Abstract
We report that Plasmodium falciparum (Pf) encodes a 912 amino acid ATP-dependent DNA ligase. Protein sequence analysis of Pf DNA Ligase I indicates a strong sequence similarity, particularly in the C-terminal region, to DNA ligase I homologues. The activity of recombinant Pf DNA Ligase I (PfLigI) was investigated using protein expressed in HEK293 cells. The PfLigI gene product is ~ 94 kDa and catalyzes phosphodiester bond formation on a singly-nicked DNA substrate. The enzyme is most active at alkaline pH (8.5) and with Mg2+ or Mn2+ and ATP as cofactors. Kinetic studies of PfLigI revealed that the enzyme has similar substrate affinity (Km 2.6 nM) as compared to human DNA ligase I and kcat (2.3×10−3s−1) and kcat/Km (8.8×105 M−1s−1) which are similar to other ATP dependent DNA ligases. PfLigI was able to join RNA-DNA substrates only when the RNA sequence was upstream of the nick, confirming that it is DNA ligase I and has no associated DNA ligase-III like activity.
1. Introduction
Malaria remains a constant global health threat, endangering close to 50% of the world’s population [1]. Plasmodium falciparum, the most pathological of all human malaria species, continues to devastate developing nations, with 1 to 3 million deaths reported annually and mortality rates expected to rise due to increases in antimalarial drug resistance. The recent completion of the P. falciparum genome [2] has broadened our understanding of the cellular and molecular pathways of the malaria parasite. However, little is known about the mechanisms that P. falciparum employs to maintain genomic integrity and stability.
The DNA base excision repair (BER) pathway is the primary conduit most organisms utilize to relieve their DNA from the daily insults it undergoes due to sources of damage such as reactive oxygen species, environmental mutagens or spontaneous base damage. BER is a series of carefully orchestrated events that begins with the actions of DNA glycosylases, lyases and endonucleases excising the damaged DNA base and the resultant baseless sugar phosphate backbone. DNA polymerases fill in the resultant apurinic/apyrimidinic (AP) site and DNA ligases complete the repair by catalyzing phosphodiester bond formation between adjacent nucleotides.
There are two distinct BER pathways available to organisms to repair DNA lesions; short-patch and long-patch BER. As their names suggest, these two pathways are delineated by the size of the excised repair patch, 1 nucleotide for short-patch and 2–11 nucleotides for long-patch [3]. Additionally, each pathway is comprised of a specific set of coordinating enzymes to accomplish this task. Short-patch BER utilizes Pol β, XRCC1 and DNA ligase III to fully repair DNA lesions, while long-patch BER is characterized by the repair activities of Pol δ/ε, FEN-1 and DNA ligase I. Among the eukaryotes studied, short-patch is the predominant BER repair pathway, constituting the removal of about 80% of damaged nucleotides [4–7].
Interestingly, the P. falciparum genome is devoid of homologues to essential genes required for DNA short-patch BER, e.g., Pol β, XRCC1 and DNA ligase III. Furthermore, it has been established previously from our laboratory, using in vitro cell free lysate assays, that P. falciparum repairs abasic lesions exclusively through a long-patch DNA BER pathway [8]. Moreover, DNA ligase I, which catalyzes the final step in long-patch BER, appears to be the only DNA ligase contained within the P. falciparum genome. By contrast, many eukaryotes express three DNA ligase isoforms, DNA ligase I, III and IV. DNA ligase I constitutes the primary ligase utilized in DNA replication within the eukaryotic nucleus and plays a critical role in DNA repair and recombination. DNA ligase III, which presently has been associated only with vertebrate cells, is involved with DNA repair in the nucleus and mitochondria [9]. DNA ligase IV has been identified in lower eukaryotes such as yeast and is involved in repairing DNA double strand breaks as well as V(D)J recombination and non-homologous end-joining (NHEJ) [10].
DNA ligases are ubiquitous enzymes that are characterized by their ability to utilize either ATP or nicotinamide adenine dinucleotide (NAD+) as a cofactor during phosphodiester bond formation. Viruses, archeabacteria and eukaryotes breakdown ATP to AMP and pyrophosphate (PPi), while eubacteria use NAD+, leaving behind AMP and nicotinamide mononucleotide (NMN). Phosphodiester bond formation from ATP-dependent DNA ligases consumes one molecule of ATP and utilizes covalent intermediates in the process. This is the basis for the three step reaction that DNA ligases employ for nick sealing. In the first step, the active site lysine nucleophile attacks the α-phosphate of ATP creating a covalent ligase-AMP intermediate and releases pyrophosphate. This reaction can occur in vitro in the presence of ATP and the absence of nicked DNA. The AMP is then transferred to the 5′-phosphate group of the nicked DNA strand. In the final step, the activated 5′ terminus is attacked by the 3′-OH in a phosphoryl transfer reaction. The nicked DNA is then sealed and the AMP molecule is eliminated [11]. DNA ligase I has a dual role within the cell: the joining of Okazaki fragments during DNA replication and nick sealing during long-patch BER. Genetic manipulation of the DNA ligase I gene has lead to the observation that knockouts are lethal in mice [12]. Furthermore, elevated expression of DNA ligase I has been linked to various human cancers and has induced genetic instability in yeast [13,14].
Here we report the identification, recombinant expression, purification and biochemical characterization of a codon-optimized DNA ligase I from P. falciparum. PfLIGI is a 2.7 kb gene on chromosome 13 that encodes for 912 amino acid polypeptide with a predicted molecular weight of 104 kDa.
2. Materials and methods
2.1 Parasite Cultures
Plasmodium falciparum FCR3/C5 strain parasites were cultured at a 4% hematocrit as described previously [15]. 5% sorbitol was used to synchronize parasite cultures according to well established methods [16]. Two sorbitol treatments 10 – 12 h apart were employed to obtain tightly synchronous cultures. Parasite cultures were harvested at the appropriate time point in the lifecycle corresponding to ring, trophozoite and schizont stages.
2.2 Whole Cell Parasite Extracts
Parasites were liberated from infected erythrocytes by incubation in 0.05% saponin/PBS (10 mM sodium phosphate, 140 mM NaCl, pH 7.6) for 30 min. at 37°C. Parasites were pelleted at 5000g for 10 min. and washed 4–5 times with ice cold PBS. The final pellet was resuspended in 1–2 pellet volumes of 20 mM Tris-HCl, pH 7.5 plus protease inhibitor cocktail (Sigma, MO). The parasites were lysed by sonication in an ice-water bath using 4 pulses of 30 sec duration with a 50% pulse rate and a power setting of 5 using a W-225 Ultrasonic processor (HeatSystems-Ultrasonic, Inc). The sonicate was centrifuged at 12,000g for 30 min. The supernatant was removed and flash frozen using liquid nitrogen. Protein concentration was determined using the BCA protein assay (Pierce, IL).
2.3 Generation of PfLigI antibody
A 16 amino acid peptide was generated (Invitrogen, CA) with the following sequence: CNKNKNIDYNDDTESE, which corresponds to the last 15 amino acids of PfLigI C-terminus. A cysteine residue was placed on the N-terminus of the peptide to aid in peptide conjugation to keyhole limpet hemocyanin (KLH). Subsequently, the KLH-conjugated peptide was used to immunize rabbits for polyclonal antibody production (Cocalico Biologicals, PA). Rabbit pre-immune sera was screened by Western Blot using parasite lysate for pre-existing cross-reactive proteins and was found to be negative (data not shown).
Rabbit sera, positive for PfLigI antibody, was purified by first adding 2 volumes of 60 mM sodium acetate buffer (pH4.0) and adjusting the final pH to 4.8. Serum proteins were precipitated from the mixture using 0.075 ml of octanoic acid per ml of the original rabbit sera volume and removed by centrifuging at 5,000g for 10 min. The supernatant was saved and an equal volume of saturated ammonium sulfate was slowly added to a final concentration of 50% to precipitate the IgG fraction. The mixture was precipitated at 4°C for 18h and centrifuged at 5,000g for 30 min. The pellet was resuspended, until the solution was translucent, in 50 mM Tris pH 7.5. The solution was loaded onto a Q-sepharose column, washed with ≥10 column volumes of 50 mM Tris pH 7.5 and eluted with 10 column volumes of elution buffer (1 M NaCl, 50 mM Tris pH7.5). The sample was then dialyzed overnight with PBS.
Affinity purification of the IgG mixture was accomplished by first coupling PfLigI to an agarose matrix using an AminoLink® Plus Immobilization Kit (Pierce, IL) per the manufacturer’s instructions. The ammonium sulfate precipitated IgG mixture was then passed over the column, PfLigI IgG was eluted and fractions were analyzed by Western Blot for positive reactivity with PfLigI protein.
2.4 Codon Optimization
The PfLIGI gene was synthesized by GENEART Inc. (Regensburg, Germany) using the cDNA sequence as reported by the completed P. falciparum genome (Gene ID# MAL13P1.22, PlasmoDB.org). The DNA sequence was optimized to shift the A–T base content from the endogenous 72% to 56%, using a human DNA codon set. The gene was synthesized minus the first 381 bp (encoding the first 127 amino acids of the N-terminus) of PfLigI. The synthesized gene was assembled from synthetic oligonucleotides and cloned into pPCR-Script (Stratagene, CA) using KpnI and SacI restriction sites. PfLIGI gene was subcloned into the pET24a (Novagen, CA) bacterial expression vector using the BamHI and XhoI restriction sites. The final constructs were verified by sequencing through GENEART Inc.
2.5 PfLig1 Expression
The PfLIGI gene was subcloned from pET24a into the pYM-v200flaghisN eukaryotic expression vector using the BamHI and XhoI restriction sites. pYM-v200flaghisN vector was constructed from pcDNA3 (Clontech, CA) by the following modifications: disruption of the original NdeI site in the CMV promoter region, insertion of a Kozak sequence, a FLAG® tag, a 6-His tag and a new NdeI site at the polylinker region. Since this vector confers a FLAG® and 6-His tag onto the N-terminus, the calculated molecular weight of PfLigI was 94 kDa. HEK 293 cells (ATCC, VA) were grown in 150 cm2 culture flasks in DMEM (Cellgro, VA) supplemented with FBS to 5% and 0.1 mg/ml Penicillin/Streptomycin. Cells were grown in a humidified chamber at 37°C and with 5% CO2. Transfections were performed by adding 40 μg of pYM-v200flaghisN/PfLigI plasmid DNA to 1.5 ml of DMEM, without FBS or antibiotics, and allowed to stand at room temperature for 5 min. 120 μg of polyethylenimine (Polysciences, PA) was then added and the mixture was incubated at 37°C for 20 min before being added to the flask. After 24 h, the HEK 293 media was changed and the cells were harvested after 48 h.
2.6 PfLigI Purification
DMEM was removed from transfected HEK 293 cells. The cells were washed twice with cold PBS and 2 ml of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 1% Triton X-100) was added to each 150 cm2 culture flask. Cells were incubated on a shaker for 20 min and the cell lysate was centrifuged at 12,000 × g for 10 min. The supernatant was filtered using a 0.2 μm filter and loaded onto a 5 ml Anti-FLAG® M2 affinity gel column (Sigma, MO). The column was washed with 10 volumes of TBS pH 7.5 and the bound sample was eluted with 0.1 M glycine pH 3.5 into 1 ml fractions containing 20 μl of 1 M Tris pH 8.8 for neutralization. Western blot analysis was performed on the flow through, wash and eluate fractions using the anti-FLAG® M2 monoclonal antibody (Sigma, MO). A typical transfection of 8, 150 cm2 flasks yielded approximately 1.5 – 2 mg of FLAG® purified PfLigI
2.7 Oligonucleotides
Gel purified oligonucleotides were purchased from Midland Certified Reagent Company (Midland, TX). 32P γ-ATP was from Perkin Elmer (Boston, MA). Polynucleotide kinase was from Roche Molecular Biochemicals (Indianapolis, IN). QIAquick columns (Qiagen, CA) were used for the purification of labeled DNA.
2.8 Ligation substrates
Oligos were modeled after previously published sequences [17]. Briefly, a double stranded 44mer with a single nick between bases 18 and 19 was created with the following oligos: (44mer) 5′-gcg tcg ggt gga cgg gtg gat tga aat tta gg ctg gca cgg tcg -3′; (18mer) 5′ Pi- cac ccg tcc acc cga cgc – 3′; (26mer) 5′-cga ccg tgc cag cct aaa ttt caa tc – 3′. 100 pmol of the 26mer oligo was 5′-end labeled using E. coli polynucleotide kinase and 32P γ-ATP for a minimum of 1 h at 37°C. Purification of labeled DNA was performed using the QIAquick nucleotide removal kit (Qiagen, CA). Samples were eluted from the column in 50 μl. To determine the percent recovery after column purification, samples were taken pre- and post- column load and were subsequently separated on a 12% SDS-PAGE gel. The gel was exposed to a PhosphorImager™ screen and scanned using a Molecular Dynamics Storm 840 Scanner. Densitometry was performed on the scanned image using ImageQuant analysis software (Molecular Dynamics). Percent recovery of the purified oligo was calculated using the following equation: (densitometric volume of the post-purification sample/densitometric volume of the pre-purification) × 100. Equimolar amounts of 44mer and 18mer were added to the eluted sample, equal to the calculated recovery of the 26mer oligo, and were brought up to 60 μl in annealing buffer (20 mM Tris-HCl pH 7.4 and 150 mM NaCl). Samples were placed into a boiling water bath for 5 min and allowed to slowly cool to room temperature overnight. RNA oligos used to determine the substrate specificity of PfLigI were labeled 44R, 18R, and 26R. The oligos were created as described for the aforementioned DNA oligos, substituting in ribonucleotides and replacing thymidine bases with uracil.
2.9 Ligation Assays
Standard reaction conditions consisted of: 20 μl reactions containing 67 mM Tris-HCl (pH 8.5), 1 mM ATP, 5 mM MgCl2, and 5 mM DTT, except for initial experiments to test for PfLigI activity in which the buffer pH was 7.5. PfLigI and nicked substrate were added at the indicated amounts (see figure legends) and the reactions were incubated at 37°C for 30 min. Reactions were stopped with an equal volume of loading buffer, 90% formamide, 0.025% bromphenol blue, 0.025% xylene cylanol and 2.6 mM Tris pH 8.0. Samples were resolved on 20% polyacrylamide, 7 M urea denaturing gels. Quantification of ligated products was achieved using a PhosphoImager™ and ImageQuant software. All assays were performed with a minimum of three replicates and error bars represent the standard error of the mean.
3. Results
3.1 PfLigI protein sequence has similarities to other eukaryotic DNA ligase I proteins
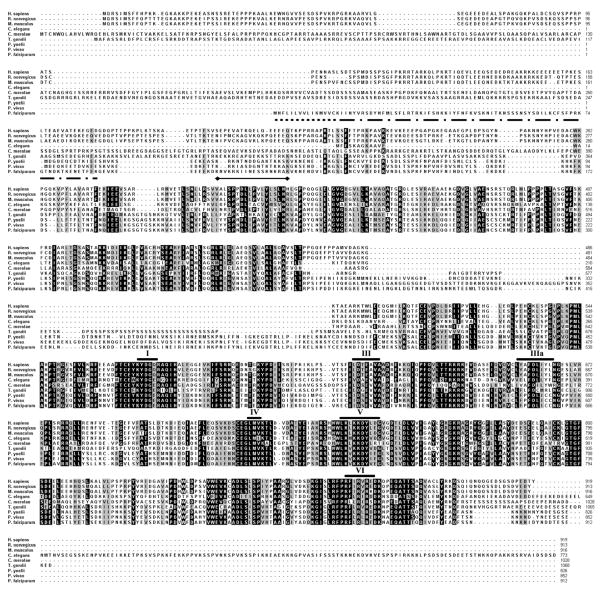
BLAST searches of the P. falciparum genome using an array of known ATP-dependent and NAD-dependent DNA ligases from viral, prokaryotic and eukaryotic organisms, yielded a single ligase gene on chromosome 13 of 2.7 kb. Translating the PfLigI ORF generated a protein with a predicted molecular weight of 104 kDa. Clustal alignment of DNA ligase I protein sequences from an array of eukaryotic species was performed, revealing a 30% overall sequence identity to human DNA ligase I and 60% identity in the C-terminal region (Fig. 1). Curiously missing from the N-terminus of PfLigI is a PCNA binding domain, which is replaced by a putative apicoplast targeting signal (Fig. 1, dashed line). A series of conserved motifs was identified previously in sequence alignments of mRNA capping enzymes and ATP-dependent ligases and constitutes a superfamily of nucleotidyl transferases. Residues from motifs I-V are found within the DNA ligase adenylation domain (AdD) and constitute the nucleotide binding pocket, which is highly conserved [10]. Motif II, which is comprised of arginine, phenylalanine and proline amino acid repeats and found in many capping enzymes of this superfamily, is absent in DNA ligases. The conserved motif VI resides within the oligo-binding domain (OBD) and together with motifs I–V comprises the “catalytic-core” of DNA ligase molecules [10].
Figure 1.
Protein sequence alignments of DNA ligase I from various eukaryotic organisms. PfLigI contains all of the conserved domains common to ATP-dependent DNA ligases (labeled I – VI). Unique to PfLigI is a putative apicoplast signal sequence, consisting of a signal peptide ( ) and a transit peptide (
) and a transit peptide ( ). Juxtaposed to the transit peptide is a bipartite nuclear localization signal (
). Juxtaposed to the transit peptide is a bipartite nuclear localization signal ( ). Accession numbers for aligned proteins are as follows: H. sapiens NP_000225, R. norvegicus NP_001019439, M. musculus NP_034845, C. elegans NP_741625, C. merolae CMK235C, T. gondii 25.m01820, P yoelii PY01533, P. vivax PV122045 and P. falciparum MAL13P1.22
). Accession numbers for aligned proteins are as follows: H. sapiens NP_000225, R. norvegicus NP_001019439, M. musculus NP_034845, C. elegans NP_741625, C. merolae CMK235C, T. gondii 25.m01820, P yoelii PY01533, P. vivax PV122045 and P. falciparum MAL13P1.22
3.2 Identification of endogenous PfLigI
Western blot analysis of P. falciparum stage specific lysates using an anti-PfLigI antibody, revealed a single protein band present only in the schizont stage. Conversely, lysate from uninfected erythrocytes showed no cross-reactive bands, indicating a parasite specific protein. The identified protein had a relative migration on SDS-PAGE that was consistent with the predicted molecular weight for PfLigI (Fig. 2A). This data is consistent with the published microarray data for PfLigI (PlasmoDB gene ID # MAL13P1.22), which shows mRNA expression increasing predominately in late stage parasites [18,19].
Figure 2.

Identification, recombinant expression, purification and nick sealing activity of PfLigI. Endogenous expression of PfLigI (A) 25 μg of uninfected erythrocyte lysate (E) and stage specific parasite lysates (R, T and S for ring, trophozoite and schizont stages, respectively) were analyzed by Western blot using anti-PfLigI affinity purified antibody at 1:2500. 0.1 μg of PfLigI was used as a control (+). PfLigI-FLAG® was expressed in HEK-293 cells and purified using an anti-FLAG® affinity column. (B) upper panel; Coomassie stained, 10% SDS-PAGE gel of FLAG® column fractions. Lane 1, molecular weight standards; lanes 2–4, pre-column, flow-through and wash fractions, respectively. Lane 5, 3μg of FLAG® column protein eluate. Western blot of FLAG® column fractions (lower panel). Fractions are identical to the Coomassie gel with the exception that 0.5 μg of protein was loaded in lane 5. Anti-FLAG®-M2 monoclonal was used at a 1:5000 dilution and the Western blot was visualized using 3,3′-diaminobenzadine. (C) Western blot of FLAG® column fractions using anti-human ligase I antibody at 1:1000 dilution. Gel was loaded as described in B, upper panel. (D) Schematic representation of the nicked DNA substrate utilized for ligation activity assays. Star denotes the location of the 5′-32P radiolabel; OH represents the 3′-OH end of the nick and P represents the 5′-phosphate end of the nick. (E) Nick sealing activity of FLAG® purified PfLigI. Ligation reactions (20 μl) containing 67 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 5 mM DTT, 1mM ATP and 1 pmol of nicked substrate were incubated for 30 min at 37°C. Lane 1, no enzyme; Lanes 2 and 3, PfLigI (0.1 and 1 pmol, respectively) and Lane 4, 1 pmol T4 DNA ligase. Upper bands represent ligated 44-mer and lower bands represent unligated radiolabeled 26-mer. Samples were separated on a 20% denaturing polyacrylamide gel.
3.3 Expression and purification of PfLigI
Due to the high A–T content of the P. falciparum genome (>70%), recombinant protein expression from this parasite has proven difficult. Initial attempts at expression of wild-type PfLIGI in prokaryotic and eukaryotic systems were unsuccessful. It was for this reason that the PfLIGI gene sequence, minus the first 381 nucleotides was submitted to GENEART Inc. for codon optimization and DNA synthesis. We hypothesized that the N-terminal amino acids 1–127, which comprise the putative apicoplast targeting signal, may have caused cell expression problems due to an enrichment in basic (KRH), asparagine and lysine residues. We also surmised that since this region lacked any homology to other known DNA ligase I molecules, it would be dispensable for ligation activity. A mammalian codon set was utilized for the optimization process and the synthesized gene was imparted with 3′ BamHI and 5′ XhoI restriction sites. The PfLIGI gene was subcloned into a mammalian expression vector, pYM-v200flaghisN, under the control of a CMV promoter and was used for transfection of HEK 293 cells. The PfLigI protein expressed in transfected cells was purified using an anti-FLAG® affinity chromatography column. The purified recombinant protein was visualized by coomassie staining (Fig. 2B upper panel) and Western blot using an anti-FLAG® antibody (Fig. 2B lower panel). Purity of the affinity column eluate was greater than 95%. Since HEK 293 cells contain endogenous ligase I, column fractions were also probed with a human DNA ligase I antibody (IMGENEX, Ca.), generated towards the N-terminal, non-homologous region of the protein. Endogenous ligase I protein was observed only in the flow through fractions of the affinity column purification and not with the purified PfLigI fraction (Fig. 2C).
3.4 Purified PfLigI has nick sealing activity
A double stranded DNA oligonucleotide containing a single nick in the phosphodiester backbone was generated to determine the activity of the purified PfLigI. The 26mer oligo upstream of the nick was radiolabeled on the 5′ end with 32P and the nicked substrate was assembled with the 18mer downstream of the nick, which contained a 5′-phosphate, and the complimentary template 44-mer oligo (Fig. 2D). 1 pmol of nicked substrate was mixed with 0.1 or 1 pmol of purified PfLigI in T4 DNA ligase buffer (67 mM Tris pH 7.5, 5 mM MgCl2, 1mM ATP, and 5 mM DTT) and incubated at 37°C for 30 min. 1 pmol of T4 DNA ligase was used as a control for the nick sealing reaction. Nick sealing was successful when a radiolabeled band was observed to migrate at the same position as the T4 DNA ligase control lane and higher than the zero enzyme control lane (Fig. 2E). Under these conditions 1 pmol of PfLigI was observed to generate similar levels of ligated product as the T4 control.
3.5 Establishing optimal conditions for PfLigI activity
Initial experiments of PfLigI nick sealing activity utilized buffer conditions that had been optimized for T4 DNA ligase. Determining the ideal conditions for phosphodiester bond formation was an essential first step in the biochemical analysis of PfLigI.
The optimal temperature for ligation was determined to be 37°C (Fig. 3A). Lowering the temperature to 25°C showed an approximate 50% reduction in product formation. Moreover, increasing the temperature to 40°C, a temperature often associated with malaria fevers, revealed a similar reduction in nick sealing activity. PfLigI was observed to have maximal activity at pH 8.5 (Fig. 3B). A sharp decline in activity was noticed as the pH of the reaction was increased. Furthermore, at lower pH values the enzyme maintained a steady level of activity, approximately half of the observed nick sealing activity at pH 8.5. PfLigI showed extreme sensitivity to monovalent salts (Fig. 3C). Increasing the concentration of NaCl to ≥20 mM led to a greater than 60% reduction in product formation. Similar inhibition was observed for KCl at 30 mM. Standard reactions contained no more than 1 mM NaCl, which was contributed from the annealing of the DNA substrate. Extreme sensitivity for monovalent ions has been reported previously for yeast and archeal ligases [20,21].
Figure 3.

PfLigI activity optimization. To determine the optimal conditions for PfLigI activity, the enzyme was assayed for nick sealing activity under various conditions. A to C used 200 fmol of nicked substrate and 40 fmol of PfLigI. Graphs represent the percent ligation of substrate in 30 min. Error bars on all graphs represents the standard error of the mean of at least 3 separate experiments. (A) Temperature dependence of PfLigI activity and (B) pH dependence of PfLigI activity (C) Effect of monovalent salts on PfLigI activity. Standard reaction conditions, minus salt, were normalized to 100% and all other values were calculated as a percentage of the zero salt sample. (D) Nucleotide cofactor specificity. Ligation reactions (20 μl) containing 67 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 5 mM DTT, 80 fmol DNA ligase, 400 fmol of substrate and 1 mM NTP or dNTP as indicated were incubated for 30 min at 37°C. (E) Divalent cation dependence of PfLigI. Nick sealing reactions (20 μl) containing 67 mM Tris-HCl (pH 8.5), 5 mM DTT, 1 mM ATP (Na salt), 200 fmol nicked substrate and 40 fmol of PfLigI were incubated for 30 min at 37°C using increasing concentrations of Mg2+, Mn2+ or Ca2+.Ligation activity is plotted as a function of ion concentration. (F) PfLigI concentration dependence. Ligation reactions containing 67 mM Tris- HCl (pH 8.5), 5 mM MgCl2, 5 mM DTT, 1 pmol of nicked DNA substrate and the indicated amount of PfLigI were incubated at 37°C. 20 μl aliquots were removed from the reaction at the indicated time points and were quenched with an equal volume of denaturing gel loading buffer (90% formamide, 5% bromophenol blue and 5% xylene cyanol). Ligation is plotted as a function of time.
3.6 Nucleotide specificity
In addition to divalent metal ions, ligation reactions also require a nucleotide cofactor. We observed that PfLigI had the greatest affinity for ATP in nick sealing reactions (Fig. 3D). The absence of ATP produced a low level of activity, which we attributed to pre-adenylated PfLigI obtained during purification of the enzyme. Ligation activity was slightly stimulated by the addition of 1 mM dATP, but no activity was observed using the same concentration of NAD, UTP, CTP or GTP (Fig. 3D).
3.7 Divalent cation specificity of PfLigI
Ligation of oligonucleotides requires divalent cations. We observed that phosphodiester bond formation was achieved in the presence of magnesium, manganese and to a modest extent, calcium. Ligation activity was not observed with cobalt, copper, nickel or zinc at either 1 mM or 10 mM concentrations (data not shown). The sodium salt of ATP was employed for these experiments, so as not to impart any divalent cations with the exception of those externally added. Concentration curves were generated for magnesium, manganese and calcium (Fig. 3E). Magnesium and manganese were able to serve as cofactors for DNA ligation at approximately the same maximum level, albeit at different concentrations. Manganese was observed to aid ligation maximally at 0.6 mM, whereas the highest level of ligation activity for magnesium was at an approximately 10-fold higher concentration. It has been reported previously that the addition of calcium into ligation reactions resulted in the accumulation of DNA-adenylate intermediates [21,22]. Furthermore, a NAD+ dependent ligase was able to fully seal DNA nicks using calcium as a cofactor [23]. PfLigI was also able to utilize calcium as a cofactor during DNA ligation, but no accumulation of DNA-adenylate intermediates was observed (data not shown). To our knowledge, PfLigI is the only ATP-dependent DNA ligase that can utilize calcium as a cofactor for the catalysis of phosphodiester bonds.
3.8 Substrate kinetics
A concentration response curve of PfLigI nick sealing activity was generated to show the proportionality of enzyme concentration to nicked substrate (Fig. 3F). Incubation of an equimolar ratio of enzyme and substrate (1 pmol) caused rapid product formation and quick saturation (Fig. 3F, triangles). Decreasing the enzyme/substrate ratio to 0.2 or 0.04 produced slower product formation (Fig. 3F, open and closed circles). Steady-state kinetic constants were determined by measuring the formation of radiolabeled product using increasing substrate concentrations (0.125 – 75 nM) with a fixed quantity of enzyme (5 fmol). Kinetic values were calculated by a direct fit of the data into the Michaelis-Menten equation. A Km of 2.6 nM nicked substrate was calculated for PfLigI. Moreover, a kcat of 2.3×10−3s−1 and a kcat/Km of 8.8×105 M−1s−1 were derived from the steady-state data (Table 1). Kinetic values are similar to those reported for human DNA ligase I, as well as other ATP dependent DNA ligases, but with a lower turnover rate and catalytic efficiency than T4 DNA ligase [24–26]. Increasing concentrations of ATP showed a rise in PfLigI activity which plateaued at about 3 μM. A Km of 1.3 μM ATP was calculated by a direct fit of the data into the Michaelis-Menten equation and the values listed (Table 1).
Table 1.
Kinetic properties of PfLigI and comparison to other DNA ligases. (A) Nicked Substrate Kinetics for PfLigI: 20 μl reactions containing 67 mM Tris-HCl (pH 8.5), 1 mM ATP, 5 mM MgCl2 5 mM DTT, 5 fmol PfLigI and increasing amounts of nicked DNA substrate (0.125 – 75 nM) were incubated at 37°C for 10 min. (B) Ligation kinetics for human DNA ligase I and the ATP-dependent ligases from N. meningitidis and Bacteriophage T4. Superscripts a, b and c refer to references 24, 25 and 26, respectively. (C) ATP Affinity of PfLigI: Buffer conditions were as described for A. Reactions were performed with increasing amounts of ATP, 1 pmol of nicked substrate, 100 fmol of PfLigI and incubated at 37°C for 10 min. Results for PfLigI represent an average of at least three experiments. Kinetic values for PfLigI were calculated by a direct fit of the data into the Michaelis-Menten equation (SigmaPlot 9.0).
| Ligation Kinetics | ||||
|---|---|---|---|---|
| A. | B. | |||
| P.falciparum | H. sapiens a | N. meningitidis b | Bacteriophage T4 c | |
| Km (M) | 2.6 × 10−9 | 3.4 × 10−9 | 2.9 × 10−8 | 1.6 × 10−7 |
| Vmax (Ms−1) | 5.8 × 10−13 | -- | -- | -- |
| kcat (s−1) | 2.3 × 10−3 | 2.6 × 10−3 | 8 × 10−3 | 4.9 |
| kcat/Km (M−1s−1) | 8.8 × 105 | 7.6 × 105 | 2.7 × 105 | 3.06 × 10 7 |
| PfLigI ATP Affinity | ||||
| C. | ||||
| Km (M) | 1.3 × 10−6 | |||
3.9 Substrate specificity
Being that PfLigI may be the sole DNA ligase present in the P. falciparum genome, we wanted to ascertain the enzyme’s ability to distinguish RNA-DNA oligo pairs. It is well established that DNA ligase I from bovine and humans can ligate RNA-DNA oligos if the RNA is upstream of the nick [27]. Any RNA, even a single base, downstream of the nick results in a dramatic decrease in ligation activity [28]. Ligase III, which is involved in the DNA short-patch repair pathway, is much more promiscuous and will ligate RNA-DNA oligos regardless of their position with respect to the nick. We observed that PfLigI has nick sealing activity on substrates that are consistent with DNA ligase I homologue activity (Fig. 4). Specifically, PfLigI is only capable of catalyzing phosphodiester bonds on the substrate which contains RNA upstream of the nick (Fig. 4, lane 2). Conversely, PfLigI, as with other LigI homologues, is unable to ligate molecules containing RNA downstream of the nick (Fig. 4 lanes 5 and 8). Other ATP dependent ligases, such as T4 DNA ligase have the capacity to ligate RNA-DNA oligos more indiscriminately (Fig. 4 lanes 3, 6 and 9).
Figure 4.
Substrate specificity of PfLigI. Substrates were labeled and annealed as described in the experimental procedures. Nick sealing reactions (20 μl) containing 67 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 5 mM DTT, 1 mM ATP, 1 pmol of indicated substrate and 20 pmol of PfLigI of T4 DNA ligase were incubated at 37°C for 30 min. Samples were separated on a 20% denaturing polyacrylamide gel. Lanes 1, 4 and 7 contain no enzyme. Lanes 2, 5 and 8 contain 20 pmol PfLigI and lanes 3, 6 and 9 contain 20 pmol T4 DNA ligase. Star denotes the location of the 5′-32P radiolabel; OH represents the 3′-OH end of the nick and P represents the 5′-phosphate end of the nick.
4. Discussion
We report here the identification, expression and characterization of DNA ligase I from P. falciparum. To our knowledge this is the first study of a DNA ligase from an apicomplexan parasite. The alignment in Figure 1 shows that PfLigI contains high homology to DNA ligase I proteins, particularly in the C-terminal “catalytic-core” region. Conversely, the N-terminus of ATP-dependent DNA ligases is more variable, which is evident by the lack of homology within the alignment across species. This region often contains intracellular targeting signals as well as interaction motifs for binding to PCNA and Pol β; however it is dispensable for ligation activity. Further downstream within the N-terminus is the DNA binding domain (DBD), which imparts most of the binding affinity for LigI and is required for efficient ligation [28].
Our data show that the production of codon optimized PfLigI in a mammalian expression system yields large quantities of the recombinant protein. We typically obtain 1–2 mg of FLAG® affinity-purified protein from 8 × 150 cm2 culture flasks of HEK cells growing in a monolayer. We determined that purified PfLigI was able to catalyze phosphodiester bond formation on nicked DNA substrates and observed that product formation was similar when compared to equimolar T4 DNA ligase (Fig. 2E, lanes 3 and 4).
The results from our experiments depicted in Figure 3 establish that PfLigI is an ATP-dependent DNA Ligase that requires a divalent metal cofactor. While magnesium is the divalent cation most often associated with ligation activity, it is worth noting that PfLigI was able to catalyze nick sealing, albeit modestly, in the presence of Ca 2+ (Figure 3E, triangles). Since Ca2+ has a larger ionic radius than Mg2+ or Mn2+ this suggests more flexibility within the “catalytic-core” of PfLigI than what has been observed for other ATP-dependent DNA ligases. Kinetically, PfLigI possesses an affinity for nicked substrates that closely resembles what has been reported for human DNA ligase I [24]. Specifically, the data listed in Table 1 shows that the turnover rate (kcat) and catalytic efficiency (kcat/Km) for PfLigI is similar to human DNA ligase I and comparable to the ATP- dependent DNA ligase from N. meningitidis [25]. Conversely, T4 DNA ligase has a greater turnover rate and catalytic efficiency then the DNA ligase homologues, but a reduced affinity for its substrate, which is reflected in its higher Km [26]. The high turnover rate and low substrate affinity of T4 ligase are indicative of its reduced fidelity [29]. This is contrary to what has been observed for human DNA Ligase I, which shows strong discrimination between correctly and mispaired bases at the nick [30]. The high homology and similar kinetic properties to human DNA ligase would suggest that PfLigI is also a high fidelity ligase.
The results from the substrate specificity experiment (Fig. 4) further show that PfLigI has activity and fidelity consistent with other DNA ligase I homologues. It was essential to determine if PfLigI had any alternative DNA ligase activity, e.g., ligase III. Since there was no activity on substrates with RNA downstream of the nick, this indicates that PfLigI does not possess an additional DNA LigIII activity. Further investigation is needed to determine if P. falciparum has an unusual, unidentified backup enzyme with DNA ligase activity or if its DNA ligase activity, essential for replication and repair, is entirely dependent on PfLigI.
The relationship of DNA ligase I with other repair/replication proteins is an integral part of understanding this enzyme’s activity in vivo. There are a number of reports concerning the interaction and stimulation of DNA ligase I by PCNA [17,31–33]. It is of interest to note that PfLigI is lacking a PCNA binding domain. Furthermore, BLAST searches of the OrthoMCL database [34] using PfLigI as the query showed that none of the seven Apicomplexa parasite DNA ligase I homologues analyzed contained a PCNA binding domain. Additionally, there has been a recent report of a novel PCNA binding domain in the archeal bacteria Pyrococccus furiosus [35], but examination of the PfLigI protein sequence reveals that it does not contain that motif either. It is conceivable that PfLigI contains a novel PCNA binding domain unique to the apicomplexa phylum. Moreover, a homologue of PCNA, PCNA2, has been identified in Plasmodium [36], which may interact with PfLigI during replication and repair. Finally, it is possible, given P. falciparum’s unusual DNA base excision repair and streamlined repair protein genome that PfLigI does not interact with PCNA or PCNA2, during replication or repair. If so, P. falciparum would be the only known organism which utilizes a PCNA-independent long-patch DNA base excision
Elucidating the inherent mechanisms of malarial molecular biology is critical to suppressing the ever escalating drug resistance of this parasite. While the focus of the drug resistance paradigm should be centered on the proteins involved in maintaining genomic fidelity, these pathways have all but been ignored in parasite biology. Although PfLigI may not be a suitable target for chemotherapy due to its high homology to host DNA ligase I, the expression and characterization of this protein represents a critical step towards understanding the molecular trappings underlying Plasmodium genomic maintenance.
Acknowledgments
Funding for this research is provided by NIH grant R01 AI48567.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization - World Malaria Report. Available at http://rbm.who.int/wmr2005.
- 2.Gardner MJ, Hall N, Fung E, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. 1995 [Google Scholar]
- 4.Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992;12:1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dianov G, Bischoff C, Piotrowski J, Bohr VA. Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J Biol Chem. 1998;273:33811–33816. doi: 10.1074/jbc.273.50.33811. [DOI] [PubMed] [Google Scholar]
- 6.Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J Biol Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- 7.Dianov GL, Thybo T, Dianova II, Lipinski LJ, Bohr VA. Single nucleotide patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J Biol Chem. 2000;275:11809–11813. doi: 10.1074/jbc.275.16.11809. [DOI] [PubMed] [Google Scholar]
- 8.Haltiwanger BM, Matsumoto Y, Nicolas E, et al. DNA base excision repair in human malaria parasites is predominantly by a long-patch pathway. Biochemistry. 2000;39:763–772. doi: 10.1021/bi9923151. [DOI] [PubMed] [Google Scholar]
- 9.Martin IV, MacNeill SA. ATP-dependent DNA ligases. Genome Biol. 2002;3:REVIEWS 3005.1–3005.7. doi: 10.1186/gb-2002-3-4-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timson DJ, Singleton MR, Wigley DB. DNA ligases in the repair and replication of DNA. Mutat Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 11.Johnson A, O’Donnell M. DNA ligase: getting a grip to seal the deal. Curr Biol. 2005;15:R90–R92. doi: 10.1016/j.cub.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 12.Bentley DJ, Harrison C, Ketchen AM, et al. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J Cell Sci. 2002;115:1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian J, Vijayakumar S, Tomkinson AE, Arnheim N. Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics. 2005;171:427–441. doi: 10.1534/genetics.105.042861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun D, Urrabaz R, Nguyen M, et al. Elevated expression of DNA ligase I in human cancers. Clin Cancer Res. 2001;7:4143–4148. [PubMed] [Google Scholar]
- 15.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 16.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 17.Tom S, Henricksen LA, Park MS, Bambara RA. DNA ligase I and proliferating cell nuclear antigen form a functional complex. J Biol Chem. 2001;276:24817–24825. doi: 10.1074/jbc.M101673200. [DOI] [PubMed] [Google Scholar]
- 18.Bozdech Z, Llinas M, Pulliam BL, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Roch KG, Zhou Y, Blair PL, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 20.Tomkinson AE, Tappe NJ, Friedberg EC. DNA ligase I from Saccharomyces cerevisiae: physical and biochemical characterization of the CDC9 gene product. Biochemistry. 1992;31:11762–11771. doi: 10.1021/bi00162a013. [DOI] [PubMed] [Google Scholar]
- 21.Sriskanda V, Kelman Z, Hurwitz J, Shuman S. Characterization of an ATP-dependent DNA ligase from the thermophilic archaeon Methanobacterium thermoautotrophicum. Nucleic Acids Res. 2000;28:2221–2228. doi: 10.1093/nar/28.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong J, Barany F, Cao W. Ligation reaction specificities of an NAD(+)-dependent DNA ligase from the hyperthermophile Aquifex aeolicus. Nucleic Acids Res. 2000;28:1447–1454. doi: 10.1093/nar/28.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong J, Cao W, Barany F. Biochemical properties of a high fidelity DNA ligase from Thermus species AK16D. Nucleic Acids Res. 1999;27:788–794. doi: 10.1093/nar/27.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranalli TA, DeMott MS, Bambara RA. Mechanism Underlying Replication Protein A Stimulation of DNA Ligase I. J Biol Chem. 2002;277:1719–1727. doi: 10.1074/jbc.M109053200. [DOI] [PubMed] [Google Scholar]
- 25.Magnet S, Blanchard JS. Mechanistic and kinetic study of the ATP-dependent DNA ligase of Neisseria meningitidis. Biochemistry. 2004;43:710–717. doi: 10.1021/bi0355387. [DOI] [PubMed] [Google Scholar]
- 26.Wu DY, Wallace RB. Specificity of the nick-closing activity of bacteriophage T4 DNA ligase. Gene. 1989;76:245–254. doi: 10.1016/0378-1119(89)90165-0. [DOI] [PubMed] [Google Scholar]
- 27.Tomkinson AE, Roberts E, Daly G, Totty NF, Lindahl T. Three distinct DNA ligases in mammalian cells. J Biol Chem. 1991;266:21728–21735. [PubMed] [Google Scholar]
- 28.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 29.Goffin C, Bailly V, Verly WG. Nicks 3′ or 5′ to AP sites or to mispaired bases, and one-nucleotide gaps can be sealed by T4 DNA ligase. Nucl Acids Res. 1987;15:8755–8771. doi: 10.1093/nar/15.21.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhagwat AS, Sanderson RJ, Lindahl T. Delayed DNA joining at 3′ mismatches by human DNA ligases. Nucl Acids Res. 1999;27:4028–4033. doi: 10.1093/nar/27.20.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardoso MC, Joseph C, Rahn HP, et al. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J Cell Biol. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin DS, Bai W, Yao N, O’Donnell M, Tomkinson AE. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci U S A. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Refsland EW, Livingston DM. Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast. Genetics. 2005;171:923–934. doi: 10.1534/genetics.105.043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiyonari S, Takayama K, Nishida H, Ishino Y. Identification of a novel binding motif in pyrococcus furiosus DNA ligase for the functional interaction with proliferating cell nuclear antigen. J Biol Chem. 2006 doi: 10.1074/jbc.M603403200. [DOI] [PubMed] [Google Scholar]
- 36.Patterson S, Whittle C, Robert C, Chakrabarti D. Molecular characterization and expression of an alternate proliferating cell nuclear antigen homologue, PfPCNA2, in Plasmodium falciparum. Biochem Biophys Res Commun. 2002;298:371–376. doi: 10.1016/s0006-291x(02)02436-1. [DOI] [PubMed] [Google Scholar]