HCHO can still result in high-quality MS3 spectra when this ion is generated with sufficient intensity in MS2scans. For example, peptide RHPYFYAPELLFFAK*R was identified by both fragmentation of the pyrylium ion (neutral loss of 3 H2O) and the furylium ion (neutral loss of 3 H2O + HCHO), as shown in Figs. 2(a) and 2(b). It is clear that the ions that carry the partially cleaved Amadori moiety (b15 and the y-ions) were either from the pyrylium-ion precursor (Fig. 2(a)) or the furylium-ion precursor (Fig. 2(b)).
Table 3.
Summary of glycated peptides identified by NLMS3- and MSA-based approaches when all potential neutral losses are considered. Glycated peptides correspond to peptides containing Amadori-modified lysines. Numbers in parentheses are the percentage value calculated with respect to the total number of modified peptides
| Approach | Total # of glycated peptides |
# of glycated peptides identified by loss of 3 H2O |
# of glycated peptides identified by loss of 3 H2O+HCHO |
# of glycated peptides identified by loss of C6H10O5 |
|---|---|---|---|---|
| NLMS3 All neutral losses | 1424 | 1262 (88.6%) | 162 (11.4%) | 0 |
| MSA All neutral losses | 2790 | 1332 (47.7%) | 716 (25.7%) | 754 (27.0%) |
Figure 2.
Product-ion spectra produced from the [M + 3H]3+ ion of peptide RHPYFYAPELLFFAK*R under different neutral-loss-triggered approaches, where * represents the Amadori adduct modification site. (a) NLMS3 triggered by neutral loss of 3 H2O; (b) NLMS3 triggered by neutral loss of 3 H2O + HCHO; (c) MSA triggered by neutral loss of 3 H2O; and (d) MSA triggered by neutral loss of 3 H2O + HCHO.
The overlap in the number of unique glycated peptides identified by the three different NLMS3 approaches (excluding neutral loss of Amadori-adduct-triggered MS3, due to ambiguity in some identified peptides when two or more lysines exist in the same sequence) is shown in Fig. 3(a). There is significant overlap among the three different approaches, although a slightly higher number of unique glycated peptides were identified by further activation of the pyrylium ion compared with the other two approaches.
Figure 3.
Venn diagrams showing the overlap of unique glycated peptides as identified by different neutral-loss-triggered approaches. (a) NLMS3-based approaches; (b) MSA-based approaches; and (c) total number of unique glycated peptides identified by all NLMS3- and MSA-based approaches. I, II and III in (a) and (b) represent neutral losses of 3 H2O, 3 H2O + HCHO, and the combination of all potential neutral losses, respectively.
Evaluation of MSA-based approaches
As described above, MSA functions by first fragmenting a precursor ion in the MS2 stage. The product ions corresponding to a specified neutral loss are then further activated and fragmented in the MS3 stage. The product ions (MS2) from the initial precursor ion and the activated neutral-loss product ions (MS3) are then simultaneously stored, and a composite spectrum is generated containing ions from all of the activated precursors. The neutral losses used in our MSA experiments were identical to those used in the NLMS3 experiments described above. The number of spectra identified in the MSA experiments is listed in Table 2. In general, many more spectra were identified by MSA compared to NLMS3. However, although the data acquisition methods and data search parameters were tailored for the identification of Amadori-modified peptides, the MSA-based approaches resulted in lower specificity compared with the NLMS3-based approaches. For example, only 19.0% of peptides were identified as glycated in the case of neutral loss of Amadori-adduct-triggered MSA, and only 56.0% were identified as glycated when all potential neutral losses were considered. As in the case of NLMS3-based approaches, neutral loss of Amadori-adduct-triggered MSA resulted in far fewer glycated peptide identifications due to the low intensity of the ions corresponding to this neutral loss.
However, unlike NLMS3-based approaches where only one product ion from the MS2 stage is selected as the precursor ion in the subsequent MS3 stage, MSA is capable of fragmenting several ions if they all meet the specified neutral losses. For example, Fig. 2(c) shows a peptide spectrum that was acquired and identified by neutral loss of 3 H2O. In theory, we should observe the furylium ion [M + 3 H − 3 H2O − HCHO]3+ at m/z 712.3. However, because this neutral loss is 28 Da at charge state 3+, it overlaps with an ion corresponding to the targeted neutral loss of 3 H2O (pyrylium ion) at charge state 2+ (27 Da). Thus, the furylium ion was also fragmented in the 2nd stage and its product ions indeed observed in this spectrum, similar to what was observed for NLMS3 (Fig. 2(b)) but with lower relative intensity (these peaks are not labeled in Fig. 2(c)). In contrast, an intense pyrylium ion [M + 3 H − 3 H2O]3+ at m/z 722.3 dominates the spectrum shown in Fig. 2(d), which was acquired and searched using furylium ion [M + 3 H − 3 H2O − HCHO]3+ parameters. This mixing effect in the combined MSA spectra was further illustrated in the data acquisition approach utilizing all potential neutral losses. Of the 2790 total identified spectra containing the Amadori modification (Table 3), there were a handful of peptides that were identified by more than one search parameter.
In general, significantly more unique glycated peptides were identified in the MSA pyrylium-ion approach compared with the other MSA methods, which is consistent with the high intensity of ions corresponding to this type of neutral loss. Also, the percentage of overlap among all three MSA approaches is less compared with the case of NLMS3(Fig. 3(b)), which indicates that annotation and interpretation of spectra from MSA is more complex, particularly when multiple fragmentation pathways coexist.
NLMS3 versus MSA
When comparing all the unique glycated peptides identified by these two data acquisition strategies, MSA (283 unique glycated peptides) identified more Amadori-modified peptides compared to NLMS3 (231 unique glycated peptides), as shown in Fig. 3(c). MSA is superior in this regard and should be utilized if the results will be used for population of a peptide database, such as with the accurate mass and time tag approach.27,28 However, NLMS3-based approaches offer the advantage of high specificity in terms of glycated peptide identifications. Almost all of the peptides identified by NLMS3 were Amadori-modified, which provides an additional level of confidence in the results. In addition, approximately 90% less time is required for database searching of NLMS3 datasets compared with MSA datasets, because only the MS3 scans are searched when using these approaches. Thus, NLMS3-based approaches are more suitable if the goal is not the identification of very low abundance proteins or if the sample is not overly complex. In general, the significant amount (178 unique glycated peptides) of overlap in identifications using these two advanced CID strategies does not clearly indicate which method should be used when sequencing Amadori-modified peptides.
CONCLUSIONS
We have evaluated different NLMS3- and MSA-based approaches for sequencing glycated peptides enriched from human serum glycated in vitro as alternatives to ETD when that technique is unavailable. Each advanced CID technique offers advantages and disadvantages. While MSA identified more glycated peptides, the time required for data processing and analysis is significantly higher compared to NLMS3. In contrast, NLMS3 resulted in extremely high identification specificity, with ∼99% of spectra corresponding to glycated peptides. Thus, the choice of which advanced CID approach to use for the identification of glycated peptides should be driven by the goals of the experiments and the informatics capabilities of each laboratory.
Supplementary Material
Acknowledgements
The authors thank Dr. Bart Haigh of the Institute for Bioanalytics for kindly providing the GlycogelTM II boronate affinity gel. This research was supported by NIH grant DK071283 to R.D.S. (PI) and T.O.M. (co-PI); portions of this research were supported through the National Center for Research Resources (RR018522) and performed at the Environmental Molecular Sciences Laboratory, a national scientific user facility located at Pacific Northwest National Laboratory (PNNL) and sponsored by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research. PNNL is operated by Battelle for the DOE under Contract No. DE-AC06-76RLO-1830.
Footnotes
SUPPORTING INFORMATION Additional supporting information may be found in the online version of this article. This comprises a table of the unique glycated peptides and corresponding proteins identified in this study.
REFERENCES
- 1.Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. Prog. Clin. Biol. Res. 1989;304:43. [PubMed] [Google Scholar]
- 2.Brownlee M. Annu. Rev. Med. 1995;46:223. doi: 10.1146/annurev.med.46.1.223. [DOI] [PubMed] [Google Scholar]
- 3.Baynes JW. Exp. Gerontol. 2001;36:1527. doi: 10.1016/s0531-5565(01)00138-3. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed N, Thornalley PJ. Diabetes Obes. Metab. 2007;9:233. doi: 10.1111/j.1463-1326.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- 5.Zheng X, Wu SL, Hancock WS. Int. J. Pharm. 2006;322:136. doi: 10.1016/j.ijpharm.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed N, Thornalley PJ. Biochem. Soc. Trans. 2003;31:1417. doi: 10.1042/bst0311417. [DOI] [PubMed] [Google Scholar]
- 7.Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A. Biochem. J. 2003;375:581. doi: 10.1042/BJ20030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. J. Am. Soc. Mass Spectrom. 2004;15:496. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, Frolov A, Tang N, Hoffmann R, van de Goor T, Metz TO, Smith RD. Rapid Commun. Mass Spectrom. 2007;21:661. doi: 10.1002/rcm.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov A, Hoffmann P, Hoffmann R. J. Mass Spectrom. 2006;41:1459. doi: 10.1002/jms.1117. [DOI] [PubMed] [Google Scholar]
- 11.Gadgil HS, Bondarenko PV, Treuheit MJ, Ren D. Anal. Chem. 2007;79:5991. doi: 10.1021/ac070619k. [DOI] [PubMed] [Google Scholar]
- 12.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. USA. 2004;101:9528. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Proc. Natl. Acad. Sci. USA. 2007;104:2193. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. Biochim. Biophys. Acta. 2006;1764:1811. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakhtiar R, Guan Z. Biotechnol. Lett. 2006;28:1047. doi: 10.1007/s10529-006-9065-z. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q, Tang N, Brock JW, Mottaz HM, Ames JM, Baynes JW, Smith RD, Metz TO. J. Proteome Res. 2007;6:2323. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, Tang N, Schepmoes AA, Phillips LS, Smith RD, Metz TO. J. Proteome Res. 2008;7:2025. doi: 10.1021/pr700763r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good DM, Wirtala M, McAlister GC, Coon JJ. Mol. Cell. Proteomics. 2007;6:1942. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Zumwalt AM, Choudhary G, Cho D, Hemenway E, Shofstahl J, Mylchreest I. Proc. 51st ASMS Conf. Mass Spectrometry and Allied Topics. Montreal, Canada: 2003. [Google Scholar]
- 20.Wolschin F, Weckwerth W. Plant Methods. 2005;1:9. doi: 10.1186/1746-4811-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. Anal. Chem. 2004;76:3590. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 22.Mann K, Olsen JV, Macek B, Gnad F, Mann M. Proteomics. 2007;7:106. doi: 10.1002/pmic.200600635. [DOI] [PubMed] [Google Scholar]
- 23.Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, Zhao R, Shen Y, Orton DJ, Moore RJ, Kelly RT, Udseth HR, Smith RD. Anal. Chem. 2008;80:294. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Available: http://pubchem.ncbi.nlm.nih.gov/omssa/
- 25.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. J. Proteome Res. 2004;3:958. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 26.Mennella C, Visciano M, Napolitano A, Del Castillo MD, Fogliano V. J. Peptide Sci. 2006;12:291. doi: 10.1002/psc.722. [DOI] [PubMed] [Google Scholar]
- 27.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Mass Spectrom. Rev. 2006;25:450. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metz TO, Qian WJ, Jacobs JM, Gritsenko MA, Moore RJ, Polpitiya AD, Monroe ME, Camp DG, 2nd, Mueller PW, Smith RD. J. Proteome Res. 2008;7:698. doi: 10.1021/pr700606w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.