Abstract
The solution structure of the oligodeoxynucleotide 5′-d(CTCGGCXCCATC)-3′·5′-d(GATGGCGCCGAG)-3′ containing the heterocyclic amine 8-[(3-methyl-3H-imidazo[4,5-f]quinolin-2-yl)amino]-2′-deoxyguanosine adduct (IQ) at the third guanine in the NarI restriction sequence, a hot spot for −2 bp frameshifts, is reported. Molecular dynamics calculations restrained by distances derived from 24 1H NOEs between IQ and DNA, and torsion angles derived from 3J couplings, yielded ensembles of structures in which the adducted guanine was displaced into the major groove with its glycosyl torsion angle in the syn conformation. One proton of its exocyclic amine was approximately 2.8 Å from an oxygen of the 5′ phosphodiester linkage, suggesting formation of a hydrogen bond. The carcinogen-guanine linkage was defined by torsion angles α′ [N9-C8-N(IQ)-C2(IQ)] of 159 ± 7° and β′ [C8-N(IQ)-C2(IQ)-N3(IQ)] of −23 ± 8°. The complementary cytosine was also displaced into the major groove. This allowed IQ to intercalate between the flanking C·G base pairs. The disruption of Watson—Crick hydrogen bonding was corroborated by chemical-shift perturbations for base aromatic protons in the complementary strand opposite to the modified guanine. Chemical-shift perturbations were also observed for 31P resonances corresponding to phosphodiester linkages flanking the adduct. The results confirmed that IQ adopted a base-displaced intercalated conformation in this sequence context but did not corroborate the formation of a hydrogen bond between the IQ quinoline nitrogen and the complementary dC.
Introduction
The browning of protein-rich foods imparts flavor during cooking. It leads to the formation of heterocyclic amines (HCA) such as 2-amino-3-methylimidazo[4,5-f]quinoline (IQ).1–4 Various HCAs, including IQ, have been identified in grilled foods at ppb levels.5,6 Daily human intakes of HCAs, estimated to be ∼60 ng/day,7 are modest; however, exposure to these compounds, which have been isolated from human urine,8 is of concern with regard to human health.
Exposure to IQ is associated with carcinogenesis. Tumors in organs of rodents and in the livers of monkeys are induced by IQ.9–11,16 In mice, exposures lead to liver, forestomach, and lung tumors.12 In rats, exposures lead to cancers in the liver, intestine, zymbal gland, clitoral gland, skin,13 mammary glands, liver, and ear ducts.14 TD50 values for in rats are 0.7 mg/kg/day, and in mice are 14.7 mg/kg/day.15 Human exposure to HCAs is associated with pancreatic,17 colon,18 prostate,19 and breast cancer.20,21
In bacterial reversion assays,22–25 HCAs are active in point and frameshift tester strains.26 IQ is one of the strongest chemical mutagens.27 It is less prevalent than 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)28 but is 200-fold more mutagenic than the latter in Salmonella reversion assays.3 IQ is an order of magnitude more mutagenic than is aflatoxin B1. In bacteria, mutations occur primarily at G:C base pairs.29,30 It exhibits frameshift mutations in CG repeats. Similar levels of mutations are seen in mammalian hprt31 and ef-232 gene assays. In mammalian cells, point mutations are observed.33–36 Sister chromatid exchanges are observed in rodent cells.37–39
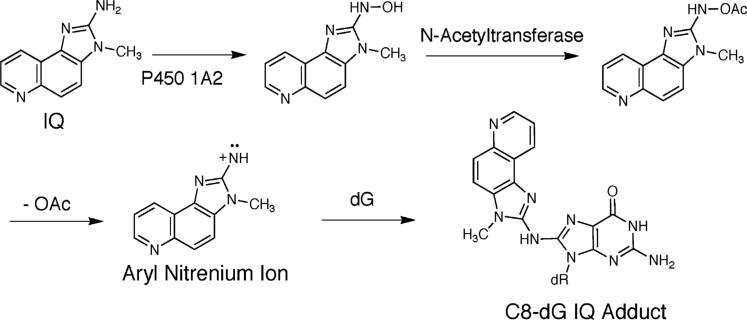
IQ is activated primarily by the enzyme CYP P450 1A2 to an N-hydroxyl oxidation product.40–43 Extra-hepatic CYP P450s oxidize HCAs with lower efficiencies.44 The N-hydroxyl oxidation product is acetylated by cellular N-acetyl transferases, particularly NAT2.45–47 The resulting nitrenium ion is the ultimate reactive electrophile.36,44 The NAT2 fast acetylator polymorphism is associated with an increased risk of colorectal cancer in humans.48,49
The C8-dG adducts of HCAs are observed both in rodents and primates, as measured by 32P postlabeling.35 The major adduct formed by IQ occurs by substitution at C8-dG (Chart 1); a minor N2-dG adduct is also formed.50 The structures of these adducts are established.51–53 The formation of the C8-dG adduct probably involves initial alkylation at N7-dG, followed by rearrangement.54 High sensitivity LC/ESI-MS55 has measured several adducts per 107 nucleotides in animal tissues.19,56 The levels of C8 and N2-dG IQ adducts measured in tissues of rats and primates using mass spectrometry57,58 are in agreement with data obtained by 32P postlabeling.
Chart 1.
Metabolic Activation of IQ
Heretofore, site-specific DNA adducts of HCAs have not been readily accessible. A synthesis of PhIP-adducted oligodeoxynucleotides involved reacting single-stranded DNA with the PhIP nitrenium ion.59 The low yield, coupled with complexities of purification, limited the approach to oligodeoxynucleotides containing a single dG. In the COS-7 site-specific mutagenesis system,60 if dC was at the 5′-flanking position to dG-C8 PhIP, incorporation of dC, the correct base, was observed. However, G → T transversions, and lesser amounts of G → A transitions and G → C transversions, were detected. If the dC 5′-flanking base was replaced by T, dA, or dG, the mutational spectra were similar, but greater mutational frequencies were observed with dC or dG than with dA 5′ to the adduct. Single-base deletions were detected only when dG or T flanked the adduct. Thus, dG-C8 PhIP was mutagenic, generating primarily G → T transversions.61
A study of the C8-dG PhIP adduct in 5′-d(CCATCXCTACC)-3′·5′-d(GGTAGCGATGG)-3′ represents the only conformational analysis of an HCA-adducted duplex.59 This yielded a base-displaced intercalated structure, in which the adducted dG was in the syn conformation and situated in the major groove. The C6-phenyl and N3-methyl groups protruded into the minor groove, widening it and compressing the major groove, resulting in DNA bending.
An efficient strategy for synthesis of C8-dG arylamine adducts involving the Buchwald-Hartwig palladium-catalyzed N-arylation reaction of a protected 8-bromo-2′-dG derivative with arylamines facilitated preparation of the C8-IQ-adducted dG nucleoside, which was incorporated into oligodeoxynucleotides using phosphoramidite chemistry.62,63 A combination of thermal melting studies, and UV and circular dichrosim spectroscopy, led to the proposal of a base-displaced intercalated conformation at the G3-position of the 5′-d(CG1G2CG3CC)-3′ recognition site of the NarI enzyme. It was proposed that this was stabilized by a hydrogen bond between the quinoline nitrogen of IQ and the complementary cytosine.63 In contrast, molecular mechanics analysis of the C8-dG IQ-modified duplex 5′-d(G1G2CX3CCA)-3′·5′-d(TGGCGCC)-3′ suggested that the favored conformation featured the modified dG in the syn conformation with IQ in the minor groove and directed 3′ with respect to the modified strand.64 This suggested that the base-displaced intercalated conformation was ∼10 kcal/mol higher in energy than the minor groove conformation.64 A study of the C8-dG IQ adduct at the nucleoside level confirmed that the adducted dG was in the syn conformation about the glycosyl bond.53
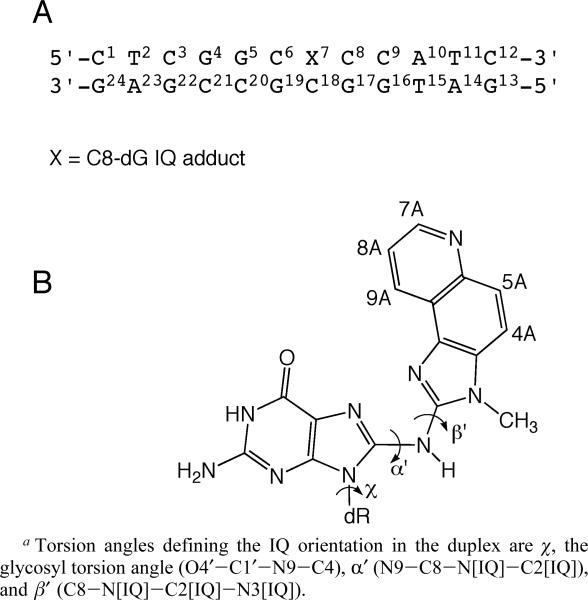
This work presents a study of the C8-dG IQ adduct in 5′-d(C1T2C3G4G5C6X7C8C9A10T11C12)-3′·5′-d(G13A14T15G16G17C18G19C20C21G22A23G24)-3′; X = 8-[(3-methyl-3H-imidazo[4,5-f]quinolin-2-yl)amino]-2′-deoxyguanosine, named the Nar1IQ3 sequence. It contains the 5′-d(CG1G2CX3CC)-3′ recognition site of the NarI restriction enzyme, in which the third guanine (G3 in the NarI sequence and X7 in this study) represents a hot spot for −2 bp frameshifts (Chart 2). The results reveal a base-displaced intercalated structure. The adducted dG adopts a syn conformation about the glycosyl bond and extrudes into the major groove, the IQ moiety intercalates into the DNA, and the complementary dC extrudes from the helix. A hydrogen bond between the IQ quinoline nitrogen and the complementary dC63 is not observed.
Chart 2.
(A) Nar1IQ3 Duplex and (B) C8-dG IQ Adducta
Materials and Methods
Sample Preparation
The oligodeoxynucleotides 5′-d(CTCGGCGCCATC)-3′ and 5′-d(GATGGCGCCGAG)-3′ were obtained from the Midland Certified Reagent Company, purified by anion exchange chromatography. The oligodeoxynucleotide 5′-d(CTCGGCXCCATC)-3′ was synthesized and purified as described.63 All oligodeoxynucleotides were characterized by MALDI-TOF mass spectrometry and enzymatic digestion, and their purities were assessed by capillary zone electrophoresis (CZE). Oligodeoxynucleotide duplexes were annealed at 70 °C. Their stoichiometry was established by 1H NMR. The duplexes were dissolved in 0.25 mL of buffer containing 0.1 M NaCl, 10 mM NaH2PO4, and 50 μM Na2EDTA (pH 7.0). The oligodeoxynucleotide concentrations were ∼0.7 mM using an extinction coefficient of 1.10 × 105 M−1 cm−1 at 260 nm.65
NMR
1H NMR spectra were obtained at 500.13, 600.20, and 800.23 MHz. COSY spectra were collected at 15, 20, 25, 30, and 35 °C in 99.996% D2O. 1H NOESY experiments in D2O were conducted at 15 °C. To obtain distance restraints, spectra were recorded at mixing times of 150, 200, and 250 ms at the 1H NMR frequency of 800.23 MHz. The data were recorded with 1024 real data points in the t1 dimension and 2048 real points in the t2 dimension. The relaxation delay was 2 s. The data in the t1 dimension were zero-filled to give a matrix of 2K × 2K real points. NOESY spectra for the exchangeable protons were recorded at 5 °C, in 90:10 H2O/D2O, using the Watergate sequence66 for water suppression and a 250-ms mixing time at a 1H NMR frequency of 600.20 MHz. Chemical shifts of proton resonances were referenced to water. Double quantum-filtered 1H correlation (DQF—COSY)67,68 and exclusive COSY (E-COSY)69 spectra were collected at 25 °C at 500.13 MHz and zero-filled to give a matrix of 1024 × 2048 real points. A skewed sine-bell square apodization function with a 90° phase shift and a skew factor of 1.0 was used in both dimensions. 1H-31P HMBC spectra70,71 were obtained at 30 °C. The data matrix was 256 (t1) × 2048 (t2) complex points. The data were Fourier transformed after zero filling in the t1 dimension, resulting in a matrix size of 512 (D1) × 2048 (D2) real points. Trimethyl phosphate was used as an external standard. NMR data were processed using the program FELIX2000 (Accelyris, Inc., San Diego, CA) on Silicon Graphics (Mountain View, CA) Octane workstations.
Experimental Restraints
(a) Distance Restraints
Footprints were drawn around cross-peaks for the NOESY spectrum measured at a mixing time of 250 ms, using the program FELIX2000. Identical footprints were applied to the cross-peaks obtained at other mixing times. Cross-peak intensities were determined by volume integration. The intensities were combined with intensities generated from a complete relaxation matrix analysis of a starting DNA structure to generate a hybrid intensity matrix.72 The program MARDIGRAS (v. 5.2)73,74 was used to refine the hybrid matrix by iteration. The molecular motion was assumed to be isotropic. The noise level was set at half the intensity of the weakest cross-peak. Calculations were performed using DNA starting structures generated using the program INSIGHT II (Accelyris, Inc.), and NOE intensities derived from experiments at three mixing times, and with three τc values (2, 3, and 4 ns), yielding 18 sets of distances. Analysis of these data yielded experimental distance restraints and standard deviations used in restrained molecular dynamics calculations. For overlapped cross-peaks, the bounds on the distances were increased. The restraints were divided into four classes, reflecting the confidence level in the data.
(b) Torsion Angle Restraints
Deoxyribose pseudorotations were estimated by monitoring the 3JHH couplings of sugar protons.75 The JH1′-H2′ and JH1′-H2″ couplings were measured from the E-COSY experiment,69 whereas the intensities of JH2″-H3′ and JH3′-H4′ couplings were determined from the DQF-COSY experiment. The data were fit to curves relating the coupling constants to pseudorotation (P), sugar pucker amplitude (ϕ), and the percentage S-type conformation. The pseudorotation and amplitude ranges were converted to the five dihedral angles ν0 to ν4.
Restrained Molecular Dynamics
Calculations were performed in vacuo using a simulated annealing protocol with the program X-PLOR.76 The force field was derived from CHARMM77 and adapted for nucleic acids. The empirical energy function treated hydrogens explicitly. The van der Waals energy term used the Lennard-Jones potential energy function. The electrostatic term used the Coulomb function, based on a full set of partial charges (−1 per residue) and a distance-dependent dielectric constant of 4r. The nonbonded pair list was updated if any atom moved more than 0.5 Å, and the cutoff radius for nonbonded interactions was 11 Å. The effective energy function included terms describing distance and dihedral restraints, in the form of square-well potentials. Sets of rMD calculations for the unmodified and Nar1IQ3 duplexes, and different starting structures of Nar1IQ3 with IQ located in the minor groove (syn), major groove (anti), and intercalated position (syn), were considered. These were generated using INSIGHT II through modification at G7 C8, followed by energy minimization using X-PLOR. Partial charges and atom types for IQ used for X-PLOR calculations were those obtained by Wu et al.64 Calculations were initiated by coupling to a heating bath, with a target temperature of 1100 K. The force constants were 25 kcal mol−1 Å−2 for empirical hydrogen bonding, 10 kcal mol−1 Å−2 for torsion angle restraints, and 50, 45, 40, and 35 kcal mol−1 Å−2 for the four classes of NOE restraints. The target temperature was reached in 10 ps and was maintained for 25 ps. The system was cooled to 300 K over 10 ps and maintained at that temperature for 25 ps of equilibrium dynamics. The force constants for the four classes of NOE restraints were scaled during 10 ps of the heating period to 200, 180, 160, and 140 kcal mol−1 Å−2 in the order of confidence factor. These weights were maintained during the remainder of the heating period and for the first 5 ps of equilibrium dynamics. They were then scaled to 100, 90, 80, and 70 kcal/mol−1 Å−2 in the order of confidence factor. The torsion angle and base pair distance force constants were scaled to 180 and 100 kcal mol−1 Å−2 during the same period as for the NOE restraints. They were scaled to 70 and 45 kcal mol−1 Å−2, also at the same time as the NOE restraints. Coordinate sets were archived every 0.1 ps, and 41 structures from the last 4.1 ps were averaged. These average rMD structures were subjected to 200 iterations of conjugate gradient energy minimization to obtain the final structures. Final structures were analyzed using X-PLOR to measure rmsd between the averaged and the converged structures. Back-calculation of NOE intensities from the emergent structures was performed using the program CORMA (v. 5.2).72 Helicoidal parameters were examined using the program 3DNA.78
Results
NMR Spectroscopy
(a) DNA Nonexchangeable Protons
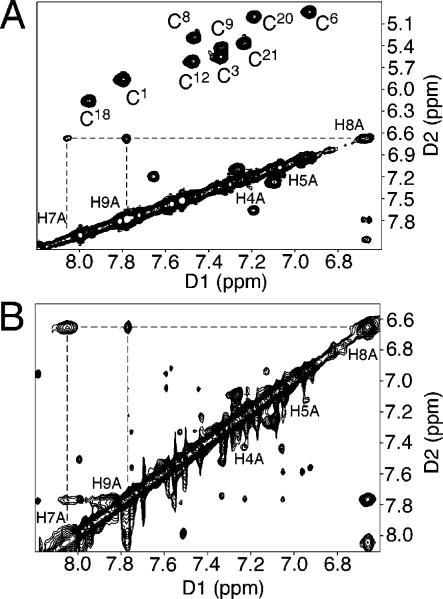
For the Nar1IQ3 duplex, sequential NOE connectivities79,80 were interrupted (Figure 1). The absence of a purine imidazole proton in the C8-IQ-dG adduct X7 precluded observation of the C6 H1′ → X7 H8 and X7 H8 → X7 H1′ NOEs. The X7 H1′ → C8 H6 NOE was of normal intensity. In the complementary strand, the G17 H1′ → C18 H6 NOE was missing. The C18 H1′ → G19 H8 sequential NOE was weak. C18 is the nucleotide opposite to X7 in the complementary strand of the Nar1IQ3 duplex. In the unmodified duplex, the G7 H8 → G7 H1′ NOE was of normal intensity and all scalar cross-peaks between deoxyribose H1′ and H2′, H2″ protons were in the anticipated 1.6−2.8 ppm chemical-shift range. In contrast, for the Nar1IQ3 duplex, the X7 H2′ resonance shifted downfield to 3.61 ppm. This was characteristic of a syn dG orientation at X7. Complete sets of sequential NOEs were observed for both strands of the unmodified duplex (Figure S1 in the Supporting Information). The resonance assignments for the nonexchangeable protons of the Nar1IQ3 and unmodified duplexes are found in Tables S1 and S2 of the Supporting Information.
Figure 1.
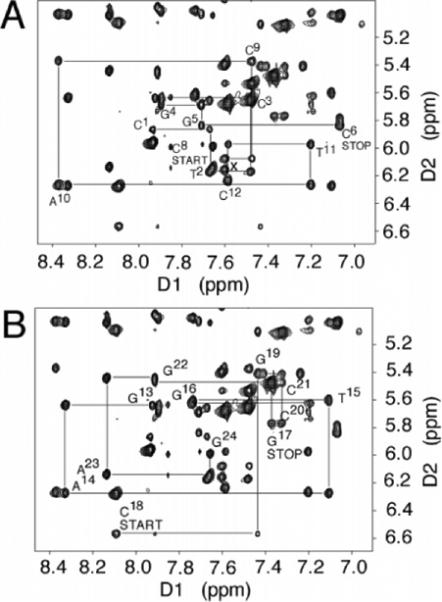
Aromatic—anomeric proton region of the 800.13 MHz NOESY spectrum for the Nar1IQ3 duplex at 15 °C at 250 ms mixing time, showing sequential NOE connectivity. (A) Nucleotides C1 → C12 of the modified strand. (B) Nucleotides G13 → G24 of the complementary strand.
(b) DNA Exchangeable Protons
The presence of the C8-dG IQ adduct resulted in chemical-shift dispersion of the imino proton resonances in the downfield region of the 1H spectrum (Figure 2; Figure S2 in the Supporting Information shows an expanded contour plot of the imino proton resonances of the unmodified duplex). Whereas for the unmodified duplex the imino resonances arising from G4, G5, G7, G16, G17, and G22 were observed between 13 and 13.4 ppm, for the Nar1IQ3 duplex, these imino protons resonated between 9.6 and 13.4 ppm. The imino protons of the Nar1IQ3 duplex were assigned from NOEs between adjacent base pairs and NOEs to their corresponding base-paired amino protons.81 Interruptions in the NOEs between Watson—Crick hydrogen-bonded amino and imino protons of the Nar1IQ3 duplex occurred between base pairs C6·G19 and X7·C18 and base pairs X7·C18 and C8·G17. All other sequential NOEs were observed. The X7 and G17 N1H resonances shifted upfield and were observed at 9.6 and 11.7 ppm, respectively. The X7 N1H proton exhibited NOEs to the X7 NH2 protons. The amino protons resonated at 6.67 and 8.83 ppm, respectively. At the 5′-adjacent G6·C19 base pair, G19 N1H showed NOEs to the C6 NH2 protons and to C6 H5. At the 3′-adjacent C8·G17 base pair, G17 N1H showed NOEs to the C8 NH2 protons and to C8 H5. The T2 N3H → A23 H2 and T11 N3H → A10 H2 NOEs were detected. The resonance assignments for the nonexchangeable protons of the Nar1IQ3 and nonmodified duplexes are found in Table S3 of the Supporting Information.
Figure 2.
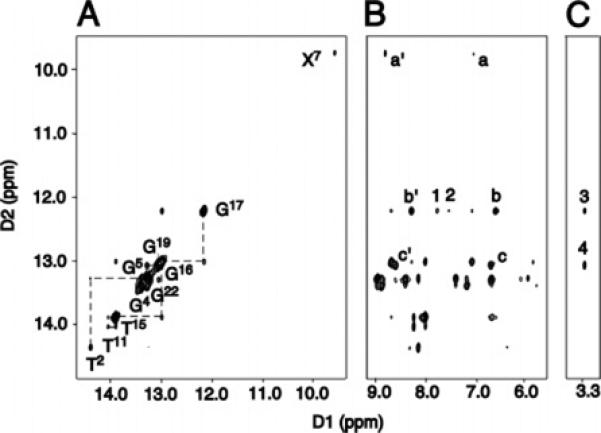
(A) Sequential NOE connectivity for the imino protons of base pairs T2·A23 → T11·A14 for the Nar1IQ3 duplex at 5 °C. The labels represent the imino proton of the designated base. (B) NOE connectivity between the imino protons and the base and amino protons. The cross-peaks involving the imino protons are labeled as: a′, a, X7 N1H → X7 NH2-2b, e; b′, b, G17 N1H → C8 NH2-4b, e; c′ and c, G19 N1H → C6 NH2-4b, e; 1, G17 N1H → X7 H4A; 2, G17 N1H → X7 H5A. (C) NOE connectivity between the imino and the IQ methyl protons. The IQ-DNA cross-peaks labeled are as: 3, G17 N1H → X7 CH3; and 4, G19 N1H → X7 CH3.
(c) IQ Protons
The resonance assignments of the IQ protons were achieved using a combination of COSY and NOESY spectra, collected at 5 °C intervals between 15 and 45 °C. The COSY IQ H4A → H5A cross-peak was observed at all of these temperatures. However, the small scalar coupling between the IQ H8A and H7A protons was not observed below 25 °C, presumably due to line broadening at the lower temperatures. The COSY cross-peaks between the IQ protons also broadened above 35 °C. This could be due to thermal melting of the duplex as the temperature was increased and might also reflect conformational exchange at higher temperatures. Figure 3 compares the NOESY spectrum collected at 15 °C with a magnitude COSY spectrum collected at 30 °C. The IQ H4A proton was assigned at 7.28 ppm on the basis of a cross-peak to the IQ methyl protons in the NOESY spectrum. The IQ H5A proton resonance was assigned at 7.05 ppm on the basis of its scalar coupling to H4A. The IQ H7A, H8A, and H9A proton resonances were distinguished on the basis of comparison of scalar couplings and chemical shifts to those of pyridine. The resonances at 8.06 and 7.79 ppm were assigned to the H7A and H9A protons, and that at 6.70 ppm was assigned to the H8A proton. The H7A resonance exhibited broadening attributed to the pyridinyl nitrogen.
Figure 3.
Expanded plots from the COSY spectrum at 30 °C and aromatic—aromatic region of the NOESY spectrum at 15 °C for the Nar1IQ3 duplex, showing assignments for the IQ protons.
IQ-DNA NOEs
There were 24 NOEs observed between the IQ moiety and DNA protons (Figure 4, and Table S4 in the Supporting Information). The IQ H4A proton exhibited NOEs to G19 H1′ and H5′, and to G17 N1H. The IQ H5A proton exhibited NOEs to G19 H1′, H5′, and H5″, G17 N1H (weak), and C18 H3′ and H4′ (weak). The IQ H9A proton exhibited NOEs to G17 H1′, H2′, H2″, and H8, and to C18 H1′, H3′, H4′, H5′, and H5″. The IQ methyl protons exhibited NOEs to X7 H1′, C8 H6, G17 N1H, G19 H8, and G19 N1H.
Figure 4.
NOE cross-peaks between nonexchangeable protons of DNA and IQ protons in the Nar1IQ3 duplex. (a—i) G17 H2′, G17 H2″, C18 H5′, H4′, H5″, H3′, G17 H1′, C18 H1′, and G17 H8 → IQ H9A; (j) C8 H6 → IQ CH3; (k—l) G19 H5′ and G19 H1′ → IQ H4A; (m—n) G19 H8 → IQ CH3 and H5A; (o—s) G19 H5″, G19 H5′, C18 H4′, C18 H3′, and G19 H1′ → IQ H5A, respectively.
Torsion Angle Analysis
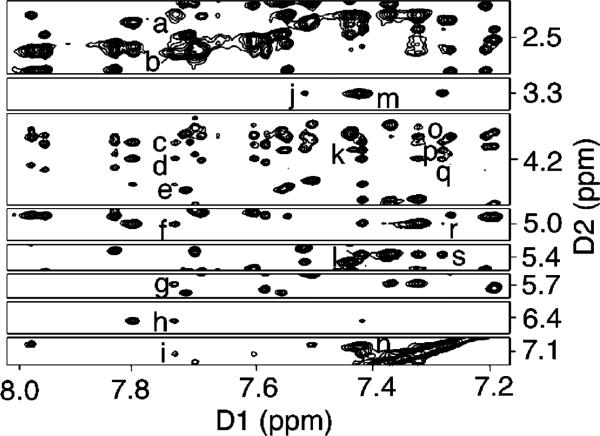
The glycosyl torsion angle, χ, was evaluated by inspection of chemical shift data at the deoxyribose H2′,H2″ protons.82,83 Expanded DQF—COSY plots identifying scalar couplings between deoxyribose H1′ (5.0−6.7 ppm) and H2′, H2″ protons in the unmodified and the Nar1IQ3 duplexes are shown in Figure 5. For the Nar1IQ3 duplex, the X7 H2′ resonance shifted downfield to 3.61 ppm. This was characteristic of the syn dG orientation at X7. Analysis of DQF-COSY spectra suggested that all of the pyrimidine pseudorotation values were in the C1′-exo range of P = 126 ± 18°, and all of the purines in the center 10 base pairs had pseudorotation values in the C2′-endo range of P = 162 ± 18°. The sugar pucker of X7 was in C2′-endo region. The glycosyl torsion angles and the deoxyribose pseudorotations for the Nar1IQ3 and unmodified duplexes are found in Table S5 of the Supporting Information.
Figure 5.
Expanded COSY spectra at 15 °C, establishing connectivity between the H1′ and H2′, H2″ protons. The H2′ and H2″ protons of the nucleotides adjacent to the lesion site are labeled. (A) Unmodified duplex. (B) Nar1IQ3 duplex.
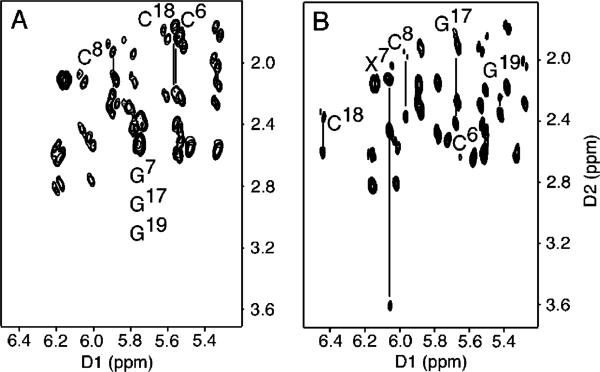
Figure 6 shows the 31P HMBC correlation spectrum for the Nar1IQ3 duplex and its unmodified counterpart, and the assignments of P6,P7,P17, and P18, the phosphodiester linkages 5′- and 3′ to the IQ adduct in the modified and complementary strands, respectively. The C8-dG IQ adduct dispersed these four 31P resonances, with the most significant change occurring at P6, the phosphodiester 5′ to X7 in the modified strand. The downfield 31P chemical shift at P6 presumably reflects conformational perturbations associated with the P6 phosphodiester.84 The small differences observed for 31P chemical shifts for P17 and P18 suggested that the phosphodiesters opposite to X7 in the complementary strand were less perturbed. The carcinogen-base linkage site at X7 residue is defined by the torsion angles α′ (N9-C8-N[IQ]-C2[IQ]) and β′ (C8-N[IQ]-C2[IQ]-N3-[IQ]). The absence of an NOE between the IQ NH and methyl protons suggested that the β′ torsion angle must be in an eclipsed conformation, placing these protons far apart. Molecular modeling confirmed four stable syn conformations with α′ and β′ at 0 and 180° in all combinations.
Figure 6.
H3′ regions of nonselective excitation 31P-1H HMBC spectra of the unmodified (upper) and the Nar1IQ3 duplexes (lower). Both spectra were collected at 15 °C.
Chemical-Shift Perturbations
The 1H NMR chemical shifts of the Nar1IQ3 dodecamer were compared with those of the unmodified duplex (Figure 7). The largest perturbations were observed for the aromatic and anomeric protons of C18 in the complementary strand, opposite to the adduct. Smaller perturbations were also observed for the G17 H8, G19 H8 and H1′, C6 H6 and H1′, X7 H1′, and C8 H6 and H1′ resonances.
Figure 7.
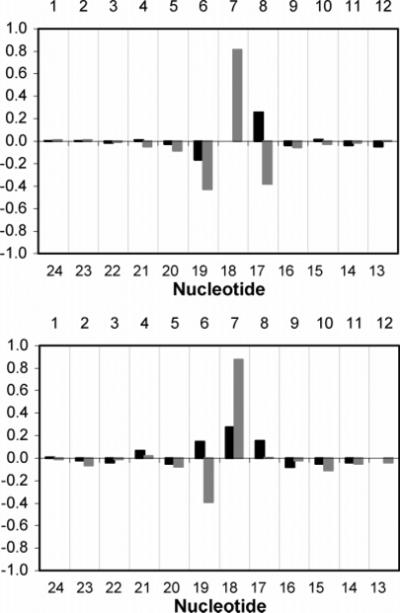
Chemical shift perturbations of (top) H6/H8 and (bottom) H1′ protons of the Nar1IQ3 duplex relative to the unmodified duplex. Dark bars represent the modified strand; light bars represent the complementary strand. Δδ = [δmodified oligodeoxynucleotide - δunmodified oligodeoxynucleotide] (ppm).
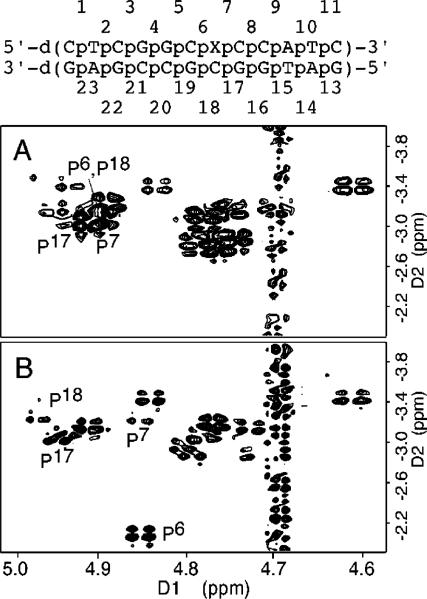
Structural Refinement
For the Nar1IQ3 duplex, a total of 488 NOE-based distance restraints were obtained, consisting of 148 inter- and 340 intra-nucleotide distances. They included 24 DNA-IQ distances. For the unmodified duplex, a total of 463 NOE-based distance restraints were obtained, consisting of 138 inter-and 325 intra-nucleotide distances. For the Nar1IQ3 duplex, the pyrimidine pseudorotation values were restrained in the C1′-exo range of P = 126 ± 18°, and the purines in the center 10 base pairs were restrained with pseudorotation values in the C2′-endo range of P = 162 ± 18°. No backbone torsion angle restraints were used for the modified strand at the lesion site. Elsewhere, the backbone angles α, β, and ξ were restrained to −60 ± 30°, 180 ± 30°, and −90 ± 30°, respectively, to allow both A- and B-like geometry.85 No empirical base-pairing restraints were used at the lesion site. Elsewhere, empirical base-pair planarity and Watson—Crick hydrogen-bonding restraints were used. These were consistent with crystallographic data.86 Their inclusion was based on data that showed that DNA maintained Watson-Crick base pairing.
The restrained molecular dynamics calculation employed a simulated annealing protocol. This began with a searching strategy guided by intermolecular IQ-DNA restraints. The DNA starting conformation was B-like except for the syn glycosyl torsion angle at X7. The orientation space was searched with 16 energy minimization trials in which the IQ angles α′ and β′ (Chart 2) were started at 0, 90, 180, and 270° in all combinations. This generated four stable conformations with the glycosyl bond in the syn conformation and the angles α′ and β′ at 0 and 180°. For the conformations with α′ ≈ 0° IQ oriented in minor groove, while for conformations with α′ ≈ 180° IQ intercalated. These were checked with respect to NOE violations. One exhibited the best fit to the NOE data of the Nar1IQ3 duplex.
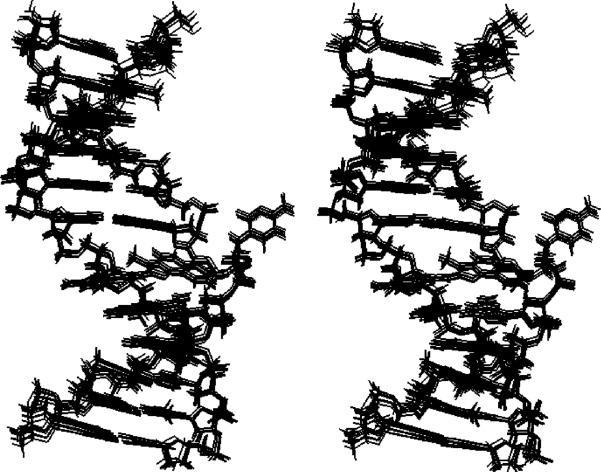
Figure 8 shows a stereoview of an ensemble of 10 structures obtained from randomly seeded calculations. Their precision was determined by pairwise rmsd measurements. These exhibited a maximum pairwise rmsd of 0.68 Å, suggesting convergence. Figure S3 of the Supporting Information shows the corresponding results for the unmodified duplex.
Figure 8.
Stereoview of 10 superimposed structures emergent from randomly seeded rMD calculations on the Nar1IQ3 duplex.
The accuracy of the refinement was determined by calculation of NOE intensities from the emergent structures using the program CORMA (v. 5.2)72 (Figure 9). The overall sixth root residual R1x for structures of the Nar1IQ3 duplex was 8.1 × 10−2. Figure S4 of the Supporting Information shows corresponding data for the unmodified duplex. For the Nar1IQ3 duplex, inter-and intraresidue R1x values were on the order of 10%. At the adduct site, the residuals were 6.2, 11, 5.2, and 6.1 (× 10−2) for C8, G17, C18, and G19, respectively. The structural statistics are found in Table 1.
Figure 9.
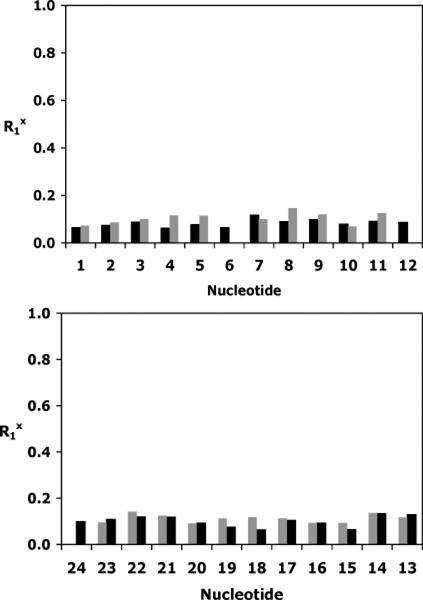
Sixth root residuals for NOE intensities of the Nar1IQ3 duplex. (Top) Nucleotides C1 → C12. (Bottom) Nucleotides G13 → G24. Dark bars represent intranucleotide values; light bars represent internucleotide values.
Table 1.
Analysis of RMD-Generated Structures of the Unmodified and Nar1IQ3 Duplexes
| NMR restraints | NarI | Nar1IQ3 |
|---|---|---|
| total no. of distance restraints | 463 | 488 |
| interresidue distance restraints | 138 | 148 |
| intraresidue distance restraints | 325 | 340 |
| DNA-IQ distance restraints | 0 | 24 |
| IQ-IQ distance restraints | 0 | 5 |
| H-bonding restraints | 33 | 30 |
| dihedral planarity restraints | 24 | 22 |
| sugar pucker restraints | 120 | 120 |
| backbone torsion angle restraints | 90 | 78 |
| structural statistics | ||
| NMR R-factor (R1x) 〈rMDRi〉 | 0.0812 ± 0.0003 | 0.0854 ± 0.0005 |
| rmsd of NOE violations (Å) | 0.00763 ± 0.00001 | 0.00798 ± 0.00002 |
| no. of NOE violations > 0.2 Å in the root-mean-square deviations from ideal geometry | 0 | 0 |
| bond length (Å) | 0.02402 ± 0.00005 | 0.02783 ± 0.00006 |
| bond angle (deg) | 2.613 ± 0.007 | 2.672 ± 0.006 |
| improper angle (deg) | 0.64 ± 0.02 | 0.79 ± 0.02 |
| pairwise rmsd (Å) over all atoms 〈rMDRi〉 vs 〈rMDav〉 | 0.65 ± 0.01 | 0.68 ± 0.02 |
Solution Conformation
The IQ moiety was inserted into the helix with the modified guanine and its complement C18 displaced in the major groove. Views normal to the helix axis and looking into the major groove of the central 5′-d(C6X7C8)-3′·5′-d(G17C18G19)-3′ segments of the unmodified and Nar1IQ3 duplexes are shown in Figure 10. The IQ ring inserted between the C6·G19 and C8·G17 base pairs by displacing the modified guanine of the syn X7 nucleotide into the major groove. The glycosyl torsion angle χ (O4′-C1′-N9-C4) of the X7 residue was calculated as 85 ± 10°.
Figure 10.
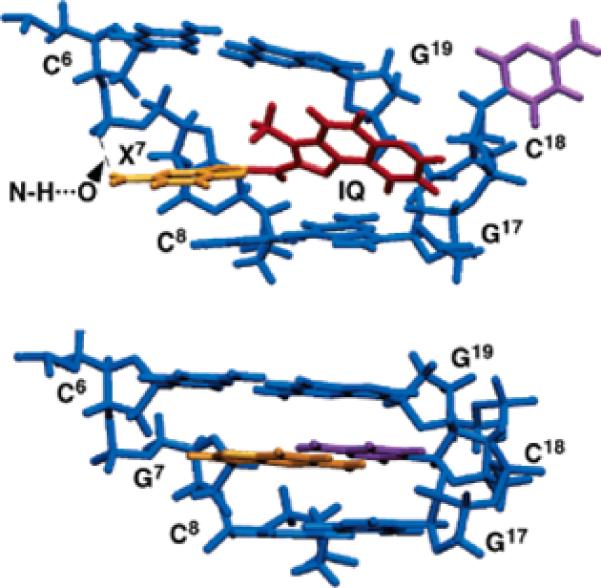
Comparison of the average structures, looking into the major groove and normal to the helix axis of the central segment. (A) Nar1IQ3 duplex. The IQ ring is shown in red and is inserted between base pairs C6·G19 and C8·G17. (B) Unmodified duplex.
The IQ methyl group faced into the helix. This placed the IQ H4A and H5A protons facing into the duplex, toward C18 and G19 in the complementary strand, whereas the IQ H7A, H8A, and H9A protons faced into the major groove in the vicinity of G17 and C18 in the complementary strand. One proton of the X7 exocyclic amino group was close to oxygen at the phosphodiester linkage P6 between C6 and X7. This yielded a N-H···O distance of 2.8 Å, suggesting the formation of a hydrogen bond. Opposite to X7, the insertion of the IQ ring into the helix resulted in the displacement of C18 into the major groove.
Views looking down the helix axis of the 5′-d(C6X7C8)-3′·5′-d(G17C18G19)-3′ segment are shown in Figure 11. The primary interaction involved base pair C8·G17, the 3′-neighboring base pair with respect to X7. The G17 imino proton was shielded by the IQ ring. However, the calculations suggested that the IQ ring tilted with respect to the DNA base pairs, presumably reducing stacking with base pairs C6·G19 and C8·G17. This might be attributed to steric hindrance from the IQ methyl group. This tilt was defined by the IQ torsion angle β′ which was measured from the refined structures as −23 ± 8°. The IQ torsion angle α′ was calculated as 158 ± 7°, resulting in the amine linkage of the IQ adduct being in plane with the C8-modified dG. The calculated glycosyl torsion angles and sugar pseudorotation P of the Nar1IQ3 duplex are found in Table S5 in the Supporting Information. The presence of the C8-dG IQ adduct opposite dC resulted in a bend at the adduct site of 23 ± 5°, and helical twist angles of −56 ± 3° and −76 ± 3° for base pair steps C6 → X7 and X7 → C8, respectively.
Figure 11.
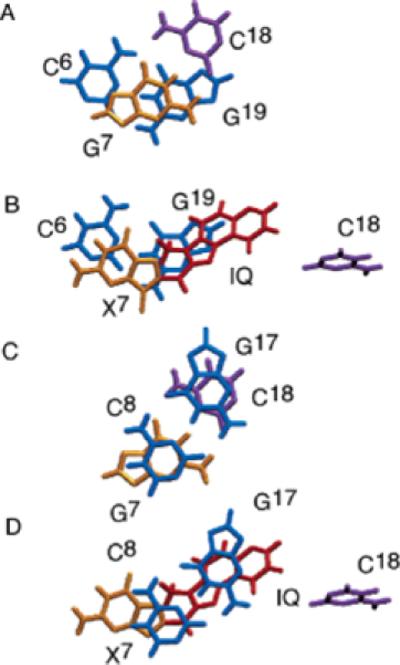
Base stacking of the Nar1IQ3 and the unmodified duplexes. (A) Unmodified duplex. Stacking of C6·G19 and G7·C18. (B) Nar1IQ3 duplex. Stacking of C6·G19 and X7·C18. (C) Unmodified duplex. Stacking of G7·C18 and C8·G17. (D) Nar1IQ3 duplex. Stacking of X7·C18 and C8· G17.
Discussion
The synthesis of site-specific C8-dG arylamine oligodeoxy-nucleotide adducts62,63 enabled high-resolution conformational studies of a site-specific C8-dG IQ adduct in the Nar1IQ3 duplex. The conformation of the C8-dG IQ adduct in DNA has been of interest because this adduct represents one of the most mutagenic HCAs found in the human diet. Both base-displaced insertion63 and minor groove64 conformations have been proposed for the C8-dG IQ DNA adduct, and indeed, the energetic differences between the two proposed conformations are likely to be modest and dependent upon sequence context.
Base-Displaced Insertion of the IQ Adduct
The present studies reveal that for the Nar1IQ3 duplex, in which the C8-dG IQ adduct is located at position G3 of the 5′-d(CG1G2CG3-CC)-3′ recognition site of the NarI enzyme, the base-displaced insertion conformation is favored. The key evidence supporting the conclusion that the X7 glycosyl torsion angle χ was in the syn conformation was the downfield chemical shift for the X7 H2′ resonance, observed at 3.61 ppm (Figure 5). This downfield shift of the H2′ resonance is a characteristic marker of the syn conformation of dG in modified duplexes.82,83 This corroborated work showing that the dG-C8 IQ adduct was in the syn conformation at the nucleoside level.53
Rotation of the glycosyl bond into the syn conformation at X7 placed the Watson—Crick hydrogen bonding edge of the modified dG into the major groove. The X7 imino and amino protons were exposed to solvent. Displacement of the modified dG into the major groove was consistent with the observed X7 imino proton chemical shift of 9.6 ppm, which was to the high field of the imino protons of other G·C base pairs. One proton of the amino group of the modified guanine was within 2.8 Å of the oxygen at the phosphodiester linkage P6 between C6 and X7. This suggested the potential for formation of a hydrogen bond that was consistent with the X7 amino proton chemical shifts of 6.67 and 8.83 ppm (Figure 2). This hydrogen bond might cause the local electrostatic potential at phosphodiester linkage P6 to be perturbed. This notion was supported by the significant 31P chemical shift perturbation observed at P6 (Figure 7). A strong X7 H1′ → C8 H6 NOE for the X7 → C8 base step (Figure 1) was consistent with a separation between these protons of 3.0 ± 0.4 Å, as measured in the intensity-refined structures of the duplex.
Evidence supporting insertion of the IQ ring between C6·G19 and C8·G17 base pairs included the upfield shift of the G17 imino proton as a result of stacking with the intercalated IQ ring (Figure 2). The IQ ring stacked primarily with G17 and G19 (Figure 11). There was no stacking between IQ and C6 or C8. The observed NOEs (Table S4 in the Supporting Information) were consistent with the IQ H4A and H5A protons being directed toward C18 and G19 in the complementary strand, and the IQ H7A, H8A, and H9A protons being directed toward G17 and C18 in the complementary strand (Figure 10). The IQ methyl protons were closer to G19 than to C8 (Figure 10), also consistent with the observed NOEs (Table S4). The absence of an NOE between the IQ amine and methyl protons was attributed to rapid exchange of the amine proton with solvent.
On the basis of a decrease of the IQ absorption, Elmquist et al.63 suggested a hydrogen bond between the IQ quinoline nitrogen and the exocyclic amine of complementary base C18. They proposed that this might stabilize the base-displaced insertion conformation with respect to a minor groove conformation. Spectroscopic evidence for this hydrogen bond was not observed. A series of rMD calculations that included this hydrogen bond as a restraint yielded structures that did not agree with the experimental NOEs. Instead, C18 was displaced into the major groove (Figure 10). This displacement of C18 resulted in a break in the 1H sequential NOE connectivity at the G17 → C18 step (Figure 1). This distance was predicted to be 7.9 ± 0.4 Å in the refined structures. The C18 amino proton resonances were not observed. This was consistent with the displacement of C18 into the major groove. These proton resonances were presumably broadened due to an intermediate rate of rotation about the C4-N4 bond and exchange with solvent.
The present results revealing a base-displaced insertion structure of the C8-dG IQ adduct in the Nar1IQ3 duplex differ from the predictions of a molecular mechanics study on the duplex 5′-d(G1G2CX3CCA)-3′·5′-d(TGGCGCC)-3′.64 The latter study predicted small energetic differences between groove-bound and base-displaced intercalated conformations of the C8-dG IQ adduct. The favored conformation as predicted by molecular mechanics contained the modified dG in the syn conformation about the glycosyl bond with the IQ in the minor groove and directed in the 3′ direction with respect to the modified strand. The calculations suggested that the base-displaced intercalated structure was ∼10 kcal/mol higher in energy than the minor groove structure.
Sequence Dependence
The small energetic differences predicted by the molecular mechanics calculations64 suggest that the conformations of C8-dG IQ adducts in duplex DNA are likely to be influenced by DNA sequence. Differing conformations may be responsible for increased genotoxicity in hotspot sequences. Sequence analysis of HCA-induced mutations in SOS-induced bacteria revealed the majority of mutations in the lac Z gene were clustered in hotspots involving iterated Gs.87 The 5′-d(CG1G2CX3CC)-3′ recognition sequence of the NarI restriction enzyme examined herein represents a hot spot for −2 frameshift mutations82 at G3 when G3 is modified by aromatic amines.88–96 Elmquist et al.63 examined the properties of the C8-dG IQ adduct located in the 5′-d(GGCAGXTGGTG)-3′·5′-d(CACCACCTGCC)-3′ duplex, bearing codon 12 of the human N-ras protooncogene (underlined). Utilizing a combination of thermal UV melting studies, UV spectroscopy, and circular dichroism, they concluded that the C8-dG IQ adduct adopted a groove-bound conformation in the ras12 sequence, similar to that predicted by Wu et al. 64 It will be of interest to examine the sequence dependence of the C8-dG IQ adduct in greater detail.
Comparison with the C8-dG PhIP Adduct
The solution structure of the C8-dG PhIP adduct was reported in 5′-d(CCATCXCTACC)-3′·5′d(GGTAGCGATGG)-3′.59 The PhIP-modified duplex with 5′-d(CXC)-3′ sequence adopted a conformation similar to that of the C8-dG IQ adduct in the Nar1IQ3 sequence. In the PhIP-modified duplex, the C8-dG PhIP adduct existed with the modified dG in the syn conformation and displaced into the major groove. The complementary dC was displaced into the major groove. The IP ring inserted into the duplex, stacking with the flanking G18 purine and the C5 and G16 rings. However, the out-of-plane geometry of the phenyl ring with respect to the IP ring in the PhIP adduct contributed to a greater unwinding and twisting of the helix as compared to the C8-dG IQ adduct. The PhIP phenyl ring was inclined out-of-plane relative to the IP ring, rotating rapidly, precluding stacking with the flanking bases. Additionally, the PhIP methyl group was positioned toward the modified strand, directed toward the minor groove edge of the DNA, whereas in the Nar1IQ3 duplex, the IQ methyl group was stacked between the flanking bases. Corresponding to the above differences, the PhIP-dG linkage site was defined by torsion angles α′ and β′ by 221.3 ± 3.0 and 132.5 ± 8.0°.
Comparison with Aminofluorene and Acetyl-amino-fluorene C8-dG Adducts
Adducts arising from the arylamines 2-aminofluorene (AF) and N-acetyl-2-aminofluorene (AAF) were extensively studied.10 The biological responses to the AF and AAF lesions differed,97 although they were structurally similar. In bacterial mutagenesis assays, the AAF adduct gave −1 and −2 frameshift mutations,88,98–101 whereas base-pair substitutions were largely observed for AF. Both the C8-dG IQ (this work) and C8-dG AAF adducts102 exhibited single base-displaced inserted structures when placed opposite dC in the 5′-d(CXC)-3′ context. NMR data for the C8-dG AF adduct in the Nar1IQ3 sequence was equivocal due to a mixture of conformers.103 An AF-intercalated conformer with the modified dG in the syn conformation and displaced with the 5′-flanking dC residue into the major groove was reported for the C8-dG AF adduct opposite −2 base deletion in the Nar1IQ3 sequence.104
Structure—Activity Relationships
The 5′-d(CG1G2CG3CC)-3′ NarI sequence represents the strongest known hotspot for frameshift mutagenesis.88,105 Within the NarI sequence, the propensity for frameshift mutagenesis is sequence-dependent. These mutations occur following adduct formation at the G3 but not the G1 or G2 positions.105,106 A single C8-dG acetylaminofluorene adduct located at position G3 induced −2 bp frameshifts more than 107-fold over background mutagenesis in Escherichia coli.107 In a study using site-specifically adducted oligonucleotides, it was found that AAF at the G3-position of the NarI sequence gave almost exclusively two-base deletion when replicated in bacteria, while giving largely base-pair substitution at the G1 and G2-positions. In COS-7 cells, the C8-AAF adduct gave base-pair substitution at all three positions.108 The −2 bp frameshift mutations induced at position G3 in the NarI sequence by the aromatic amine AAF109,110 arose via AAF-induced stabilization of a transient strand slippage intermediate during trans-lesion replication,110–112 and it is thought that the −2 bp frameshifts induced by the PhIP C8-dG adduct arise via the same mechanism.61 Crystallographic analysis of the bypass polymerase Dpo4 from Sulfolobus solfataricus involving complexes with damaged DNA templates supports the notion that error-prone lesion bypass can involve the formation of transient slippage intermediates.113–115 Koffel-Schwartz and Fuchs demonstrated that the dinucleotide repeat GCGC was essential for the −2 bp frameshifts in the NarI sequence, whereas the flanking nucleotides NaGCGCNb, particularly Nb, modulated the relative mutagenic strength of the sequence.107 In the case of AAF, it is thought that the 3′-neighboring base Nb forms favorable stacking interactions with the fluorene ring that stabilize the transient two-base strand slippage intermediate.116
The base-displaced intercalated structure may also play a role in modulating the repair of the C8-dG IQ adduct. Turesky et al.117 proposed that differences in the accumulation and rates of removal of C8-dG IQ and N2-dG IQ adducts in rodents and nonhuman primates may be attributable to differences in conformation about the glycosyl bond in the two classes of adducts. Adducts in the syn form are proposed to create greater distortions of the DNA duplex and, hence, be more easily recognized and excised. In contrast, adducts in the anti conformation are proposed to be more refractory toward repair. Turesky et al.117 observed a preferential removal of the C8-dG IQ adduct, whereas the N2-dG IQ adduct was more persistent. The latter existed in the anti conformation about the glycosyl bond at the nucleoside level.
Structural Coordinates Available
For the unmodified Nar1 duplex, the PDB ID code is 2HKB. For the IQ-modified Nar1IQ3 duplex, the PDB ID code is 2HKC.
Supplementary Material
Acknowledgment
We thank Drs. Thomas M. and Constance M. Harris for helpful discussions and suggestions. We thank Ms. Pamela Tamura and Ms. Albena Kozekova for assistance with the oligodeoxynucleotide synthesis and mass spectrometry and Mr. Markus Voehler for assistance with NMR experiments. This work was supported by NIH Grant CA-55678 (M.P.S.). The Vanderbilt Center for Molecular Toxicology is funded by a center grant from the National Institute of Environmental Health Sciences (ES-00267). J.S.S. was supported by NIH pre-doctoral traineeship ES-07028. Funding for the NMR spectrometers was supplied by NIH Grants RR-05805, ES-00267, and Vanderbilt University.
Footnotes
Supporting Information Available: Tables S1 and S2, nonexchangeable proton chemical shifts of the Nar1IQ3 and unmodified duplexes; S3, exchangeable proton chemical shifts for the Nar1IQ3 and unmodified duplexes; S4, comparison of experimental intermolecular distance restraints with those observed for intensity-refined structures of the Nar1IQ3 duplex; and S5, pseudorotation and glycosyl torsion angles for the Nar1IQ3 and unmodified duplexes. Figures S1, contour plots of the anomeric to aromatic region of the 1H NOESY spectrum for the unmodified duplex; S2, contour plot of the imino protons of the unmodified duplex; S3, stereoview of 10 superimposed structures emergent from rMD calculations on the unmodified duplex; and S4, R1x values for refined structures of the unmodified duplex calculated using the program CORMA (v. 5.2).72 This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wakabayashi K, Nagao M, Esumi H, Sugimura T. Cancer Res. 1992;52:2092–2098. [PubMed] [Google Scholar]
- 2.Layton DW, Bogen KT, Knize MG, Hatch FT, Johnson VM, Felton JS. Carcinogenesis. 1995;16:39–52. doi: 10.1093/carcin/16.1.39. [DOI] [PubMed] [Google Scholar]
- 3.Sugimura T. Mutat. Res. 1997;376:211–219. doi: 10.1016/s0027-5107(97)00045-6. [DOI] [PubMed] [Google Scholar]
- 4.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kataoka H, Nishioka S, Kobayashi M, Hanaoka T, Tsugane S. Bull. Environ. Contam. Toxicol. 2002;69:682–689. doi: 10.1007/s00128-002-0115-5. [DOI] [PubMed] [Google Scholar]
- 6.Felton JS, Knize MG, Salmon CP, Malfatti MA, Kulp KS. Environ. Mol. Mutagen. 2002;39:112–118. doi: 10.1002/em.10070. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi M, Hanaoka T, Nishioka S, Kataoka H, Tsugane S. Mutat. Res. 2002;506–507:233–241. doi: 10.1016/s0027-5107(02)00170-7. [DOI] [PubMed] [Google Scholar]
- 8.Ushiyama H, Wakabayashi K, Hirose M, Itoh H, Sugimura T, Nagao M. Carcinogenesis. 1991;12:1417–1422. doi: 10.1093/carcin/12.8.1417. [DOI] [PubMed] [Google Scholar]
- 9.Ohgaki H, Hasegawa H, Kato T, Suenaga M, Ubukata M, Sato S, Takayama S, Sugimura T. Environ. Health Perspect. 1986;67:129–134. doi: 10.1289/ehp.8667129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heflich RH, Neft RE. Mutat. Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 11.Thorgeirsson UP, Snyderwine EG, Gomez DE, Adamson RH. In Vivo. 1996;10:145–152. [PubMed] [Google Scholar]
- 12.Ohgaki H, Kusama K, Matsukura N, Morino K, Hasegawa H, Sato S, Takayama S, Sugimura T. Carcinogenesis. 1984;5:921–924. doi: 10.1093/carcin/5.7.921. [DOI] [PubMed] [Google Scholar]
- 13.Takayama S, Nakatsuru Y, Masuda M, Ohgaki H, Sato S, Sugimura T. Gann. 1984;75:467–470. [PubMed] [Google Scholar]
- 14.Tanaka T, Barnes WS, Williams GM, Weisburger JH. Jpn. J. Cancer Res. 1985;45:570–576. [PubMed] [Google Scholar]
- 15.Sugimura T, Nagao M, Wakabayashi K. Complex Factors Pertinent to Human Hazard and Risk. Wiley; New York: 2000. pp. 349–359. [Google Scholar]
- 16.Adamson RH, Thorgeirsson UP, Snyderwine EG, Thorgeirsson SS, Reeves J, Dalgard DW, Takayama S, Sugimura T. Jpn. J. Cancer Res. 1990;50:10–14. doi: 10.1111/j.1349-7006.1990.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson KE, Hammons GJ, Kadlubar FF, Potter JD, Kaderlik KR, Ilett KF, Minchin RF, Teitel CH, Chou HC, Martin MV, Guengerich FP, Barone GW, Lang NP, Peterson LA. Carcinogenesis. 1997;18:1085–1092. doi: 10.1093/carcin/18.5.1085. [DOI] [PubMed] [Google Scholar]
- 18.Lang NP, Butler MA, Massengill J, Lawson M, Stotts RC, Hauer-Jensen M, Kadlubar FF. Cancer Epidemiol. Biomarkers Prev. 1994;3:675–682. [PubMed] [Google Scholar]
- 19.Shirai T, Sano M, Tamano S, Takahashi S, Hirose M, Futakuchi M, Hasegawa R, Imaida K, Matsumoto K, Wakabayashi K, Sugimura T, Ito N. Cancer Res. 1997;57:195–198. [PubMed] [Google Scholar]
- 20.Snyderwine EG. Cancer. 1994;74:1070–1077. doi: 10.1002/1097-0142(19940801)74:3+<1070::aid-cncr2820741515>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 21.Ronco A, De Stefani E, Mendilaharsu M, Deneo-Pellegrini H. Int. J. Cancer. 1996;65:328–331. doi: 10.1002/(SICI)1097-0215(19960126)65:3<328::AID-IJC9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Josephy PD, Evans DH, Parikh A, Guengerich FP. Chem. Res. Toxicol. 1998;11:70–74. doi: 10.1021/tx970171q. [DOI] [PubMed] [Google Scholar]
- 23.Josephy PD, Gruz P, Nohmi T. Mutat. Res. 1997;386:1–23. doi: 10.1016/s1383-5742(96)00041-5. [DOI] [PubMed] [Google Scholar]
- 24.Oda Y, Yamazaki H, Watanabe M, Nohmi T, Shimada T. Mutat. Res. 1995;334:145–156. doi: 10.1016/0165-1161(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 25.Oda Y, Aryal P, Terashita T, Gillam EM, Guengerich FP, Shimada T. Mutat. Res. 2001;492:81–90. doi: 10.1016/s1383-5718(01)00154-1. [DOI] [PubMed] [Google Scholar]
- 26.Nagao M. In: Food-Borne Carcinogens: Heterocyclic Amines. Nagao M, Sugimura T, editors. Wiley; New York: 2000. pp. 31–71. [Google Scholar]
- 27.Sugimura T, Sato S. Cancer Res. 1983;43:2415s–2421s. [PubMed] [Google Scholar]
- 28.Hecht SS. Environ. Mol. Mutagen. 2002;39:119–126. doi: 10.1002/em.10071. [DOI] [PubMed] [Google Scholar]
- 29.Kosakarn P, Halliday JA, Glickman BW, Josephy PD. Carcinogenesis. 1993;14:511–517. doi: 10.1093/carcin/14.3.511. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe M, Ohta T. Carcinogenesis. 1993;14:1149–1153. doi: 10.1093/carcin/14.6.1149. [DOI] [PubMed] [Google Scholar]
- 31.Terada M, Nagao M, Nakayasu M, Sakamoto H, Nakasato F, Sugimura T. Environ. Health Perspect. 1986;67:117–119. doi: 10.1289/ehp.8667117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson LH, Tucker JD, Stewart SA, Christensen ML, Salazar EP, Carrano AV, Felton JS. Mutagenesis. 1987;2:483–487. doi: 10.1093/mutage/2.6.483. [DOI] [PubMed] [Google Scholar]
- 33.Felton JS, Fultz E, Dolbeare FA, Knize MG. Food Chem. Toxicol. 1994;32:897–903. doi: 10.1016/0278-6915(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 34.Felton JS, Knize MG, Dolbeare FA, Wu RW. Environ. Health Perspect. 1994;102:201–204. doi: 10.1289/ehp.94102s6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schut HA, Snyderwine EG. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 36.Turesky RJ. Drug Metab. Rev. 2002;34:625–650. doi: 10.1081/dmr-120005665. [DOI] [PubMed] [Google Scholar]
- 37.Thompson LH, Carrano AV, Salazar E, Felton JS, Hatch FT. Mutat. Res. 1983;117:243–257. doi: 10.1016/0165-1218(83)90125-8. [DOI] [PubMed] [Google Scholar]
- 38.Aeschbacher HU, Ruch E. Carcinogenesis. 1989;10:429–433. doi: 10.1093/carcin/10.3.429. [DOI] [PubMed] [Google Scholar]
- 39.Tohda H, Oikawa A, Kawachi T, Sugimura T. Mutat. Res. 1980;77:65–69. doi: 10.1016/0165-1218(80)90121-4. [DOI] [PubMed] [Google Scholar]
- 40.Yamazoe Y, Shimada M, Kamataki T, Kato R. Cancer Res. 1983;43:5768–5774. [PubMed] [Google Scholar]
- 41.Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR. Cancer Res. 1996;56:2979–2984. [PubMed] [Google Scholar]
- 42.Boobis AR, Lynch AM, Murray S, de la Torre R, Solans A, Farre M, Segura J, Gooderham NJ, Davies DS. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- 43.Hammons GJ, Milton D, Stepps K, Guengerich FP, Tukey RH, Kadlubar FF. Carcinogenesis. 1997;18:851–854. doi: 10.1093/carcin/18.4.851. [DOI] [PubMed] [Google Scholar]
- 44.Guengerich FP. Drug Metab. Rev. 2002;34:607–623. doi: 10.1081/dmr-120005663. [DOI] [PubMed] [Google Scholar]
- 45.Hickman D, Pope J, Patil SD, Fakis G, Smelt V, Stanley LA, Payton M, Unadkat JD, Sim E. Gut. 1998;42:402–409. doi: 10.1136/gut.42.3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hein DW, Doll MA, Rustan TD, Gray K, Feng Y, Ferguson RJ, Grant DM. Carcinogenesis. 1993;14:1633–1638. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- 47.Minchin RF, Reeves PT, Teitel CH, McManus ME, Mojarrabi B, Ilett KF, Kadlubar FF. Biochem. Biophys. Res. Commun. 1992;185:839–844. doi: 10.1016/0006-291x(92)91703-s. [DOI] [PubMed] [Google Scholar]
- 48.Le Marchand L, Hankin JH, Pierce LM, Sinha R, Nerurkar PV, Franke AA, Wilkens LR, Kolonel LN, Donlon T, Seifried A, Custer LJ, Lum-Jones A, Chang W. Mutat. Res. 2002;506–507:205–214. doi: 10.1016/s0027-5107(02)00167-7. [DOI] [PubMed] [Google Scholar]
- 49.Ishibe N, Sinha R, Hein DW, Kulldorff M, Strickland P, Fretland AJ, Chow WH, Kadlubar FF, Lang NP, Rothman N. Pharmacogenetics. 2002;12:145–150. doi: 10.1097/00008571-200203000-00008. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y, Chladek S, Romano LJ. J. Org. Chem. 1994;59:556–563. [Google Scholar]
- 51.Snyderwine EG, Roller PP, Adamson RH, Sato S, Thorgeirsson SS. Carcinogenesis. 1988;9:1061–1065. doi: 10.1093/carcin/9.6.1061. [DOI] [PubMed] [Google Scholar]
- 52.Nagaoka H, Wakabayashi K, Kim SB, Kim IS, Tanaka Y, Ochiai M, Tada A, Nukaya H, Sugimura T, Nagao M. Jpn. J. Cancer Res. 1992;52:1025–1029. doi: 10.1111/j.1349-7006.1992.tb02716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turesky RJ, Rossi SC, Welti DH, Lay JO, Jr., Kadlubar FF. Chem. Res. Toxicol. 1992;5:479–490. doi: 10.1021/tx00028a005. [DOI] [PubMed] [Google Scholar]
- 54.Guengerich FP, Mundkowski RG, Voehler M, Kadlubar FF. Chem. Res. Toxicol. 1999;12:906–916. doi: 10.1021/tx990094u. [DOI] [PubMed] [Google Scholar]
- 55.Gorlewska-Roberts K, Green B, Fares M, Ambrosone CB, Kadlubar FF. Environ. Mol. Mutagen. 2002;39:184–192. doi: 10.1002/em.10060. [DOI] [PubMed] [Google Scholar]
- 56.Takayama K, Yamashita K, Wakabayashi K, Sugimura T, Nagao M. Jpn. J. Cancer Res. 1989;49:1145–1148. doi: 10.1111/j.1349-7006.1989.tb01644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gangl ET, Turesky RJ, Vouros P. Anal. Chem. 2001;73:2397–2404. doi: 10.1021/ac0100401. [DOI] [PubMed] [Google Scholar]
- 58.Soglia JR, Turesky RJ, Paehler A, Vouros P. Anal. Chem. 2001;73:2819–2827. doi: 10.1021/ac010218j. [DOI] [PubMed] [Google Scholar]
- 59.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moriya M. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shibutani S, Fernandes A, Suzuki N, Zhou L, Johnson F, Grollman AP. J. Biol. Chem. 1999;274:27433–27438. doi: 10.1074/jbc.274.39.27433. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Rizzo CJ. Org. Lett. 2001;3:565–568. doi: 10.1021/ol006968h. [DOI] [PubMed] [Google Scholar]
- 63.Elmquist CE, Stover JS, Wang Z, Rizzo CJ. J. Am. Chem. Soc. 2004;126:11189–11201. doi: 10.1021/ja0487022. [DOI] [PubMed] [Google Scholar]
- 64.Wu X, Shapiro R, Broyde S. Chem. Res. Toxicol. 1999;12:895–905. doi: 10.1021/tx990108w. [DOI] [PubMed] [Google Scholar]
- 65.Borer PN. Handbook of Biochemistry and Molecular Biology. CRC Press; Cleveland, OH: 1975. [Google Scholar]
- 66.Piotto M, Saudek V, Sklenar V. J. Biomol. NMR. 1992;6:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 67.Piantini U, Sorensen OW, Ernst RR. J. Am. Chem. Soc. 1982;104:6800–6801. [Google Scholar]
- 68.Rance M, Sorensen OW, Hodenhausen G, Wagner G, Ernst RR, Wuthrich K. Biochem. Biophys. Res. Commun. 1983;177:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 69.Griesinger G, Sorensen OW, Ernst RR. J. Am. Chem. Soc. 1985;107:6394–6396. [Google Scholar]
- 70.Sklenar V, Bax A, Zon G. FEBS Lett. 1986;208:94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- 71.Sklenar V, Miyashiro H, Zon G, MIles HT, Bax A. FEBS Lett. 1986;208:94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- 72.Keepers JW, James TL. J. Magn. Reson. 1984;57:404–426. [Google Scholar]
- 73.Borgias BA, James TL. J. Magn. Reson. 1990;87:475–487. [Google Scholar]
- 74.Liu H, Tonelli M, James TL. J. Magn. Reson. B. 1996;111:85–89. doi: 10.1006/jmrb.1996.0064. [DOI] [PubMed] [Google Scholar]
- 75.Salazar M, Fedoroff OY, Miller JM, Ribeiro NS, Reid BR. Biochemistry. 1993;32:4207–4215. doi: 10.1021/bi00067a007. [DOI] [PubMed] [Google Scholar]
- 76.Brunger AT. X-Plor. Version 3.1. A System for X-ray Crystallography and NMR. Yale University Press; New Haven: 1992. [Google Scholar]
- 77.Nilsson L, Karplus M. J. Comput. Chem. 1986;7:591–616. [Google Scholar]
- 78.Lu XJ, Olson WK. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reid BR. Q. Rev. Biophys. 1987;20:2–28. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 80.Patel DJ, Shapiro L, Hare D. Q. Rev. Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 81.Boelens R, Scheek RM, Dijkstra K, Kaptein R. J. Magn. Reson. 1985;62:378–386. [Google Scholar]
- 82.Fuchs RP, Schwartz N, Daune MP. Nature. 1981;294:657–659. doi: 10.1038/294657a0. [DOI] [PubMed] [Google Scholar]
- 83.Norman D, Abuaf P, Hingerty BE, Live D, Grunberger D, Broyde S, Patel DJ. Biochemistry. 1989;28:7462–7476. doi: 10.1021/bi00444a046. [DOI] [PubMed] [Google Scholar]
- 84.Gorenstein DG. Phosphorus-31 chemical shifts and spin–spin coupling constant principles and empirical observations. In: Gorenstein DG, editor. Phosphorus-31 NMR Principles and Application. Academic Press; New York: 1984. pp. 7–56. [Google Scholar]
- 85.Tjandra N, Tate S-I, Ono A, Kainosho M, Bax A. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 86.Saenger W. Principles of Nucleic Acid Structure. Springer; New York: 1984. [Google Scholar]
- 87.Solomon MS, Morgenthaler PM, Turesky RJ, Essigmann JM. J. Biol. Chem. 1996;271:18368–18374. doi: 10.1074/jbc.271.31.18368. [DOI] [PubMed] [Google Scholar]
- 88.Burnouf D, Koehl P, Fuchs RPP. Proc. Natl. Acad. Sci. U.S.A. 1989;86:4147–4151. doi: 10.1073/pnas.86.11.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belguise-Valladier P, Fuchs RPP. Biochemistry. 1991;30:10091–10100. doi: 10.1021/bi00106a005. [DOI] [PubMed] [Google Scholar]
- 90.Rodriguez H, Loechler EL. Carcinogenesis. 1993;14:373–383. doi: 10.1093/carcin/14.3.373. [DOI] [PubMed] [Google Scholar]
- 91.Rodriguez H, Loechler EL. Biochemistry. 1993;32:1759–1769. doi: 10.1021/bi00058a009. [DOI] [PubMed] [Google Scholar]
- 92.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 93.Shukla R, Jelinsky S, Liu T, Geacintov NE, Loechler EL. Biochemistry. 1997;36:13263–13269. doi: 10.1021/bi971195z. [DOI] [PubMed] [Google Scholar]
- 94.Shukla R, Liu T, Geacintov NE, Loechler EL. Biochemistry. 1997;36:10256–10261. doi: 10.1021/bi970541+. [DOI] [PubMed] [Google Scholar]
- 95.Page JE, Zajc B, Oh-hara T, Lakshman MK, Sayer JM, Jerina DM, Dipple A. Biochemistry. 1998;37:9127–9137. doi: 10.1021/bi980273v. [DOI] [PubMed] [Google Scholar]
- 96.Fernandes A, Liu T, Amin S, Geacintov NE, Grollman AP, Moriya M. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 97.Beland FA, Kadlubar FF. Environ. Health Perspect. 1985;62:19–30. doi: 10.1289/ehp.856219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fuchs RPP. J. Mol. Biol. 1983;177:173–180. doi: 10.1016/0022-2836(84)90063-9. [DOI] [PubMed] [Google Scholar]
- 99.Bichara M, Fuchs RPP. J. Mol. Biol. 1985;183:341–351. doi: 10.1016/0022-2836(85)90005-1. [DOI] [PubMed] [Google Scholar]
- 100.Melchior WB, Jr., Marques MM, Beland FA. Carcinogenesis. 1994;15:889–899. doi: 10.1093/carcin/15.5.889. [DOI] [PubMed] [Google Scholar]
- 101.Tebbs RS, Romano LJ. Biochemistry. 1994;33:8998–9006. doi: 10.1021/bi00196a018. [DOI] [PubMed] [Google Scholar]
- 102.O'Handley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 103.Mao B, Hingerty BE, Broyde S, Patel DJ. Biochemistry. 1998;37:95–106. doi: 10.1021/bi972258g. [DOI] [PubMed] [Google Scholar]
- 104.Mao B, Gorin A, Gu Z, Hingerty BE, Broyde S, Patel DJ. Biochemistry. 1997;36:14479–14490. doi: 10.1021/bi972205z. [DOI] [PubMed] [Google Scholar]
- 105.Broschard TH, Koffel-Schwartz N, Fuchs RP. J. Mol. Biol. 1999;288:191–199. doi: 10.1006/jmbi.1999.2667. [DOI] [PubMed] [Google Scholar]
- 106.Burnouf DY, Miturski R, Fuchs RP. Chem. Res. Toxicol. 1999;12:144–150. doi: 10.1021/tx9801920. [DOI] [PubMed] [Google Scholar]
- 107.Koffel-Schwartz N, Fuchs RP. J. Mol. Biol. 1995;252:507–513. doi: 10.1006/jmbi.1995.0515. [DOI] [PubMed] [Google Scholar]
- 108.Tan X, Suzuki N, Grollman AP, Shibutani S. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 109.Lambert IB, Napolitano RL, Fuchs RPP. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1310–1314. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garcia A, Lambert IB, Fuchs RP. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5989–5993. doi: 10.1073/pnas.90.13.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Milhe C, Dhalluin C, Fuchs RP, Lefevre JF. Nucleic Acids Res. 1994;22:4646–4652. doi: 10.1093/nar/22.22.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Milhe C, Fuchs RP, Lefevre JF. Eur. J. Biochem. 1996;235:120–127. doi: 10.1111/j.1432-1033.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 113.Ling H, Boudsocq F, Woodgate R, Yang W. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 114.Ling H, Boudsocq F, Woodgate R, Yang W. Mol. Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 115.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 116.Roy D, Hingerty BE, Shapiro R, Broyde S. Chem. Res. Toxicol. 1998;11:1301–1311. doi: 10.1021/tx980106w. [DOI] [PubMed] [Google Scholar]
- 117.Turesky RJ, Box RM, Markovic J, Gremaud E, Snyderwine EG. Mutat. Res. 1997;376:235–241. doi: 10.1016/s0027-5107(97)00048-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.