Abstract
Activating BRAF kinase mutations arise in about 7% of all human tumors, and pre-clinical studies have validated the RAF-MEK-ERK signaling cascade as a potentially important therapeutic target in this setting. Selective RAF kinase inhibitors are currently undergoing clinical development, and based on the experience with other kinase-targeted therapeutics, it is expected that clinical responses to these agents, if observed, will lead to the eventual emergence of drug resistance in most cases. Thus, it is important to establish molecular mechanisms underlying such resistance in order to develop effective therapeutic strategies to overcome or prevent drug resistance. To anticipate potential mechanisms of acquired resistance to RAF inhibitors during the course of treatment, we established drug-resistant clones from a human melanoma-derived cell line harboring the recurrent V600E activating BRAF mutation, which exhibits exquisite sensitivity to AZ628, a selective RAF kinase inhibitor. We determined that elevated CRAF protein levels account for the acquisition of resistance to AZ628 in these cells, associated with a switch from BRAF to CRAF dependency in tumor cells. We also found that elevated CRAF protein levels may similarly contribute to primary insensitivity to RAF inhibition in a subset of BRAF mutant tumor cells. Interestingly, AZ628-resistant cells demonstrating either primary drug insensitivity or acquired drug resistance exhibit exquisite sensitivity to the HSP90 inhibitor geldanamycin. Geldanamycin effectively promotes the degradation of CRAF, thereby revealing a potential therapeutic strategy to overcome resistance to RAF inhibition in a subset of BRAF-mutant tumors.
INTRODUCTION
Genetic alterations that contribute to tumorigenesis can give rise to proteins that are essential for maintaining the enhanced growth and survival properties of tumor cells. Such “addiction” to individual oncogenic proteins appears to explain the exquisite clinical sensitivity of some tumors to various molecularly-targeted kinase inhibitors (1). Thus, imatinib is highly effective in chronic myelogenous leukemia (CML) cells that harbor the BCR-ABL translocation and gastrointestinal stromal tumors (GIST) with activating c-KIT or PDGF receptor mutations (2). Similarly, most non-small cell lung cancers (NSCLCs) harboring an activating EGFR kinase domain mutation are sensitive to the selective EGFR tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib (3-5).
As a result of cancer genome re-sequencing efforts, activating somatic mutations in BRAF have been identified in 60% of melanomas, 40% of thyroid cancers and 20% of colon cancers (6). The most common BRAF mutation leads to a substitution of glutamic acid for valine at position 600 (V600E) within the activation segment of the BRAF kinase domain, which results in elevated kinase activity and stimulation of downstream MEK-ERK signaling, consequently promoting tumor cell survival and proliferation (6-8). Therefore, inhibition of the BRAF pathway is considered to be a promising strategy for treating melanoma and other BRAF mutant cancers, and several selective kinase inhibitors that target the BRAF-MEK-ERK pathway are currently being developed (9, 10). In pre-clinical studies, inhibition of the MEK kinase effectively and specifically inhibits the growth of human tumor cell lines harboring activating BRAF mutations (9). Similarly, in a high-throughput tumor cell line profiling study, we have recently reported that AZ628, a selective and potent investigational small molecule RAF kinase inhibitor, is remarkably effective at inhibiting the growth of a specific subset of human cancer cell lines derived from melanomas, thyroid cancers, and colorectal cancers that harbor the BRAF V600E mutation (11).
While various targeted kinase inhibitors have demonstrated both pre-clinical and clinical activity, the application of these agents to large patient populations has clearly demonstrated that while initial clinical responses can be dramatic, rapid acquisition of drug resistance is a major limitation to the overall therapeutic efficacy of these drugs. Therefore, one of the major challenges associated with the broader use of these inhibitors is the elucidation of drug resistance mechanisms and the development of strategies to overcome or prevent resistance.
In CML, GIST, and NSCLC, acquired resistance to kinase inhibitors is frequently associated with either secondary kinase domain mutations, amplification of the gene encoding the target kinase, or mutational activation of genes encoding components of alternative survival pathways (12-18). Notably, each of these identified resistance mechanisms has been succesfully modeled in cell culture using appropriate drug-treated cancer cell lines, indicating that such cell culture modeling can provide an effective system for identifying mechanisms of acquired drug resistance that are likely to arise clinically (16, 19, 20). This is important because the development of strategies to overcome drug resistance, which will generally require considerable time, first requires the identification of relevant resistance mechanisms. Therefore, the ability to anticipate clinical mechanisms of acquired resistance to targeted kinase inhibitors is likely to greatly accelerate the development of strategies to overcome or prevent acquired drug resistance (21), and to reduce the current temporal gap between initial clinical successes and subsequent disease progression in the absence of available secondary treatment options.
Selective inhibitors of the RAF and MEK kinases are currently undergoing early Phase clinical testing (22-24). To anticipate potential mechanisms of acquired resistance to RAF inhibitors that could arise during the course of treatment, we established drug-resistant clones from a human melanoma-derived cell line that harbors the V600E activating BRAF mutation, and exhibits exquisite sensitivity to AZ628, a selective RAF kinase inhibitor. In a subset of these clones, significantly increased expression of the BRAF-related CRAF protein appears to account for the acquisition of resistance to AZ628. Interestingly, the resistant clones, which have shifted their dependency from BRAF to CRAF, acquire substantial sensitivity to the HSP90 inhibitor geldanamycin. Geldanamycin effectively promotes the degradation of CRAF, thereby revealing a potential therapeutic strategy to overcome this resistance mechanism.
MATERIALS AND METHODS
Cell culture and reagents
The human melanoma cell line M14 expressing the V600E BRAF mutation was kindly provided by Daphne Bell. M14 is a cell line from the NCI-60 cell line panel that has been extensively characterized (25). Cells were maintained at 37°C in a humidified atmposphere at 5% CO2 grown in RPMI 1640 (Cellgro; Mediatech Inc., Herndon, CA) supplemented with 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin and 2nM glutamine. AZ628 resistant M14 clones were maintained in the above-mentioned medium and 2 μM of AZ628, except where otherwise indicated. The BRAF inhibitor AZ628 was synthesized by Astra Zeneca (Waltham, MA). It demonstrates IC50 values of approximately 30 nM for BRAF V600E and wild-type CRAF, and 100 nM for wild-type BRAF, and strong selectivity for RAF kinases among a panel of 150 tested kinases (L.D., unpublished observations). Additional details regarding the structure and properties of AZ628 will be reported separately. The MEK inhibitor, U0126, was purchased from Promega (Madison, WI). Geldanamycin was acquired from Biomol International (Plymouth, PA). The additional inhibitors were either obtained from the MGH pharmacy or were synthesized at the Dana Farber Cancer Institute, based on published structures.
Cellular Proliferation Assay
Approximately 50,000 or 25,0000 cells were seeded in 12 or 24 - well plates, respectively, in medium supplemented with 5% FBS. After overnight incubation, the cells were treated with various concentrations of each drug. Fresh medium and drug was replaced every 2 days until the untreated control wells reached confluence. At this time-point, the media was removed and the cells were fixed in 4% formaldehyde in PBS (Boston Bioproducts, Worcester, MA) for 20 minutes at room temperature. Cells were then washed twice with PBS and stained with a 1:5000 solution of the fluorescent nucleic acid stain Syto60 (Molecular Probes, Carlsbad, CA). Quantitation of fluorescent signal intensity was carried out at 700nm, using an Odyssey Infrared Imager (Li-Cor Biosciences). Each experiment was performed in quadruplicate and the results shown represent the average of the four values compared to untreated wells. Error bars represent standard deviation of the 4 values from the mean. High-throughput cell growth / viability assays were performed as previously reported (11).
Protein detection
To collect protein lysates, cells were washed with PBS, scraped in lysis buffer (150mM NaCl, 1% NP-40, 50mM Tris, 2mM EDTA, 10% glycerol, 5 μg/ml each of aprotinin, leupeptine and pepstatin, and 1mM each of NaF, Na3VO4 and PMSF) and incubated on ice for 40 min. The lysates were centrifuged at 14,000 RPM for 20 min and the supernatant collected. Protein concentration was measured with the bicinchoninic acid protein assay (Pierce, Rockford, IL) and proteins were resolved by SDS-PAGE. The gels were electroblotted onto PVDF membranes (Hybond-P, Amersham). Antibody detection was performed with a chemiluminescence kit (Supersignal, Pierce). The ERK1/2, phospho-ERK1/2(T202/Y204), MEK, and ARAF antibodies were from Cell Signaling Technology (Beverly, MA). The CRAF and BRAF antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). All antibodies were used at a 1:1,000 dilution.
Generation of AZ628-resistant M14 clones
Approximately 105 M14 cells were plated in each of three 10cm dishes. After overnight incubation, the medium was removed and fresh medium was added, together with AZ628 at 2 μM. Fresh medium containing drug was added to the cells every three days. Five weeks after drug selection, approximately 10 clones per dish appeared. Two to three clones per dish were isolated using cloning cylinders and propagated progressively in 3, 6, and 10 cm dishes, maintained in 2 μM AZ628. All of the clones were confirmed to be resistant to AZ628 in a follow-up cell proliferation assay with Syto60 staining and quantitation.
DNA sequencing
Genomic DNA was isolated from the M14 parental cell line and AZ628-resistant clones using the Gentra purification system according to the manufacturer's protocol. BRAF was amplified from genomic DNA by PCR. PCR products were purified using exonuclease I and shrimp alkaline phosphatase (United States Biochemical, Cleveland, OH) followed by bidirectional sequencing using BigDye v1.1 (Applied Biosystems, Foster City, CA) in combination with an ABI3100 sequencer (Applied Biosystems). Primers used for sequencing of BRAF are listed in Supplemental Table 1. Electropherograms were analyzed using Sequence Navigator software (Applied Biosystems). All mutations were confirmed by at least two independent PCR amplifications.
Fluorescence in Situ Hybridization
Two-color fluorescence in situ hybridization (FISH) was performed on 3:1 methanolacetic acid fixed cell lines using the following probes, according to the manufacturer's protocols: SpectrumOrange labeled CRAF (BAC clones RP11-148M13 and CTD-2163C15, 3p25.1) and BRAF (BAC clones RP11-1065D4 and CTD-2516J12, 7q34), a SectrumGreen labeled control probe for CRAF (BAC RP11-36I6, 3q13.2) and a SpectrumAqua labeled centromeric probe for chromosome 7 (Vysis, cat no. 32-131007) that serves as a control probe for BRAF. Images were captured using an Olympus BX61 fluorescent microscope equipped with a CCD camera, and analysis was performed with Cytovision software (Applied Imaging, San Jose, CA).
shRNA constructs and lentiviral infection
Two shRNA species targeting sequences for BRAF and two for CRAF were expressed from the pLKO.1 lentiviral vector (BRAF target sequences: 5′GCAGATGAAGATCATCGAAAT3′ and 5′CAGCAGTTACAAGCCTTCAAA3′. CRAF target sequences: 5′CATGAGTATTTAGAGGAAGTA3′ and 5′GCTTCCTTATTCTCACATCAA3′). Cells were inoculated in 96-well plates and 6 cm dishes and incubated for 16 hours until cells reach about 80% confluency. Cells were then infected with shRNA lentiviruses or control vector (PLKO.1) in the presence of polybrene 8 μg/ml (hexadimethrine bromide, Sigma-Aldrich) under 1,200 × g gravity at 32 °C for 60 minutes. Under this condition, approximately 5 M.O.I. of infection efficiency was achieved. After infection cells were maintained in the presence of 2μg/ml of puromycin (Invitrogen, Carlsbad, CA) for an additional 4-6 days. A cell line resistant to pharmacologic RAF inhibition (A549) was used to demonstrate infection efficiency and specificity.
Q-PCR
Total RNA was isolated from M14 and M14BRR2 cells using RNeasy kit (QIAGEN). One-step reverse transcription and real-time PCR was performed using FastLane Cell kit (QIAGEN). The amount of CRAF amplicons was determined using the AB7500 qPCR system (Applied Biosystems) with SYBR Green I as the fluorescence reporter dye and ROX as the passive reference dye. The amount of GAPDH amplicons was determined similarly using QuantiTect primers (QIAGEN). The qPCR thermal profile was 50 °C for 30 minutes for cDNA synthesis, 95 °C for 15 minutes for hot-start activation of the antibody-neutralized DNA polymerase, followed by 45 cycles of denaturation (94 °C, 15 sec), annealing (60 °C, 30 sec), and elongation/data acquisition (72 °C, 35 sec). After completion of the thermal cycling, PCR product melting curves were obtained using the standard protocol of the AB7500 system. The Ct values were determined using manual settings recommended by the manufacturer of the FastLane reagent.
All specimens were analyzed in triplicate. CRAF primers used are as follow: Forward #1: 5′GCACGGAGATGTTGCAGTAA3′; Reverse#1: 5′GCTACCAGCCTCTTCATTGC; Forward #2: 5′CTGTTTCCAGGATGCCTGTT; Reverse#2: 5′GCTACTGGACAGGGCTGAAG.
CRAF Expression Construct
The CRAF 22W cDNA coding sequence within a pBABE retrovirus plasmid was a kind gift from Channing Der (University of North Carolina, Chapel Hill) (26). Transfection of Phoenix Morpho cells with the retroviral vector was performed using the Fugene Transfection Reagent (Invitrogen, Carlsbad, CA). The collected retrovirus was used to infect M14 cells in the presence of polybrene 8 μg/ml under 1,200 × g gravity at 32 °C for 60 minutes. Cells were selected in puromycin 2μg/ml and resistant clones were used for analysis. M14 cells infected with an empty pBABE puro plasmid were used as control.
RESULTS
Generation of melanoma cell line clones with acquired resistance to the RAF kinase inhibitor AZ628
To identify potential mechanisms of acquired resistance to a selective RAF kinase inhibitor, we utilized the M14 human melanoma-derived cell line. These cells harbor the V600E BRAF mutation and are exquisitely sensitive to the potent and selective RAF inhibitor AZ628 (11). Five weeks after continuously exposing M14 cells to 2 μM AZ628, a concentration that rapidly promotes growth inhibition and cell death in the vast majority of treated cells, single cell-derived drug-resistant clones emerged at a frequency of approximately 1 in 104 cells. Six of these clones were isolated and expanded for further characterization.
Morphologically, the M14-derived AZ628 resistant (M14BRR) clones are flat and epithelial-like when compared to the parental M14 cell line (Supplementary Figure 1). Their growth properties are otherwise indistinguishable from the parental cells. A drug titration assay of cell viability demonstrated that AZ628-resistant clones are approximately 100-fold more resistant to AZ628 than the parental cell line, exhibiting an IC50 of approximately 10 μM, compared with 0.1 μM for the parental cell line (Figure 1A). Similar results were observed with an alternative RAF-selective inhibitor that is currently undergoing clinical development (data not shown). DNA sequence analysis of the AZ628-resistant clones excluded the presence of any secondary mutations in BRAF, a potential mechanism reported to contribute to acquired resistance to other kinase inhibitors in other tumor types (12-14). Furthermore, the presence of the V600E activating BRAF mutation in the resistant clones (not shown) confirmed that they had not arisen from a contaminating sub-population of cells.
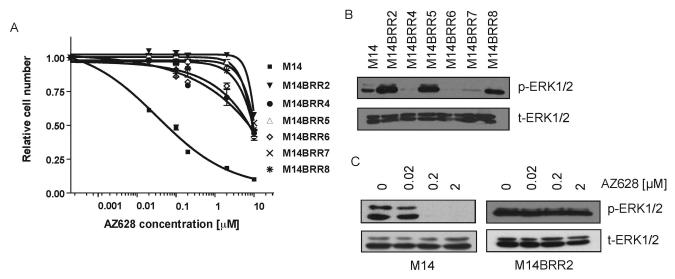
Figure 1. M14-derived AZ628-resistant (M14BRR) clones display elevated levels pERK1/2 and uncoupling of ERK signaling from BRAF.
(A) Dose-response curves of M14 and six M14BRR clones treated with the indicated concentrations of AZ628. The fraction of viable cells is expressed relative to untreated controls. Error bars represent the standard deviation from the mean.
(B) Immunoblots with the indicated antibodies demonstrating that M14BRR clones exhibit elevated basal activation of ERK1/2 compared to parental M14 cells. Cell lysates from M14 and six independently-generated M14BRR clones maintained in the presence of 2 μM AZ628 were collected.
(C) Immunoblots with the indicated antibodies demonstrating that M14BRR cells maintain ERK phosphorylation in the presence of AZ628. Cell lysates from M14 and AZ628-resistant clone (M14BRR2) were collected following 2 hour treatment with the indicated concentrations of AZ628.
Resistance to AZ628 is associated with elevated levels of the RAF downstream effector phospho-ERK1/2
To further investigate the mechanism underlying acquired AZ628 resistance in these cells, we performed immunoblotting studies of established RAF downstream effectors in the parental cell line and the resistant clones (Figure 1B). In three of the resistant clones (M14BRR2, 5 and 8) basal activation of the downstream effector ERK1/2 was significantly increased relative to levels seen in the parental cell line. Therefore, we hypothesized that ERK1/2 might play a pivotal role in the mechanism of acquired resistance to AZ628, in at least a subset of cases, and these three resistant clones were further characterized.
We next compared the effect of AZ628 treatment on RAF-dependent signaling on the M14 cells and drug-resistant derivatives. As we have previously reported, sensitivity to AZ628 is correlated with suppression of the downstream effector phospho-ERK1/2 following treatment of various melanoma-derived cell lines harboring BRAF activating mutations (11). Effective suppression of p-ERK1/2 levels was observed in the M14 parental cell line following treatment with increasing concentrations of AZ628. In contrast, p-ERK1/2 activity persisted at high levels in the resistant clones following AZ628 exposure, suggesting that sustained activation of ERK1/2 signaling may be critical to the maintenance of cell proliferation and survival in these cells, and may play a role in conferring resistance to AZ628 (Figure 1C and Figure 2B).
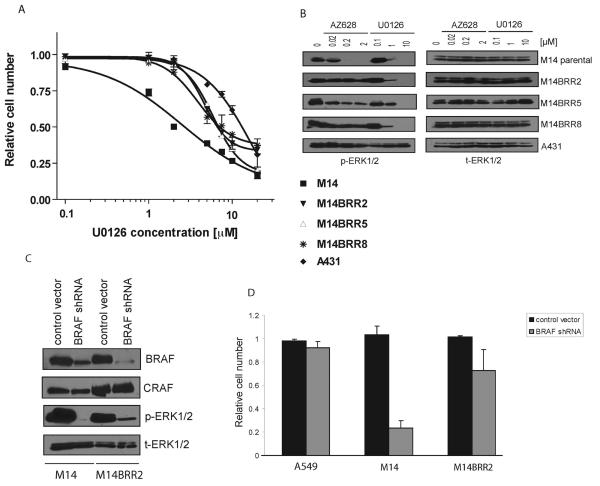
Figure 2. Proliferation of M14 AZ628-resistant (M14BRR) clones is dependent on MEK but not BRAF.
(A) Dose-response curves of M14 and three M14BRR clones treated with the indicated concentrations of the MEK inhibitor U0126. The percentage of viable cells is expressed relative to untreated controls. Error bars represent the standard deviation from the mean. The A431 cell line survival curve is shown as a negative control.
(B) AZ628-4resistant cells retain sensitivity to U0126. Cell lysates from M14, AZ628-resistant clones, and A431 (negative control) were collected following treatment with the indicated concentrations of AZ628 or U0126 for 2 hours. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
(C) Effective depletion of BRAF protein by shRNA. M14 and M14BRR2 cells were infected with lentivirus containing control (PLKO.1 empty vector) or BRAF-specific shRNA. Cells were puromycin-selected and protein lysates were collected 4 days after the infection. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
(D) Reduced dependency on BRAF in AZ628-resistant cells. Proliferation assay corresponding to cells in (C). Control or BRAF-specific BRAF shRNAs were introduced in A549, M14, and M14BRR2 cells by lentiviral infection and a cell proliferation assay with Syto60 was performed 7 days later. The fraction of viable cells is expressed relative to untreated control. Error bars represent the standard deviation from the mean.
ERK1/2 activation in AZ628-resistant clones is mediated by MEK
To determine whether activation of ERK1/2 in AZ628-resistant clones is mediated by the upstream ERK activator MEK, we assessed the sensitivity of the resistant clones to the selective MEK inhibitor, U0126 (Figure 2A). The IC50 of the resistant clones for U0126 ranged from 3 μM to 7 μM, and was very similar to that of the M14 parental cell line (2 μM). Biochemical analysis revealed that sensitivity to U0126 was consistent with the suppression of signaling to the downstream effector ERK1/2 in both the M14 parental cells and resistant clones (Figure 2B). An unrelated human tumor cell line, A431 (negative control), was relatively insensitive to U0126 (IC50 of 10 μM), and consistent with this, there was no detectable attenuation of p-ERK1/2 levels upon treatment with the inhibitor (Figure 2B). Thus, persistent ERK1/2 activation in the AZ628-resistant clones appears to be mediated by MEK.
Sustained proliferation of AZ628-resistant clones is largely independent of BRAF kinase activity
We next explored the mechanism of persistent MEK-mediated ERK1/2 activation in AZ628-resistant clones in the presence of drug. Although we had excluded the role of a secondary BRAF mutation as a resistance mechanism, it remained possible that reduced bioavailability of the inhibitor was involved in mediating resistance. A second possibility was that MEK-ERK activation was no longer driven by BRAF kinase activity, but instead, involved another aberrantly activated intracellular pathway, as previously described in the setting of acquired resistance to other selective kinase inhibitors (16). To distinguish between these possibilities, we utilized lentivirus-mediated delivery of BRAF short hairpin (sh) RNAs to down-regulate the expression of BRAF in the resistant clones. Immunoblotting demonstrated successful and specific depletion of BRAF protein using BRAF-directed shRNA in both the parental M14 cells and the M14BRR2 AZ628-resistant clone (Figure 2C). As expected, knockdown of BRAF in the parental M14 cells resulted in a substantial decrease in cell viability, consistent with dependency on BRAF kinase activity in these cells (Figure 2D). However, depleting BRAF protein in the M14BRR2 cell line, or in the unrelated A549 tumor cell line (negative control) only had a very mild effect on cell viability, suggesting that these cells exhibited substantially reduced dependency on BRAF for their sustained survival (Figure 2D). Moreover, while down-regulation of BRAF protein in the M14 parental cells resulted in complete abrogation of ERK1/2 activation, in the AZ628-resistant clones, p-ERK1/2 was only partially suppressed (Figure 2C), suggesting an uncoupling of ERK1,2 from BRAF in the resistant clones. Taken together, these results strongly suggest that sustained survival of the AZ628-resistant clones is mediated through an alternative activated pathway that is largely independent of BRAF kinase activity.
AZ628-resistant clones express elevated CRAF
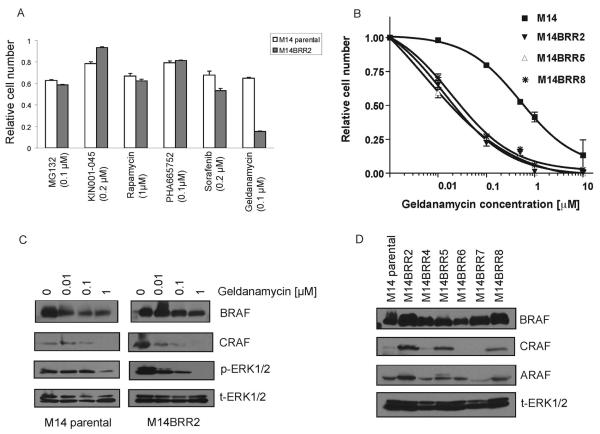
To identify the activated pathway that contributes to cell survival via sustained MEK-ERK signaling in the AZ628-resistant clones, we examined the sensitivity of these cells to a variety of small molecule inhibitors of cellular signaling pathways implicated in cancer (Figure 3A). Among the many tested inhibitors, the resistant clones exhibited significantly increased sensitivity to the HSP90 inhibitor geldanamycin relative to the parental M14 cell line (Figure 3A and 3B). HSP90 is a chaperone protein required for conformational stability of various proteins, including mutant V600E BRAF and the BRAF-related RAF family member CRAF (27-30). Therefore, we examined levels of the three RAF isoforms in these cells following geldanamycin treatment. As previously reported (28, 29), we observed that geldanamycin promotes a reduction in BRAF protein levels; although, the decline in CRAF protein levels in these cells was notably more substantial, associated with a virtually complete elimination of CRAF protein in both geldanamycin-treated M14 parental cells and the resistant clones (Figure 3C). A role for CRAF in the AZ628-resistant cells was further supported by the observation that some of the resistant clones expressed significantly elevated basal levels of CRAF relative to the M14 parental cells (Figure 3D), whereas BRAF and ARAF protein expression was not detectably changed. As expected, the AZ628-resistant clones that expressed elevated CRAF protein levels (M14BRR2, 5 and 8) correspond to the clones demonstrating increased p-ERK1/2 levels (Figure 1B).
Figure 3. M14 AZ-628 resistant clones express elevated CRAF and exhibit geldanamycin sensitivity.
(A) Increased sensitivity of AZ628-resistant cells to geldanamycin. Survival curves of M14 and AZ628-resistant (M14BRR) clones treated with the indicated concentrations of the indicated drugs. MG132, proteasome inhibitor; NFκB inhibitor; KIN001-045, src inhibitor; rapamycin, mTOR inhibitor; PHA-665752, MET kinase inhibitor; sorafenib, multi-kinase inhibitor; geldanamycin, HSP90 inhibitor.
(B) AZ628-resistant clones exhibit increased geldanamycin sensitivity. Dose-response of M14 and three M14BRR clones treated with the indicated concentrations of geldanamycin. The fraction of viable cells is expressed relative to untreated controls. Error bars represent the standard deviation from the mean.
(C) Geldanimycin causes BRAF reduction and CRAF depletion in M14 and AZ628-resistant clones. Cell lysates from M14 and M14BRR2 cells were collected following treatment with the indicated concentrations of geldanamycin for 24 hours. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
(D) Elevated CRAF expression in a subset of AZ628-resistant M14-derived clones. Cell lysates from M14 and six M14BRR clones maintained in the presence of 2 μM of AZ628 were collected. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
FISH (fluorescent in situ hybridization) analysis of interphase chromosomal spreads revealed that the increased expression of CRAF in the AZ628-resistant cells was not associated with CRAF gene amplification (Supplementary Figure 2). Similarly, RT-qPCR analysis demonstrated indistinguishable CRAF mRNA levels in both the parental cell line and the resistant clones (Supplementary Figure 3). Taken together, these data suggest that resistance to AZ628 was associated with increased CRAF protein levels that cannot be attributed to gene amplification or increased CRAF gene transcription. Thus, changes in CRAF protein levels in the resistant cell lines appear to reflect a post-transcriptional regulatory mechanism.
Survival of AZ628-resistant cells is dependent on CRAF
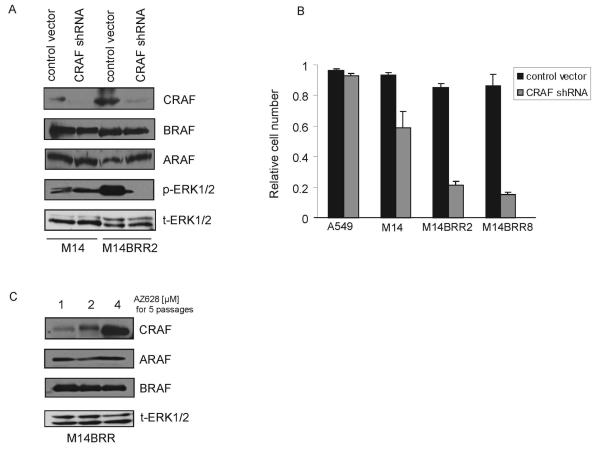
To confirm that proliferation and survival of the AZ628-resistant clones is truly mediated by CRAF, we used a RNA interference approach. M14 cells and the AZ628-resistant clones were infected with a lentiviral vector expressing shRNA designed to specifically target CRAF. Immunoblotting demonstrated specific and effective down-regulation of CRAF following infection (Figure 4A). We observed a considerable reduction in cell proliferation following CRAF down-regulation in the AZ628-resistant clones (M14BRR 2 and 8), whereas the effect on M14 cells was significantly less (Figure 4B). Furthermore, ERK1/2 activation was completely suppressed by CRAF depletion in the resistant clones (M14BRR2), but not in the M14 parental cells (Figure 4A), suggesting that ERK1/2 activation is tightly coupled to CRAF in the AZ628-resistant cells.
Figure 4. Proliferation of AZ628-resistant M14 clones is dependent on CRAF.
(A) Down-regulation of CRAF in AZ628-resistant clones results in reduced p-ERK1/2. M14 and M14BRR2 cells were infected with a lentivirus control (PLKO.1 empty vector) or a virus expressing CRAF-specific shRNA. Cells were puromycin-selected and protein lysates were collected 4 days after the infection. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
(B) AZ628-resistant M14 cells are dependent on CRAF. Cell viability assay corresponding to (A). Control or CRAF-specific shRNAs were introduced into A549, M14, M14BRR2, and M14BRR8 cells and cell proliferation assays with Syto60 were performed 5 days later. The fraction of cells relative to untreated controls is expressed. Error bars represent the standard deviation from the mean.
(C) CRAF levels in AZ628-resistant cells vary proportionately to the concentration of AZ628 in which cells are maintained. Cell lysates from M14BRR2 cells growing in the indicated concentrations of AZ628 for several passages were collected. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
During the course of characterizing the AZ628-resistant clones, we observed that varying the concentration of AZ628 in which cells were propagated led to a corresponding change in their expression of CRAF protein. Thus, after five passages in 4 μM AZ628, a significant increase in CRAF protein expression was observed (relative to cells propagated in 2 μM AZ628), whereas there was a relative decrease in CRAF levels in cells growing in 1 μM AZ628 (Figure 4C). Significantly, ARAF and BRAF protein levels were unchanged under these conditions. Such a correlation between AZ628 concentration and CRAF protein levels is consistent with a “compensatory” regulation of CRAF expression to permit cell growth and survival in the presence of AZ628.
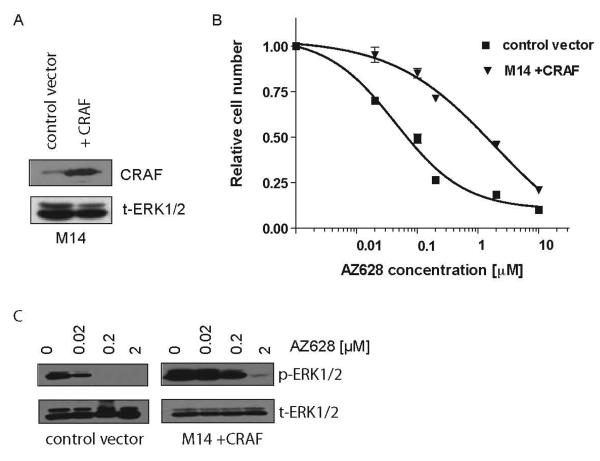
To confirm a causal role for increased CRAF expression in the AZ628-resistant phenotype, we determined that stable over-expression of CRAF in the M14 parental cells confers significant resistance to AZ628 (IC50 2 μM) when compared to M14 cells transfected with a vector control (IC50 100nM) (Figure 5A-C). However, we note that CRAF-overexpressing M14 cells were not as resistant to AZ628 as the clones that were initially selected in 2 μM AZ628 (IC50 10 μM). Taken together, these results suggest that elevated CRAF expression is a potential mechanism of acquired resistance to continuous AZ628 exposure, leading to sustained activation of ERK1/2.
Figure 5. CRAF overexpression can confer resistance to RAF inhibition.
(A) CRAF cDNA or control (pBABE empty vector) were introduced into M14 parental cells. Cell lysates were collected and immunobloting analysis was performed using antibodies directed against CRAF and t-ERK 1/2 (loading control).
(B) M14 cells expressing exogenous CRAF exhibit reduced sensitivity to AZ628. Dose-response curves corresponding to cells in (D). M14+CRAF and M14+pBABE control vector cells were treated with the indicated concentrations of AZ628. The fraction of viable cells is expressed relative to untreated controls. Error bars represent the standard deviation from the mean.
(C) M14 cells expressing exogenous CRAF exhibit reduced suppression of ERK1,2 activation following AZ628 treatment. Cell lysates from M14+pBABE control vector and M14+CRAF were collected following treatment with the indicated concentrations of AZ628 for 2 hours. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
Elevated CRAF can confer primary insensitivity to a RAF inhibitor
In a high-throughput profiling study of AZ628 sensitivity across 500 human tumor-derived cell lines, we recently demonstrated that while sensitivity to AZ628 is highly correlated with the presence of the activating BRAF V600E mutation, a small subset of cell lines exhibit primary insensitivity to AZ628 despite harboring the V600E BRAF mutation (11). To determine if elevated CRAF expression could potentially account for insensitivity to BRAF inhibitors in such lines, we further characterized the three most AZ628-insensitive cell lines harboring BRAF V600E mutations, A2058, Sw1417 and Wm1552C (Figure 6A). p-ERK1/2 activity was not significantly suppressed by exposure to AZ628 in one of these three AZ628-insensitive cell lines (Wm1552C) suggesting that, as in the M14-derived AZ628-resistant clones, activated ERK1/2 was uncoupled from BRAF in this cell line (Figure 6A).
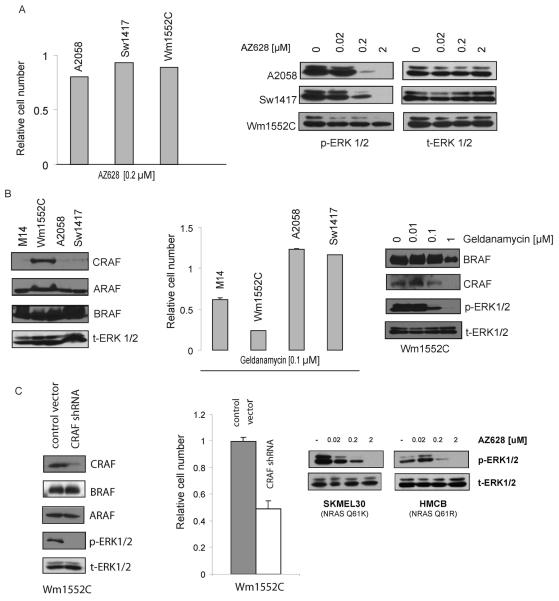
Figure 6. CRAF overexpression can confer insensitivity to RAF inhibition.
(A) (Left) AZ628-insensitivity among three BRAF mutant tumor cell lines. Three AZ628-insensitive BRAF mutant cell lines (A2058, Sw1417, and Wm1552C) were treated with 0.2 μM AZ628, and a proliferation assay with Syto60 was performed 72 hours later. The fraction of viable cells is expressed relative to untreated controls. Error bars represent the standard deviation from the mean. (Right) AZ628 fails to suppress p-ERK1,2 in Wm1552C cells. Cell lysates from three AZ628-insensitive cell lines were collected following treatment with the indicated concentrations of AZ628 for 2 hours. Immunobloting analysis was performed using antibodies directed against p-ERK1/2 and t-ERK1/2 (loading control).
(B) (Left) CRAF levels are relatively high in Wm1552C cells. Cell lysates from M14 and three AZ628-insensitive cell lines were collected. Immunobloting analysis was performed using antibodies directed against the indicated proteins. (Middle) Wm1552C cells exhibit geldanamycin sensitivity. M14 and three AZ628-insensitive cell lines (Wm1552C, A2058, Sw1417) were treated with 0.1 μM geldanamycin, and a proliferation assay with Syto60 was performed 72 hours later. The percentage of viable cells is expressed relative to untreated controls. Error bars represent the standard deviation from the mean. (Right) Geldanimycin treatment causes CRAF depletion and suppresses ERK1,2 activation in Wm1552C cells. Cell lysates from Wm1552C were collected following treatment with the indicated concentrations of geldanamycin for 24 hours. Immunobloting analysis was performed using antibodies directed against the indicated proteins.
(C) (Left) CRAF depletion by shRNA suppresses ERK1,2 activation in Wm1552C cells. Wm1552C cells were infected with a lentivirus control (PLKO.1 empty vector) or a virus expressing CRAF-specific shRNA. Cells were puromycin-selected and protein lysates were collected 4 days after the infection. Immunobloting analysis was performed using antibodies directed against the indicated proteins. (Middle) CRAF depletion by shRNA suppresses ERK1,2 inhibits cell growth in Wm1552C cells. Cell viability corresponding to cells in (F). Control or CRAF-specific shRNAs were introduced in Wm1552C cells and a cell proliferation assay with Syto60 was performed 5 days later. The fraction of viable cells is expressed relative to untreated controls. Error bars represent standard deviation. (Right) Immunoblots demonstrating that AZ628 effectively suppresses ERK phosphorylation in two different melanoma cell lines that harbor activated NRAS alleles.
Significantly, immunoblot analysis revealed relatively high levels of CRAF protein in the Wm1552C cell line (Figure 6B). Wm1552C is a melanoma-derived cell line that harbors the BRAF V600E mutation and is highly refractory to AZ628 treatment (IC50 10 μM), as well as to an alternative RAF inhibitor (data not shown). Interestingly, in a high-throughput screen of 500 tumor cell lines for geldanamycin sensitivity, Wm1552C scored as being exquisitely sensitive to geldanamycin when compared to the other AZ628-insensitve cell lines, consistent with a potential role for elevated CRAF in maintaining cell proliferation in Wm1552C cells (Figure 6B and data not shown). Consistent with this possibility, immunoblot analysis of geldanamycin-treated Wm1552C cells demonstrated markedly reduced CRAF protein levels and suppression of ERK1/2 activation (Figure 6B).
To confirm that Wm1552C cells are in fact dependent on CRAF, we utilized the lentiviral-mediated CRAF shRNA to reduce CRAF protein in these cells. After efficient depletion of CRAF in Wm1552C cells, a significant decrease in viability was observed (Figure 6C). Taken altogether, our findings suggest that elevated CRAF levels may also represent a mechanism that confers primary insensitivity to RAF kinase inhibition in a subset of tumor cells harboring the BRAF V600E mutation. Since it has been reported that in NRAS mutant melanomas, ERK activation is mediated largely through CRAF (8), we also examined the ability of AZ628 to suppress ERK phosphorylation in two different melanoma lines that harbor activated NRAS alleles. We found that, unlike in the AZ628-resistant M14 cells in which AZ628 fails to suppress ERK activation, AZ628 treatment efficiently attenuates ERK activation in the NRAS mutant melanoma cells, suggesting distinct roles for CRAF in the activation of ERK in these two settings (Figure 6C).
DISCUSSION
Kinase-targeted drugs have emerged as an important new class of cancer therapeutics, with demonstrated clinical efficacy in multiple tumor contexts. However, treatment with such agents is invariably associated with the eventual emergence of drug resistance, which remains the most significant limitation of such therapies. Therefore, it is important to identify mechanisms underlying acquired drug resistance, as well as to utilize pre-clinical models to reveal resistance mechanisms that are likely to be observed clinically in order to accelerate the development of strategies to overcome or prevent such resistance. Here, we have described a cell culture model of acquired resistance to RAF inhibition in a BRAF V600E mutated melanoma cell line with exquisite sensitivity to the selective RAF kinase inhibitor AZ628. Inhibitors of this class are currently undergoing early clinical testing, and consequently, it will be quite some time before mechanisms of acquired drug resistance can be established from clinical specimens—assuming that these agents produce clinical benefit. Our findings have revealed a potential mechanism underlying acquired resistance to a selective RAF kinase inhibitor, and furthermore, have demonstrated that HSP90 inhibition may be an effective therapeutic strategy to overcome such resistance. Since HSP90 inhibitors are currently undergoing clinical evaluation as cancer therapeutics (30-32), our findings may prompt another potential application of such agents. Notably, a recent report demonstrated that HSP90 inhibition effectively promotes tumor regression in a mouse transgenic model of EGFR in which the T790M TKI-resistance mutation was expressed (33). Such findings, together with our findings, implicate HSP90 inhibitors as potentially important drugs in a variety of drug resistance settings.
Previous pre-clinical cell culture-based studies have demonstrated that most BRAF mutant tumor cells are in fact “addicted” to BRAF, and that signaling to the downstream effector ERK, via the MEK kinase, is a critical pathway by which BRAF drives cell proliferation and survival in such tumors (6, 7, 9, 34). Our finding that M14 melanoma cells are growth inhibited by both BRAF and MEK kinase inhibitors is consistent with those observations. Moreover, the sustained ERK activation seen in drug-treated M14-derived AZ628-resistant cells is also consistent with a critical role for this pathway in these melanoma cells. Notably, not all of the individual clones exhibit the same signaling properties, suggesting that multiple distinct mechanisms of acquired resistance can be established through such modeling, and further study will certainly be required to reveal such alternative resistance mechanisms in those clones.
We focused on the subset of AZ628-resistant clones in which ERK signaling is sustained in the presence of the inhibitor. Interestingly, these cells exhibit significantly elevated expression of CRAF, which appears to involve a post-transcriptional mechanism. Although this finding suggests that these cells may have switched their dependency from BRAF to CRAF, it is noteworthy that AZ628 inhibits the BRAF and CRAF isoforms somewhat equally in vitro (L.D., unpublished observation). Therefore, it is possible that increased CRAF protein levels decrease the bioavailability of drug within cells by virtue of increased intracellular concentration of drug-binding targets. Similarly, increased expression of the BCR-ABL kinase, frequently as a result of specific gene amplification, in imatinib-resistant cases of CML may contribute to resistance through a similar drug-titration mechanism (13).
The increased expression of CRAF in the AZ628-resistant cells suggests that these cells may have shifted their dependency from BRAF to CRAF. Notably, this switch does not necessarily require CRAF kinase activity, as previous reports have demonstrated kinase-independent functions for CRAF (35, 36). The fact that the AZ628-resistant cells can be growth inhibited by shRNA-mediated knockdown of CRAF, but much less so by knockdown of BRAF, is consistent with a shift in their dependency from BRAF to CRAF. Previous studies have demonstrated a physical interaction between BRAF and CRAF, with BRAF promoting CRAF-mediated ERK activation through the formation of BRAF:CRAF heterodimers, thereby demonstrating an intimate relationship between these two proteins in regulating the ERK pathway (8, 37, 38). Moreover, recent studies have revealed a role for cAMP signaling in a switch from BRAF to CRAF dependency for MEK-ERK signaling in BRAF mutant melanoma cells (39). Thus, it is possible that the acquisition of AZ628 resistance in M14 melanoma cells similarly involves altered cAMP signaling. It is worth noting that the parental M14 cells also display some CRAF dependency, as revealed by gene knockdown studies, and that the M14 cells transfected with CRAF are not as resistant to AZ628 as the selected clones. Thus, the collective data point to a critical balance between BRAF and CRAF in parental M14 cells that drives ERK signaling, which is substantially impacted by the relative levels of these proteins. However, the precise mechanism by which CRAF levels are increased to disrupt this balance in the drug-resistant clones is currently unknown.
Molecular mechanisms of acquired resistance to kinase inhibitors can also contribute to primary insensitivity to such treatment in some cases. For example, the T790M EGFR mutation that appears to account for acquired resistance to EGFR TKIs in about half of TKI-responsive patients who subsequently relapse has also been detected in a small subset of untreated tumors (40-42). Similarly, amplification of the gene encoding the MET tyrosine kinase, which occurs in a small percentage of EGFR TKI-treated NSCLCs, has also been detected in NSCLCs demonstrating primary insensitivity to treatment with EGFR TKIs (11). Based on our pre-clinical findings, CRAF overexpression may similarly represent both a mechanism of acquired drug resistance as well as primary drug insensitivity in a subset of cases. The fact that tumor cell lines demonstrating elevated CRAF protein are highly geldanamycin-sensitive, irrespective of whether they were derived through acquired resistance or not, suggests that elevated CRAF protein may constitute a tumor biomarker that predicts response to HSP90 inhibition, as well as a lack of response to BRAF kinase inhibitors, in both of these clinical settings.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of the Settleman laboratory for helpful discussions. This work was supported by NIH RO1CA115830 and a V Foundation award to J.S. C.M. was supported by Instituto de Salud Carlos III Grant BA07/90064 and Grant from Sociedad Española de Oncología Médica (Seom) - Roche Farma España 2006.
REFERENCES
- 1.Sharma SV, Settleman J. Oncogene addiction: Setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21(24):3214–31. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 7.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 8.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6(4):313–9. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439(7074):358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 11.McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104(50):19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 14.Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat Rev Drug Discov. 2004;3(12):1001–10. doi: 10.1038/nrd1579. [DOI] [PubMed] [Google Scholar]
- 15.Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24(29):4764–74. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 16.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 17.Debiec-Rychter M, Cools J, Dumez H, et al. Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology. 2005;128(2):270–9. doi: 10.1053/j.gastro.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 18.Donato NJ, Wu JY, Stapley J, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101(2):690–8. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 19.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112(6):831–43. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 20.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116(10):2695–706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azam M, Daley GQ. Anticipating clinical resistance to target-directed agents: the BCR-ABL paradigm. Mol Diagn Ther. 2006;10(2):67–76. doi: 10.1007/BF03256446. [DOI] [PubMed] [Google Scholar]
- 22.Lorusso PM, Adjei AA, Varterasian M, et al. Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J Clin Oncol. 2005;23(23):5281–93. doi: 10.1200/JCO.2005.14.415. [DOI] [PubMed] [Google Scholar]
- 23.Strumberg D, Clark JW, Awada A, et al. Safety, pharmacokinetics, and preliminary antitumor activity of sorafenib: a review of four phase I trials in patients with advanced refractory solid tumors. Oncologist. 2007;12(4):426–37. doi: 10.1634/theoncologist.12-4-426. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Boerner SA, Winkler JD, LoRusso PM. Clinical experience of MEK inhibitors in cancer therapy. Biochim Biophys Acta. 2007;1773(8):1248–55. doi: 10.1016/j.bbamcr.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Ross DT, Scherf U, Eisen MB, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24(3):227–35. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 26.Stanton VP, Jr., Nichols DW, Laudano AP, Cooper GM. Definition of the human raf amino-terminal regulatory region by deletion mutagenesis. Mol Cell Biol. 1989;9(2):639–47. doi: 10.1128/mcb.9.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maloney A, Clarke PA, Workman P. Genes and proteins governing the cellular sensitivity to HSP90 inhibitors: a mechanistic perspective. Curr Cancer Drug Targets. 2003;3(5):331–41. doi: 10.2174/1568009033481822. [DOI] [PubMed] [Google Scholar]
- 28.da Rocha Dias S, Friedlos F, Light Y, Springer C, Workman P, Marais R. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65(23):10686–91. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- 29.Grbovic OM, Basso AD, Sawai A, et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci U S A. 2006;103(1):57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp S, Workman P. Inhibitors of the HSP90 molecular chaperone: current status. Adv Cancer Res. 2006;95:323–48. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- 31.Workman P, Burrows F, Neckers L, Rosen N. Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann N Y Acad Sci. 2007;1113:202–16. doi: 10.1196/annals.1391.012. [DOI] [PubMed] [Google Scholar]
- 32.Solit DB, Rosen N. Hsp90: a novel target for cancer therapy. Curr Top Med Chem. 2006;6(11):1205–14. doi: 10.2174/156802606777812068. [DOI] [PubMed] [Google Scholar]
- 33.Regales L, Balak MN, Gong Y, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS ONE. 2007;2(8):e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikenoue T, Hikiba Y, Kanai F, et al. Functional analysis of mutations within the kinase activation segment of B-Raf in human colorectal tumors. Cancer Res. 2003;63(23):8132–7. [PubMed] [Google Scholar]
- 35.Ehrenreiter K, Piazzolla D, Velamoor V, et al. Raf-1 regulates Rho signaling and cell migration. J Cell Biol. 2005;168(6):955–64. doi: 10.1083/jcb.200409162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baccarini M. Second nature: biological functions of the Raf-1 “kinase”. FEBS Lett. 2005;579(15):3271–7. doi: 10.1016/j.febslet.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 37.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20(6):963–9. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Rushworth LK, Hindley AD, O'Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26(6):2262–72. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumaz N, Hayward R, Martin J, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66(19):9483–91. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 40.Kozuki T, Hisamoto A, Tabata M, et al. Mutation of the epidermal growth factor receptor gene in the development of adenocarcinoma of the lung. Lung Cancer. 2007;58(1):30–5. doi: 10.1016/j.lungcan.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 41.Soh J, Toyooka S, Ichihara S, et al. EGFR mutation status in pleural fluid predicts tumor responsiveness and resistance to gefitinib. Lung Cancer. 2007;56(3):445–8. doi: 10.1016/j.lungcan.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37(12):1315–6. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.