Abstract
Cardiac and skeletal muscle development are controlled by evolutionarily conserved networks of transcription factors that coordinate the expression of genes involved in muscle growth, morphogenesis, differentiation, and contractility. In addition to regulating the expression of protein-coding genes, recent studies have revealed that myogenic transcription factors control the expression of a collection of microRNAs, which act through multiple mechanisms to modulate muscle development and function. In some cases, microRNAs fine-tune the expression of target mRNAs, whereas in other cases they function as “on-off” switches. MicroRNA control of gene expression appears to be especially important during cardiovascular and skeletal muscle diseases, in which microRNAs participate in stress-dependent remodeling of striated muscle tissues. We review findings that point to the importance of microRNA-mediated control of gene expression during muscle development and disease, and consider the potential of microRNAs as therapeutic targets.
Introduction
The development of cardiac and skeletal muscle is orchestrated by evolutionarily conserved networks of transcription factors that regulate the expression of genes involved in muscle growth, differentiation, and contractility. The most ancient of these myogenic transcription factors is the MADS (MCM1, Agamous, Deficiens, Serum response factor) box transcription factor myocyte enhancer factor-2 (MEF2), which directly activates the majority of muscle genes through combinatorial interactions with other transcription factors [1]. In cardiac muscle, MEF2 and another MADS-box transcription factor, serum response factor (SRF), cooperatively activate cardiac gene expression by association with GATA, T-box, and Nkx2.5 transcription factors, as well as the myocardin family of transcriptional co-activators, whereas in skeletal muscle MEF2 activates the myogenic differentiation program in conjunction with the basic-helix-loop-helix (bHLH) transcription factors, MyoD and myogenin [2,3]. Recent studies have revealed that, in addition to activating genes involved in muscle differentiation and muscle contraction, these myogenic transcription factors activate the expression of a set of conserved microRNAs (miRNAs) that function to ‘fine-tune’ the output of these transcriptional networks, resulting in precise cellular responses to developmental, physiologic, and pathologic signals. The integration of miRNAs into the core muscle transcriptional program expands the precision and complexity of gene regulation in muscle cells because individual miRNAs are capable of regulating hundreds of mRNAs, and individual mRNAs can be targeted by many miRNAs. In this review, we discuss recent publications that have illuminated the roles of miRNAs in muscle development and disease.
miRNA Biogenesis and Function
miRNAs are small, evolutionarily conserved noncoding RNAs that are transcribed by RNA polymerase II as long pri-miRNAs encoding one or more miRNAs. Most miRNAs are transcribed as independent transcripts, but approximately a third are embedded within introns of protein-coding genes and processed following splicing of pre-messenger RNAs [4]. Pri-miRNAs are processed in the nucleus by the proteins Drosha and DGCR8, which produce an ~70 nucleotide hairpin RNA, termed the pre-miRNA, which is subsequently exported to the cytoplasm where it is processed by Dicer to yield a duplex RNA ~22 nucleotides long. This duplex is released from Dicer and the single-stranded mature miRNA is incorporated into the RNA-induced silencing complex (RISC) where it associates with complementary target mRNAs [4,5].
miRNAs regulate gene expression through sequence-specific interactions with the 3′ untranslated regions (UTRs) of target mRNAs resulting in translational repression or mRNA destabilization [5]. The primary determinant of binding specificity to complementary target mRNAs is determined by Watson-Crick base-pairing of nucleotides 2 to 8 at the 5′ end of the miRNA, referred to as the “seed sequence”. However, nucleotides outside of this region also influence mRNA repression, as does the secondary structure of the surrounding regions of the mRNA 3′ UTR sequence and the location of the miRNA target site relative to the open reading frame [6]. miRNA target identification and validation remains a major challenge in the field, as different bioinformatic prediction programs frequently predict remarkably different mRNA targets for individual mRNAs, and it is not uncommon for predicted targets to show little or no regulation by a predicted miRNA when tested biochemically. Another limitation to the validation of miRNA targets in vivo is that most studies to date have involved over-expression of miRNAs and analysis of their effects on artificial reporters linked to the 3′ UTRs of predicted mRNA targets. Even in these cases of miRNA over-expression, the effects of individual miRNAs on mRNA targets are often quite modest (~2-fold). While some miRNAs may exert their effects through pronounced repression of a relatively small number of mRNA targets, more commonly, miRNAs regulate dozens or hundreds of targets by relatively subtle changes in expression. Thus, the summation of small changes in multiple mRNAs may be responsible for phenotypic effects of miRNAs. Proteomic methods to analyze global changes in protein expression in response to miRNA deletion represents a powerful approach to define the functions of miRNAs, but even in this case, it can be difficult to determine with certainty which of the changes in mRNA expression are responsible for the functions of the miRNA [7*,8*]. A recent study also demonstrated the ability of a miRNA to be capable of enhancing translation, raising interesting new possibilities for the regulatory actions of miRNAs [9]. In addition, there is also evidence that small RNAs can directly control transcription through sequence-specific interactions with promoter elements of target genes [10]. Perhaps, miRNAs can also function in a similar setting by directly conferring transcriptional, as well as post-transcriptional regulation on target genes.
Muscles without miRNAs
An essential role for miRNAs in mouse development was shown by a loss of function mutation in the miRNA-generating enzyme, Dicer, which results in embryonic lethality by day 7.5 [11]. In order to circumvent the lethality associated with the deletion of Dicer and to study the roles of Dicer in specific tissues, several groups have generated conditional null alleles of Dicer. Deletion of Dicer in cardiomyocytes using a Cre recombinase ‘knocked-in’ to the endogenous Nkx2.5 locus, which directs expression in cardiac progenitors from embryonic day (E) 8.5, leads to embryonic lethality by E12.5 due to pericardial edema and a poorly developed ventricular myocardium [12**]. In addition, post-natal deletion of Dicer in cardiomyocytes results in a loss of cardiac contractility, cardiac function, and premature death [13,14]. The deletion of Dicer from post-natal cardiomyocytes was shown to result in down-regulation of miRNAs, as predicted. However, intriguingly, a set of miRNAs was also up-regulated in these mutant hearts [14]. The molecular basis for this observation is unclear, but it may reflect compensatory changes in miRNA expression in non-myocyte cell populations of the heart.
Deletion of a conditional Dicer allele in embryonic skeletal muscle using a MyoD Cre recombinase transgene also results in perinatal lethality due to skeletal muscle hypoplasia [15]. While the above studies illustrate the importance of miRNA processing for muscle development and function, they do not indicate whether this essential function of Dicer reflects requisite roles of specific miRNAs or multiple miRNAs in these processes.
Muscle Specific miRNAs
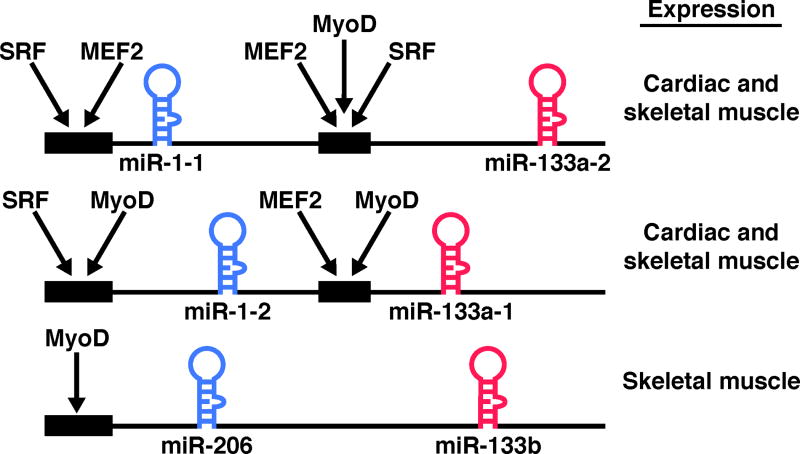
Several individual miRNAs are specifically expressed in cardiac and skeletal muscle (Table 1) [16]. Of these, the most widely studied are members of miR-1/206 and miR-133a/133b families, which originate from bicistronic transcripts on three separate chromosomes (Figure 1) [17*]. miR-1-1 and miR-1-2 are identical and differ from miR-206 by 4 nucleotides, and miR-133a-1 and miR-133a-2 are identical and differ from miR-133b by 2 nucleotides. Cardiac and skeletal muscle specific transcription of miR-1-1/133a-2 and miR-1-2/133a-1 in vertebrates appears to be controlled by two separate enhancers, one upstream and the other intronic (Figure 1) [18*–20]. The myogenic transcription factors SRF, MEF2, and MyoD control the expression of miR-1 and miR-133a in cardiac and skeletal muscle. In the case of the miR-1-2/133a-1 locus, SRF directs cardiac specific expression through the upstream enhancer and MEF2 directs ventricular expression through the intronic enhancer [18*,20]. In addition, a miRNA microarray showed miR-1 and miR-133a to be among the most significantly down-regulated miRNAs in Srf deficient hearts [21].
Table 1.
Muscle Specific miRNAs
| miR | Target Gene(s) | Function | References |
|---|---|---|---|
| miR-1 | Hdac4 | Myoblast Differentiation | 17 |
| Irx5, Connexin 43, Kcnj2, Hcn2, Hcn4 | Cardiac Conduction System | 12, 29, 30 | |
| Hand2 | Cardiac Proliferation | 12, 20 | |
| Dll1 | Cardiac Mesoderm Differentiation | 24 | |
| Mef2 | Neuromuscular Synapse Function | 28 | |
| miR-133 | Srf | Myoblast Proliferation, Smooth Muscle Gene Expression | 17, 23 |
| Cyclin D2 | Cardiac Proliferation | 23 | |
| Rhoa, Cdc42, Whsc2 | Cardiac Hypertrophy | 55 | |
| Ether-a-go-go, Hcn2 | Cardiac Conduction System | 30, 31 | |
| miR-206 | Pola1, Connexin 43, Fst1, Utrn | Myoblast Differentiation | 22, 32, 33 |
| miR-208 | Thrap1 | β-MHC Gene Expression | 39 |
Figure 1. Three bicistronic clusters of muscle-specific miRNAs.
Three bicistronic gene clusters each encoding two miRNAs are shown. miR-1-1, -1-2 and -206 are nearly identical in sequence, as are miR-133a-2, -133-a-1 and -133b. Cis-regulatory elements that direct muscle-specific expression of each locus are indicated by black boxes, and the transcription factors that act through these elements are shown.
The third cluster of muscle specific miRNAs encoding miR-206 and miR-133b is expressed specifically in skeletal muscle [17*]. Skeletal muscle specific transcription of the miR-206/133b transcript is thought to be controlled by an up-stream regulatory region that is enriched for MyoD binding in ChIP-on-chip assays using chromatin from C2C12 muscle cells (Figure 1) [19]. Also, using a MyoD deficient fibroblast cell line, MyoD was shown to directly activate transcription of miR-206/133b [22].
In vitro and in vivo studies have demonstrated that miR-1 and miR-133a regulate fundamental aspects of muscle biology such as differentiation and proliferation (Figures 2 and 3). In C2C12 skeletal muscle cells, miR-1 represses the expression of histone deacetylase 4 (HDAC4), a negative regulator of differentiation and a repressor of the MEF2 transcription factor [17*]. Thus, the repression of HDAC4 by miR-1 establishes a positive feed-forward loop in which the up-regulation of miR-1 by MEF2 causes further repression of HDAC4 and increased activity of MEF2, which drives myocyte differentiation [3] (Figure 2). In mice, over-expression of miR-1 in embryonic cardiomyocytes results in decreased proliferation of cardiomyocytes, a phenotype that has been attributed to a decrease in the expression of Hand2, a transcription factor that promotes cardiomyocyte proliferation [20]. Consistent with this model, miR-1-2 deficient mice have an increase in Hand2 expression and an increased number of proliferating cardiomyocytes (Figure 3) [12**].
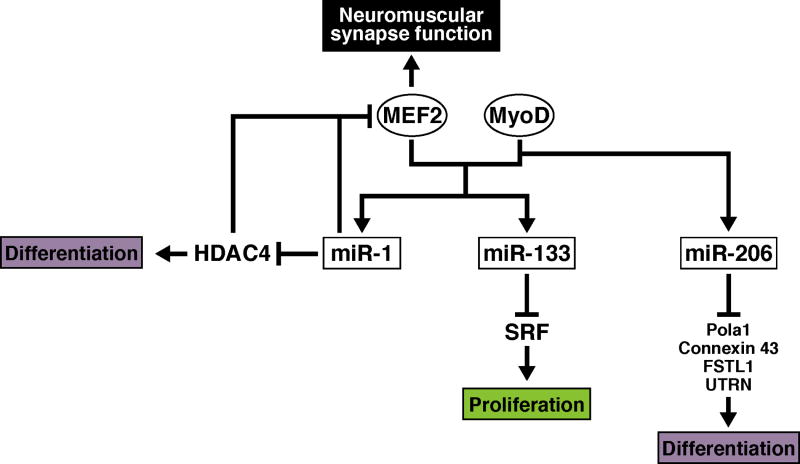
Figure 2. miRNA-transcription factor circuits involved in skeletal muscle development.
MEF2 and MyoD control expression of miR-1, miR-133 and miR-206 in skeletal muscle. Targets for repression by these miRNAs, and the processes they regulate during skeletal muscle development, are shown.
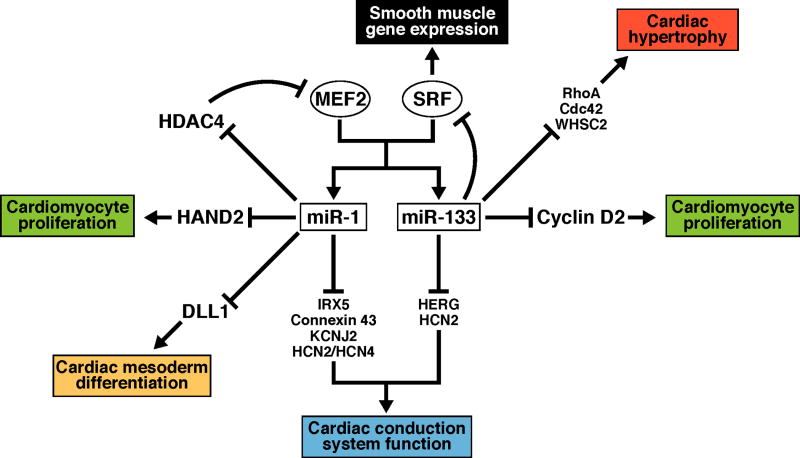
Figure 3. miRNA-transcription factor circuits involved in cardiac growth and development.
Expression of miR-1 and miR-133 in cardiac muscle is controlled by MEF2 and SRF. Targets for repression by miR-1 and miR-133, and the processes they regulate during cardiac growth and development, are shown.
In C2C12 myoblasts, miR-133a promotes proliferation, at least in part, by repressing SRF [17*]. Genetic deletion of both miR-133a-1 and miR-133a-2 showed that SRF is a direct target of miR-133a in vivo and suggested that miR-133a suppresses proliferation of cardiomyocytes by repressing cyclin D2 expression (Figure 3) [23**]. The genetic interaction between miR-133a and SRF constitutes a negative feedback loop in which the up-regulation of miR-133a by SRF results in increased repression of SRF (Figure 2). Indeed, the in vitro and in vivo results seem conflicting regarding a role for miR-133a in regulating proliferation; however, SRF has previously been shown to be capable of functioning as an activator of proliferation and differentiation depending on its association with co-factors such as myocardin, HOP, and Elk-1 [2].
Viral expression of miR-1 and miR-133a in mouse and human embryonic stem (ES) cells also showed that both of these miRNAs promote the differentiation of ES cells into mesodermal progenitors; however, only miR-1 is capable of inducing the differentiation of mesodermal progenitors to the muscle lineage [24**]. An especially fascinating finding of the latter study was the ability of miR-1 to substitute for SRF and promote mesoderm formation in SRF-null ES cells, which are otherwise unable to form mesoderm. This function was attributed to, at least in part, the repression of the Notch ligand Delta-like 1 (DLL1) [24**].
Together, these results suggest that the primary function of miR-1 is to induce the differentiation of mesodermal and muscle lineages and the function of miR-133a is to regulate proliferation and differentiation; albeit in a cell type- and signal-specific manner. Given the ability of miR-1 to induce mesoderm formation in stem cells, it is curious that global deletion of Dicer in mice or zebrafish does not prevent the early specification of muscle cells, but instead seems to act at later steps in muscle development [11,25].
Other functions have been attributed to miR-1 and miR-133a such as regulation of apoptosis in cardiomyocytes and alternative splicing during myoblast differentiation by regulation of the alternative splicing factor nPTB [26,27]. Recently, miR-1 was shown to regulate neuromuscular synapse function in C. elegans through the inhibition of the MEF2 transcription factor [28*]. miR-1 and miR-133a have also been shown to have functions within the vertebrate cardiac conduction system. miR-1-2 deficient mice exhibit electrophysiological defects such as a shortened PR interval and prolonged QRS complex, which have been attributed to the increase in expression of IRX5 [12**]. Manipulation of miR-1 expression in adult ischemic rat hearts through the delivery of oligonucleotides demonstrated that over-expression of miR-1 is capable of causing arrhythmias, while the knock-down of miR-1 expression is capable of suppressing arrhythmias through the inhibition of Connexin 43 and KCNJ2 expression [29]. In addition miR-1 and miR-133 were shown to suppress the expression of other mRNAs encoding components of the cardiac conduction system, such as ether-a-go-go related gene (HERG) and the pacemaker channel genes Hcn2 and Hcn4 [30,31].
Like miR-1, miR-206 has been shown to promote differentiation of C2C12 myoblasts in vitro [32,33]. miR-206 induces muscle differentiation by repressing the expression of a subunit of DNA polymerase alpha (Pola1), connexin 43 (Cx43); as well as follistatin-like 1 (Fstl1) and utrophin (Utrn) [22,32,33]. Also, miR-206 was reported to repress estrogen receptor alpha (ERα) protein expression in ER negative breast cancer cells [34]. The function of miR-206 and miR-133b in skeletal muscle is currently unknown, but many functions have been proposed such as a regulator of slow muscle fiber identity, a mediator of skeletal muscle hypertrophy, and a regulator of skeletal muscle regeneration [35–37].
Although expressed in a similar manner in response to various developmental and stress signals, the sequences of miR-1 and miR-133 are quite divergent (sharing only 4 nucleotides). Indeed, miR-1 and miR-133 are only predicted to share approximately 38 target genes (~10% of total) compared to 480 total predicted target genes for miR-1 and 351 total predicted target genes for miR-133 using the microRNA target prediction program TargetScan [6]. The bioinformatically predicted targets and experimentally determined functions for miR-1 and miR-133 suggest that these two miRNAs share divergent and overlapping functions.
MyomiRs
Proper function of the heart depends on the expression of myosin heavy chain (MHC) proteins MYH6 (α-MHC) and MYH7 (β-MHC). Cardiac injury results in a decrease in the expression of the adult myosin gene, Myh6, and an increase in the expression of the embryonic myosin gene, Myh7. Recently, it was discovered that these myosin genes also encode a family of miRNAs, called MyomiRs, which have important functions in regulating myosin content and stress-dependent cardiac remodeling [38].
α-MHC, the major contractile myosin in the adult murine heart co-expresses the heart specific miRNA, miR-208. miR-208 deficient mice are viable but show an inability to up-regulate the expression of β-MHC in response to injury and hypothyroidism [39**] (Figure 4). In addition to the significant attenuation of β-MHC expression, hearts from miR-208 deficient animals have increased expression of fast skeletal genes, less fibrosis, and maintain cardiac contractility and cardiac function following stress compared to wild-type animals. This function of miR-208 was attributed to the repression of protein levels of the thyroid hormone receptor coregulator 1 (THRAP1), which exerts positive and negative effects on the thyroid hormone receptor depending on genomic context (Figure 4). Loss- and gain-of-function experiments with THRAP1 in the heart have not yet been reported, but will be of particular interest.
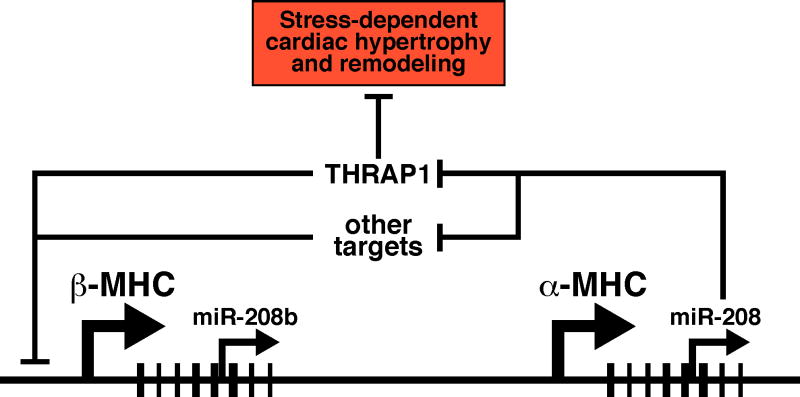
Figure 4. Control of cardiac remodeling by MyomiRs.
The alpha- and beta-MHC genes encode miR-208 and miR-208b, respectively. miR-208 represses expression of the thyroid hormone receptor-associated protein, THRAP1, which acts as a positive and negative regulator of thyroid hormone receptor-dependent transcription. miR-208 is required for up-regulation of beta-MHC expression and for hypertrophy and fibrosis in response to cardiac stress.
β-MHC, the major contractile protein in slow skeletal muscle and in the embryonic and failing murine heart encodes miR-208b. Since α-MHC and β-MHC are expressed in an opposing manner during cardiac development and remodeling, miR-208 and miR-208b are also co-expressed in an opposing manner. Since miR-208b differs from miR-208 by only 3 nucleotides, this type of regulation maintains a constant level of ‘miR-208 activity’ in the heart [38]. The third member of the MyomiR family, miR-499, is co-expressed with Myh7b. The precise functions of miR-208b and miR-499 are currently unknown and determination of their roles in the control of stress induced remodeling of cardiac and skeletal muscle is a key challenge for the future.
The functions of MyomiRs in the human heart also remain to be defined. Since α-MHC represents a very minor fraction of the total myosin content in the human heart, whereas β-MHC is highly abundant, it is conceivable that miR-208b, encoded by the β-MHC gene, fulfills the functions of miR-208 in the rodent heart.
miRNAs with Functions in Muscle
In addition to muscle-specific miRNAs, there are also examples of ubiquitously expressed miRNAs that have been shown to have functions in muscle. In zebrafish, miR-214 was shown to act as a positive regulator of the slow muscle phenotype, by targeting suppressor of fused (Su(fu)), a negative regulator of hedgehog signaling [40]. miR-138 was reported to contribute to chamber-specific gene expression patterns in the zebrafish heart by targeting aldehyde dehydrogenase-1a2 (RALDH2), a regulator of retinoic acid biosynthesis [41]. The miR-181 isoforms, miR-181a and miR-181b, were shown to be induced upon the initiation of myogenesis and regulate myoblast differentiation by repressing HoxA-11 protein levels [42].
miRNAs in Muscle Disease
Several important studies have indicated that miRNA expression is dysregulated in cardiac and skeletal muscle disease and in some cases individual miRNAs have been shown to cause or alleviate disease. The first series of such studies focused on the profiling of miRNAs in hypertrophic murine and human hearts and revealed a common set of miRNAs that are elevated in hypertrophic hearts [43–46]. One of these miRNAs, miR-195, was shown to be sufficient to induce heart failure and death due to loss of cardiac muscle cells when over-expressed in the hearts of transgenic mice under control of a cardiac-specific promoter [46]. Through profiling of miRNAs dysregulated following myocardial infarction (MI) in mice, we found that the miR-29 family of miRNAs is down-regulated following MI and this miRNA family functions to control cardiac fibrosis through the direct regulation of collagen expression [47*].
Further studies demonstrated an induction of fetal miRNAs in adult human failing hearts compared to healthy hearts, a subset of which are sufficient to induce fetal gene reprogramming in cultured cardiomyocytes [48]. Additionally, global profiling of miRNAs in different forms of human heart disease demonstrated that miRNA gene expression signatures are diagnostic for distinct but related forms of heart disease such as dilated cardiomyopathy, ischemic cardiomyopathy, and aortic stenosis [49]. In this regard, miRNA-based diagnostics are likely to emerge as a useful means of molecularly defining specific forms of heart disease and disease progression.
Several studies have also focused on the identification of miRNAs dysregulated in skeletal muscle regeneration and muscular dystrophy. Kunkel and colleagues profiled miRNAs in muscle samples from a variety of human patients with primary muscle disorders and found a collection of miRNAs commonly dysregulated among patients with different types of muscle disorders [50]. The muscle-specific miRNA, miR-206, was found to be up-regulated in the diaphragm of dystrophin deficient (mdx) mice, a model of muscular dystrophy [37]. Another recent study demonstrated that miR-206 is up-regulated in the skeletal muscle of mdx mice and also upon injection of cardiotoxin, a potent inducer of muscle regeneration; however, the expression of miR-1 and miR-133a were not changed [51]. Also, a positive feed-forward mechanism involving genetic interactions between the transcription factors NF-κB and YYI and the miRNA, miR-29, demonstrated that miR-29 regulates myoblast differentiation and functions as a tumor suppressor in rhabdomyosarcomas, a tumor resulting from the dysregulated proliferation of muscle progenitor cells [52*].
Collectively, these studies demonstrate that the expression patterns of miRNAs are dramatically and distinctly altered during various types of cardiac and skeletal muscle disease and that the manipulation of disease-associated miRNAs represents a potentially powerful diagnostic and therapeutic approach to treat muscle disease.
Therapeutic manipulation of miRNAs with antagomiRs and miR mimics
Therapeutic manipulation of miRNAs represents a potentially powerful approach to treat cardiovascular and skeletal muscle diseases. In this regard, cholesterol-modified antisense oligonucleotides, referred to as antagomiRs, show remarkable efficacy in the inhibition of miRNAs following intravenous delivery in vivo [53]. Engelhardt and colleagues showed that delivery of an antagomiR to miR-21, a miRNA up-regulated in failing hearts, preserved cardiac function and reduced cardiac fibrosis following pressure overload, which they proposed to be mediated by up-regulation of sprouty homolog 1 (SPRY1), an inhibitor of MAP kinase signaling [54**]. Delivery of an antagomiR that blocks the activity of miR-133 was shown to induce cardiac hypertrophy, which was proposed to be mediated by the derepression of mRNAs encoding proteins involved in cytoskeletal rearrangements such as Cdc42 and RhoA [53,55]. However, genetic deletion of miR-133a-1 and miR-133a-2 did not result in cardiac hypertrophy [23**].
Why might an antagomiR evoke a different phenotype than genetic deletion of a miRNA? One possibility is that antagomiRs do not completely eliminate the targeted miRNA, in contrast to a genetic deletion. Thus, residual levels of a miRNA might be sufficient for certain functions. Alternatively, genetic deletion eliminates the expression of a miRNA throughout the embryogenesis and the lifetime of an organism, which might allow for compensatory pathways that are not operative in the transient setting of antagomiR-based inhibition. Finally, antagomiRs do not affect the accumulation of pre-miRNAs or miRNA passenger strands (also referred to as star strands), whereas most gene deletion strategies eliminate these sequences. Perhaps, these molecular intermediates have additional functions that are not completely understood. In this regard, the regulation of the processing and subcellular localization of miRNAs is beginning to be recognized as additional layers of potential regulation [56–59]. It will be interesting to determine if these additional types of miRNA regulation have any function in the settings of muscle biology and disease.
Alternatively, one could reconstitute the expression of a miRNA that is down-regulated in a disease through the delivery of a miR mimic, an oligonucleotide that contains a mature miRNA sequence and can function to repress endogenous target genes. Future studies will need to improve the pharmacodynamic properties and delivery of these modified oligonucleotides to achieve successful over-expression and knock-down of miRNAs in vivo.
Conclusions
Although many important findings have been made within the past few years, our current knowledge about the function of miRNAs in muscle development and disease is still quite limited given the multitude of possible interactions between transcription factors, miRNAs, and their target mRNAs. Future studies will need to focus on characterizing the in vivo functions of individual vertebrate miRNAs followed by the identification of their downstream target mRNAs. Identification of the miRNA expression signatures of human diseases represents a rich area of future research with the potential to identify new therapeutic targets. The sequence-specific actions of miRNAs coupled with recent advances in the delivery of modified oligonucleotides suggests that modulation of miRNA expression may constitute a new therapeutic approach to treat cardiac and skeletal muscle disease in the near future.
Acknowledgments
We apologize to the many researchers whose work was not cited in this review due to space limitations. We thank Jose Cabrera for graphics and Jennifer Brown for editorial assistance. Work in the laboratory of E.N.O. is supported by grants from the NIH, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Leducq Foundation, the Sandler Foundation for Asthma Research, and the Robert A. Welch Foundation. A.H.W. is supported by a training grant from the National Heart, Lung, and Blood Institute (T32HL007360). N.L. and E.v.R. were supported by grants from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*Publications of Special Interest
**Publications of Outstanding Interest
- 1.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 3.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 4.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.*.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. 7* and 8* These papers demonstrate that the over-expression or deletion of individual miRNAs evokes widespread but modest changes in protein expression. These authors conclude that miRNAs function to ‘fine-tune’ the expression of target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.*.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. See anotation to [7*]. [DOI] [PubMed] [Google Scholar]
- 9.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 12.**.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. This paper demonstrates that mice lacking Dicer in embryonic cardiomyocytes die due to defects in cardiac morphogenesis and contractility. The authors also demonstrate that mice lacking miR-1-2 display defects in cardiac myocyte proliferation and in the cardiac conduction system. [DOI] [PubMed] [Google Scholar]
- 13.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 15.O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarthy JJ. MicroRNA-206: The skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.*.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. This paper demonstrates a functional role for miR-1 and miR-133 in regulating skeletal myoblast differentiation and proliferation, respectively. The authors also show that these functions can be attributed to the repression of HDAC4 by miR-1 and SRF by miR-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.*.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. This paper identifies a MEF2-dependent intronic enhancer that directs muscle-specific expression of miR-1 and miR-133a. The authors show that MEF2 and SRF regulate cardiac expression whereas MEF2 and MyoD regulate skeletal muscle expression of miR-1 and miR-133a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 21.Niu Z, Iyer D, Conway SJ, Martin JF, Ivey K, Srivastava D, Nordheim A, Schwartz RJ. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci U S A. 2008;105:17824–17829. doi: 10.1073/pnas.0805491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.**.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008 doi: 10.1101/gad.1738708. This paper demonstrates that miR-133a-1 and miR-133a-2 function redundantly to repress smooth muscle gene expression and proliferation in cardiac myocytes. Genetic deletion of miR-133a-1 and miR-133a-2 leads to dilated cardiomyopathy and sudden death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.**.Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. This paper demonstrates that miR-1 and miR-133 are capable of inducing the differentiation of murine and human embryonic stem cells into the mesodermal lineage. The authors also show that the reconstitution of expression of miR-1 can partially rescue the mesodermal differentiation defects seen in Srf-deficient stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 26.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 28.*.Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. This paper identifies a role for miR-1 in regulating neuromuscular synapse function in C. elegans. The authors demonstrate that miR-1 can directly repress the expression of MEF2, which regulates the expression of a retrograde neurotrophic factor secreted from muscle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 30.Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem. 2008;283:20045–20052. doi: 10.1074/jbc.M801035200. [DOI] [PubMed] [Google Scholar]
- 31.Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, Yang B, Wang Z. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282:12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 32.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 35.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293:C451–457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 38.van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 39.**.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. This paper demonstrates that the cardiac-specific miRNA, miR-208, regulates stress-dependent cardiac hypertrophy. Mice lacking miR-208 are resistant to cardiac hypertrophy, fibrosis, and the up-regulation β-MHC gene expression in response to stress signals. [DOI] [PubMed] [Google Scholar]
- 40.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 43.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 45.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.*.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. This paper identifies the expression profile of miRNAs in hearts subjected to myocardial infarction. The authors demonstrate that miR-29 is down-regulated upon myocardial infarction and suppresses the expression of collagens, implicating miR-29 as a regulator of tissue fibrosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 49.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 50.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuasa K, Hagiwara Y, Ando M, Nakamura A, Takeda S, Hijikata T. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct. 2008;33:163–169. doi: 10.1247/csf.08022. [DOI] [PubMed] [Google Scholar]
- 52.*.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. This paper defines the expression and function of miR-29 during myoblast differentiation and in rhabdomyosarcomas. The authors demonstrate that miR-29 is down-regulated in rhabdomyosarcomas and that re-expression of miR-29 can inhibit tumor growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 54.**.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008 doi: 10.1038/nature07511. This paper demonstrates that the expression of miR-21 is up-regulated in failing human hearts and miR-21 is enriched in cardiac fibroblasts. The authors demonstrate that delivery of an antagomiR against miR-21 preserves cardiac function and prevents fibrosis following pressure overload in mice. [DOI] [PubMed] [Google Scholar]
- 55.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 56.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 57.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 59.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]