Abstract
In vitro experiments led to a simple model in which basal transcription factors sequentially assembled with RNA Polymerase II to generate a preinitiation complex (PIC). Emerging evidence indicates that PIC composition is not universal, but promoter-dependent. Active promoters are occupied by a mixed population of complexes, including regulatory factors such as NC2, Mot1, Mediator, and TFIIS. Recent studies are expanding our understanding of the roles of these factors, demonstrating that their functions are both broader and more context dependent than previously realized.
Introduction
In vitro studies have shown that transcription initiation by RNA Polymerase II (RNApII) minimally requires the basal initiation factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH [1,2]. These so-called general transcription factors (GTFs) mediate promoter recognition and unwinding, and together with RNApII and promoter DNA comprise the Pre-Initiation Complex (PIC). Given the strong conservation of RNApII and its initiation factors over evolution, it is rather surprising that no single DNA sequence element is found at all promoters [3•]. This implies that there must be multiple modes of promoter recognition, which in turn leads to two corollaries. First, there are likely to be multiple types of PICs (see figure) and second, that the GTFs may not be “general” in their functions. For this reason, we will refer to the initiation factors as “basal” rather than “general” transcription factors.
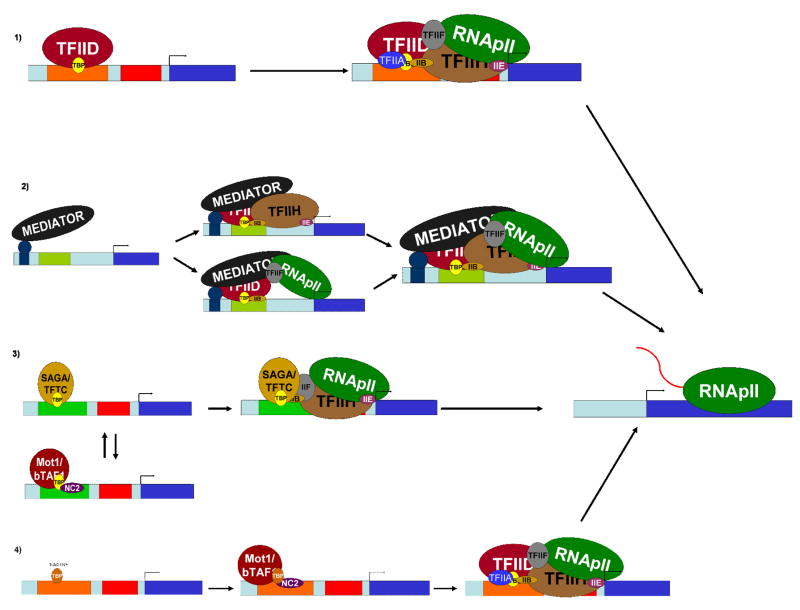
Figure. Many Paths to the PIC.
The factors and assembly pathways used to form transcriptionally competent preinitiation complexes can be promoter dependent [3,73]. 1) TBP assembling onto promoter regions via TFIID leads to recruitment of the other basal initiation factors, as outlined in the stepwise assembly pathway [1]. In S. cerevisiae, this pathway is most often utilized at TATA-less genes. At some mammalian promoters, histone H3K4 trimethylation helps to recruit the TFIID complex [5]. 2) Mediator bridges interactions between activators and the basal initiation machinery, and can stimulate basal transcription as well. At some promoters Mediator can recruit TFIIH and TFIIE independently of RNApII [45]. 3) TBP can also be brought to promoters by the SAGA complex. In S. cerevisiae, this pathway is most utilized at TATA containing promoters. The Mot1 and NC2 complexes can repress this pathway by actively removing TBP from the TATA element [73]. 4) Mot1 and NC2 can also have a positive role in transcription by removing nonproductive TBP complexes from DNA, thereby allowing functional PICs to form [60,65,77].
Different pathways to basal promoter recognition
Early studies identified the TATA element as a common basal promoter element. This sequence is recognized by the TATA-Binding Protein (TBP) subunit of TFIID, the first basal factor to engage the promoter. Indeed, TBP can support basal transcription in vitro without any of the other TAF subunits. However, as attention expanded beyond a small set of strong promoters it became clear that many promoters do not have a recognizable TATA element. Other basal elements identified include the downstream promoter element (DPE), TFIIB recognition elements (BREs), and the Initiator element (INR) [4]. Any given promoter may have one or more of these elements, but rarely are they all seen together. The other subunits of TFIID (the TBP-associated factors or TAFs) appear to interact with INR and DPEs. Besides promoter elements, specific chromatin modifications may play a role in basal factor recruitment. Recent work suggests that at some promoters, trimethylation at the lysine 4 residue of histone H3 stimulates binding of TFIID [5••]. Surprisingly, TAF1 promoter occupancy and gene expression levels correlate at only ~75% of genes [6], casting doubt upon the simple assumption that TFIID binding necessarily leads to transcription.
In higher eukaryotes, there are multiple genes encoding TBP-related factors (TRFs) and certain TAFs [1,7]. It is presumed that these subunits change the promoter specificity of the different TFIID variants, a model consistent with the fact that many of them are expressed in specific cell types or developmental stages. Deato et al. characterized a particularly striking example of TFIID variation [8••,9••]. They showed that differentiated muscle cells have very low levels of canonical TFIID and instead found that several muscle-specific genes instead utilize a complex consisting only of the TBP-like protein TRF3 and TAF3. TRF3 is also necessary for development of the hematopoietic lineage in zebrafish, where it activates key differentiation genes [10••]. It will be necessary to characterize the DNA binding properties of the different TFIID variants to see if their gene specificity is tied to specific promoter elements. Alternatively, they could be targeted using specific upstream activators or somehow compete with each other for similar sequences.
Another mechanism for delivering TBP may be via the SAGA complex, a factor better known as a histone acetyltransferase complex that is recruited to upstream activating sequences [11,12]. Both genetic and co-immunoprecipitation experiments suggested an interaction between SAGA and TBP [13,14], but a stable complex has not been isolated. However, recent crosslinking experiments support a direct interaction between the Spt3 subunit of SAGA and TBP in vivo [15••]. SAGA and TFIID have several subunits in common and are somewhat similar in overall shape [16,17], suggesting they may have evolved from a common ancestor. Gene expression studies in S. cerevisiae mutants indicate that ~10% of genes are dependent upon SAGA rather than TFIID for expression [18]. Interestingly, highly inducible genes with clear TATA boxes tend to be SAGA-dependent while TFIID appears to be preferentially used at housekeeping genes without recognizable TATA sequences [19].
The expanding PIC
The minimal set of basal factors does not respond to activators and is insufficient for transcription of chromatin templates, a finding that led to the discovery of a multitude of co-activators and chromatin-modifying complexes. To help define factors directly associated with the basal transcription machinery, a proteomics study of S. cerevisiae PICs was performed to find factors dependent upon TBP for promoter association. Most known components of the RNApII transcription machinery were found, as well as several novel components [20]. A new subunit of basal factor TFIIH (Tfb5) was discovered, and this protein is necessary for efficient transcription initiation and transcription-coupled repair (TCR) [21,22]. Surprisingly, PICs also contained the elongation factor TFIIS (discussed below) [23•]. In vitro studies of PIC assembly have been complemented by chromatin immunoprecipitation coupled with DNA microarray hybridization or deep sequencing (ChIP-ChIP or ChIP-Seq). As expected, basal factors are greatly enriched at active promoters [24•,25], although the correlation is not perfect [6]. Importantly, other factors such as TFIIS, NC2, and Mot1 also show correlation between promoter occupancy and transcription activity (discussed below).
In vitro assembly and in vivo crosslinking experiments have led to suggestions that different promoters utilize distinct subsets of basal factors, or that factors that inhibit transcription are paradoxically found in PICs and must therefore act positively. However, it should be noted that both of these experimental approaches analyze a complex mixture. Just because two factors both bind to immobilized templates or crosslink to the same promoter in vivo, this does not necessarily mean they are present at the same time in the same complex. Multiple complexes are dynamically assembling and disassembling, so promoters may be enriched for certain basal factors because of different rate limiting steps in PIC assembly. Transcription inhibitors may be present at promoters in complexes that are not on the reaction pathway to productive transcription.
Mediator in Activator-Independent Transcription
The Mediator complex was isolated as a co-activator that bridges regulatory factors and the basal machinery to allow high levels of activator-dependent transcription [1,26]. However, mounting evidence suggests that Mediator also contributes to basal (activator-independent) transcription, leading to a debate about whether Mediator should be classified as a GTF [27–33]. Some genomewide location analyses in S. cerevisiae and S. pombe found Mediator upstream of almost all active genes and some inactive genes [34,35], but under different growth conditions Mediator did not localize to many active promoters [36]. Thus, unlike the basal initiation factors and RNApII, the correlation between Mediator presence and transcription activity is less clear and Mediator functions may be promoter-specific.
Mediator is not required for basal transcription in purified systems, but can stimulate transcription in these systems even in the absence of activators (see [37] and references therein). It directly interacts with RNApII and its binding to immobilized in vitro templates is TBP-stimulated, suggesting it assembles with basal factors as a component of PICs. Although in vivo crosslinking suggests Mediator can be recruited to promoters prior to basal factors and RNApII [38–40] this early association is presumably mediated by activators bound at upstream sites followed by transfer of Mediator to the PIC at the basal promoter. The precise pathway of assembly could be promoter-dependent, explaining the variability in Mediator crosslinking. At some promoters, interdependent recruitment of TFIIB, Pol II, and Mediator in vitro suggests these factors are cooperatively recruited [41]. Indeed, “holoenzyme” forms of Pol II have been co-purified with various Mediator components and basal factors (see [1] and references therein). However, in other in vitro systems Mediator binding is required for TFIIB recruitment but not vice versa [31]. Future work is still needed to determine if specific promoter elements, cofactors, or growth conditions drive a given assembly pathway.
Irrespective of how it is recruited, how does Mediator promote basal transcription? One mechanism may involve stimulating phosphorylation of the RNApII largest subunit C-terminal domain (CTD) by TFIIH kinase [42]. Although CTD phosphorylation is not required for initiation, this modification leads to release of RNApII from Mediator and so may promote escape into elongation [43•]. Mediator may also directly stabilize PIC assembly intermediates [31,44]. Yeast strains with conditional mutations of the essential Mediator subunit Med11 have reduced RNApII occupancy, but normal levels of TFIIE and TFIIH at several constitutively active promoters [45••], suggesting these promoters may not follow the classical stepwise assembly pathway in which TFIIE and TFIIH are dependent upon RNApII for incorporation into the PIC (see [1] and references therein). A different point mutation of Med11 decreased the occupancy of the TFIIK submodule of TFIIH at some but not all active promoters, suggesting that Mediator’s role in PIC recruitment could be promoter dependent as well [45••]. Importantly, Mediator has been shown to stabilize a subcomplex of basal factors at the promoter after initiation in vitro. This “Scaffold” would then promote subsequent reinitiations [44,46]. The imbalance seen in Med11 strains between RNApII and basal factors TFIIE and TFIIH might be due to an inability to utilize Scaffolds for reinitation in vivo.
Importantly, multiple forms of Mediator can exist within cells, each bearing slightly different subunit compositions and stoichiometries [26]. Future research will focus on where each of these specific forms is recruited, and how they differ in function. B-Med, a form of Mediator isolated from mammalian cell extracts, has been shown to specifically regulate basal transcription in vitro [29,30], although it remains to be seen if this is a physiologically relevant form of Mediator in vivo.
TFIIS: An Elongation Factor’s New Role at the Promoter
TFIIS is a well-characterized transcription elongation factor that allows arrested RNApII elongation complexes to backtrack via RNA cleavage and generation of a new 3′ transcript end [47]. Surprisingly, a growing body of evidence indicates this protein also plays a role in initiation. In vitro, TFIIS was found in complexes containing RNApII and basal factors TFIIB and TFIIE [48,49], it can directly interact with the promoter-associated factors Spt8 and Med13 [50], and it associated with in vitro assembled PICs [23•]. In vivo, deletion of TFIIS is synthetic lethal with loss of subunits from Mediator and the Swi/Snf chromatin remodeling complex [51,52]. Furthermore, TFIIS was recruited to the promoter of the galactose-inducible gene Gal1 dependent upon Mediator and SAGA but not RNApII. Loss of TFIIS resulted in reduced recruitment of TBP and Pol II to the GAL1 promoter in vivo [53].
Yeast TFIIS stimulates in vitro PIC formation on the HIS4 promoter [23•]. This initiation function requires the TFIIS polymerase interaction domain [54], but is independent of transcript cleavage activity [23•]. The TFIIS N-terminal domain, which may interact with Mediator and SAGA subunits, also contributes to PIC assembly in vitro. In agreement, although deletion of TFIIS and Med31 are synthetically lethal, this can be complemented by expression of a TFIIS truncation that interacts with RNApII but does not stimulate elongation [55•]. A single point mutation in the RNApII interaction domain of TFIIS decreases polymerase recruitment to three promoters tested in vivo. In mammalian cells, transcriptional activators are required for TFIIS-mediated induction of some reporter genes, but not others [56], however whether this is related to TFIIS’ role in initiation is unclear.
Interestingly, TFIIS has also recently been shown to promote accurate transcription initiation by RNA Polymerase III (RNApIII), although it appears transcript cleavage activity may be important in this case [57••]. It is unexpected that the RNApIII system would involve TFIIS because the RPC11 subunit of this polymerase is thought to be homologous to TFIIS [58]. Mammalian genomes contain several TFIIS orthologues, some of which are expressed in a tissue specific manner [47]. It’s interesting to speculate about whether multiple TFIIS molecules function in initiation and elongation independently.
NC2 and BTAF1/Mot1: Repressors at the PIC
Two transcription repressors, Mot1/BTAF1 and NC2, act through direct interactions with TBP. Mot1/BTAF1 is a Snf2 family ATPase that removes TBP from promoters. It also behaves genetically as a repressor. NC2 is a heterodimer that blocks TFIIA and TFIIB from associating with the TBP-TATA complex. Its genetic properties are also consistent with transcription repression (see [59,60] and references therein).
Paradoxically, there have been indications that Mot1/BTAF1 and NC2 can positively affect gene expression in some contexts. Genomewide crosslinking analyses in yeast show that Mot1, NC2 and TBP are all found at most active promoters [25,61–65]. In mammalian cells, genomewide occupancy of the NC2α subunit correlates with gene activity as well [66•]. The correlation between Mot1 and NC2 is particularly high (>97%) [65••], and the proteins can physically interact [67], suggesting they might function together. Microarray expression analyses in yeast suggests that about 10% of genes are upregulated by Mot1 [63], and about 8% of genes by NC2 [61].
To explain the apparent paradox of inhibitory complexes binding to active promoters, it has been proposed that these repressors (particularly Mot1) displace TBP from cryptic TATA sequences or other inappropriate genomic locations in order to make it available to weaker promoters [62,63,68]. Several recent reports support this model. NC2 alters the conformation of the TBP/DNA complex, allowing it to slide along DNA away from TATA boxes [69•]. TBP mutants with decreased ability to form PICs suppress the gene expression defects seen in a mot1 mutant [70•]. FRAP experiments show that the rapid exchange of TBP associated with chromatin is dependent upon Mot1 [71].
Further supporting the promoter redistribution model is the observation that basal promoter sequence strongly affects response to the repressors. In yeast, NC2 and Mot1 repress TATA-containing and activate TATA-less promoters [62,72,73•]. In metazoans, NC2 binding is antagonized by the presence of INR [74•] and BREs [66•,75•]. In Drosophila extracts, NC2 stimulates transcription from DPE-containing promoters, but represses TATA -containing promoters [76]. Manipulation of TBP, Mot1, and NC2 levels in vivo also show opposing effects on TATA versus DPE promoters, leading to the suggestion that NC2 and Mot1 stimulate DPE-dependent transcription by removing TBP from these promoters [77••]. The exact biochemical function of Mot1 and NC2 in this context remains unclear but one interesting possibility to be explored is whether removal of TBP could allow binding of alternative TBP-related factors or TAF complexes to these promoters.
In addition to facilitating transfer of TBP between promoters, NC2 and Mot1 could upregulate expression by displacing transcriptionally inactive forms of TBP from promoters. A dynamic exchange of positive and negative complexes may allow rapid response to physiological signals rapidly [65••,73•]. ChIP experiments at Mot1-dependent promoters show reduced PIC assembly despite increased TBP levels following loss of Mot1 function [78]. NC2 mutants show similar decreases of PIC components at active promoters, although its unclear if the mechanism is related to the removal of inactive TBP from these promoters [79]. Recent RNA sequencing studies have shown that most eukaryotic promoters produce the expected transcripts but also a set of short unstable transcripts synthesized in the opposite direction [80–83•]. This suggests that basal promoter regions often contain multiple TBP-binding sites or are largely bidirectional [84•]. It has recently been shown that Mot1 can remove TBP bound in the “wrong” direction to free the promoter for productive TBP binding [85•].
Of course, it remains possible that NC2 and Mot1 directly participate in PIC formation. Although these repressors are not typically found in complexes with basal factors other than TBP, ChIP experiments suggest Mot1 can co-occupy promoters with TFIIB and Pol II under heat stress conditions [64]. Mot1 acts in conjunction with SAGA to remodel chromatin at the Gal1 promoter [86] and can physically interact with Mediator and several other chromatin remodeling complexes [87]. Although these observations are not easy to reconcile with structural studies, future experiments may reveal new surprises.
Conclusion and Future Prospects
It is becoming increasingly clear that transcription initiation at basal promoters is not a simple linear reaction. Recent genome and proteome scale analyses of active promoters implicate multiple factors that can both positively and negatively regulate initiation. Assembly pathways may be branched with several non-productive complexes leading to transcription inhibition. In the future it will be important to consider the dynamics of PIC assembly, since chromatin immunoprecipitation and proteomic studies do not provide temporal resolution for distinguishing multiple complexes that can occupy promoters in a population of cells. While non-basal factors such as TFIIS, NC2 and Mot1 are found at most promoters in vivo, loss of function only affects a small subset of genes. Progress is being made in defining the promoter sequences that determine responsiveness. As there appear to be many varieties of basal sequence elements, it will not be surprising to find heterogeneity in the factors present.
Table 1.
Complexes Involved in RNApII PIC assembly
| Protein Complex | Functions |
|---|---|
| RNApII | 12 Subunits; catalyzes transcription of all mRNAs and a subset of noncoding RNAs including snoRNAs and miRNAs |
| TFIIA | 2–3 subunits; functions to counteract repressive effects of negative cofactors like NC2; acts as a coactivator by interacting with activators and components of the basal initiation machinery |
| TFIIB | Single subunit; stabilizes TFIID-Promoter binding; aids in recruitment of TFIIF/Pol II to the promoter; directs accurate start site selection |
| TFIID | 14 subunits including TBP and TBP Associated Factors (TAFs); nucleates PIC assembly either through TBP binding to TATA sequences or TAF binding to other promoter sequences; coactivator activity through direct interaction of TAFs and gene specific activators |
| TFIIE | 2 subunits; helps recruit TFIIH to promoters; stimulates helicase and kinase activities of TFIIH; binds ssDNA and is essential for promoter melting |
| TFIIF | 2–3 subunits; tightly associates with RNApII; enhances affinity of RNApII for TBP-TFIIB-promoter complex; necessary for recruitment of TFIIE/TFIIH to the PIC; aids in start site selection and promoter escape; enhances elongation efficiency |
| TFIIH | 10 subunits; ATPase/helicase necessary for promoter opening and promoter clearance; helicase activity for transcription coupled DNA repair; kinase activity required for phosphorylation of RNApII CTD; facilitates transition from initiation to elongation |
| Mediator | At least 24 subunits; bridges interaction between activators and basal factors; stimulates both activator dependent and basal transcription; required for transcription from most RNApII dependent promoters |
| SAGA | 20 subunits; interacts with activators, histone H3, and TBP; histone acetyltransferase activity; deubiquitinating activity |
| Trf1 | TBP related factor identified in Drosophila; upregulated in CNS and gonads during development; can bind TATA sequences; mostly found at RNApIII dependent promoters as part of TFIIIB but also required at a subset of RNApII dependent promoters |
| Trf2 | TBP related factor identified in all metazoans; cannot bind TATA sequences; important for histone gene expression in Drosophila |
| Trf3 | TBP related factor identified in vertebrates; can bind TATA sequences; important for differentiation of muscle cells in mammals and for haematopoietic cell development in zebrafish. |
| TFIIS | 1 subunit; stimulates intrinsic transcript cleavage activity of RNApII allowing backtracking to resume RNA synthesis after transcription arrest; stimulates PIC assembly at some promoters |
| NC2 | 2 subunits; binds TBP/DNA complexes and blocks PIC assembly; can have both positive and negative effects on transcription |
| Mot1/bTAF1 | 1 subunit; induces dissociation of TBP/DNA complexes in ATP dependent manner; can have both positive and negative effects on transcription |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Reese JC. Basal transcription factors. Curr Opin Genet Dev. 2003;13:114–118. doi: 10.1016/s0959-437x(03)00013-3. [DOI] [PubMed] [Google Scholar]
- 3•.Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. This review discusses the large variety of promoter types found throughout the genome and provide models for how diverse sets of promoter recognition factors are utilized to match these core promoter elements. [DOI] [PubMed] [Google Scholar]
- 4.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. This work shows that the TAF3 subunit of TFIID can selectively bind to histone H3 trimethylated at lysine 4, and loss of this modification reduces TFIID binding to and transcription from a subset of promoters in vivo. [DOI] [PubMed] [Google Scholar]
- 6.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reina JH, Hernandez N. On a roll for new TRF targets. Genes Dev. 2007;21:2855–2860. doi: 10.1101/gad.1623207. [DOI] [PubMed] [Google Scholar]
- 8••.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. These papers demonstrate that the loss of TFIID and its replacement by the novel TAF3/TRF3 core promoter complex is required for activation of MyoD responsive genes and the differentiation of muscle cell precursors into myotubes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. This work shows the TBP related factor TRF3 is required for development of the haematopoietic system in the zebrafish embryo and provides a pathway from TRF3 to hematopeoietic specific gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel JA, Grant PA. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res. 2007;618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laprade L, Rose D, Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–2017. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 15••.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. The authors insert a nonnatural photoreactive amino acid at specific sites on TBP to map direct protein-protein interactions in the context of the PIC. Using this technique they identify a direct interaction between TBP and Spt3, and demonstrate that the interaction is necessary for optimal activation at SAGA dependent promoters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. Mapping histone fold TAFs within yeast TFIID. Embo J. 2002;21:3424–3433. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 19.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 20.Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R. The study of macromolecular complexes by quantitative proteomics. Nat Genet. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 21.Ranish JA, Hahn S, Lu Y, Yi EC, Li XJ, Eng J, Aebersold R. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet. 2004;36:707–713. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- 22.Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NG, Raams A, Argentini M, van der Spek PJ, et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 23•.Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci U S A. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. The authors identify TFIIS as a component of PICs in a proteomic screen. TFIIS stimulates PIC assembly and transcription in vitro, and IIS cleavage activity is dispensable for this function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Venters BJ, Pugh F. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009 doi: 10.1101/gr.084970.108. The data presents a genomewide localization map of a diverse set of proteins involved in transcription initiation, including sequence specific activators, chromatin remodelers, mediator, and basal factors, in the context of nucleosome positions. The authors suggest a model for PIC assembly and concurrent nucleosome removal at promoters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89:1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci U S A. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 29.Mittler G, Kremmer E, Timmers HT, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu SY, Zhou T, Chiang CM. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol Cell Biol. 2003;23:6229–6242. doi: 10.1128/MCB.23.17.6229-6242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 32.Lewis BA, Reinberg D. Promoter activation when the ChIPs are down. Nat Struct Mol Biol. 2006;13:96–97. doi: 10.1038/nsmb0206-96. [DOI] [PubMed] [Google Scholar]
- 33.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 34.Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 37.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Bhoite LT, Yu Y, Stillman DJ. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 40.Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell. 2001;7:1213–1220. doi: 10.1016/s1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 41.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 43•.Max T, Sogaard M, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282:14113–14120. doi: 10.1074/jbc.M701345200. This work shows that phosphorylation of the CTD of RNApII is sufficient to dissociate the RNApII complex from Mediator in vitro. [DOI] [PubMed] [Google Scholar]
- 44.Reeves WM, Hahn S. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol Cell Biol. 2003;23:349–358. doi: 10.1128/MCB.23.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. This work provides evidence for a direct interaction between Mediator and TFIIH, and suggests that at some promoters, TFIIH and TFIIE can be recruited independently of RNA Polymerase II. [DOI] [PubMed] [Google Scholar]
- 46.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 47.Wind M, Reines D. Transcription elongation factor SII. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 49.Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 50.Wery M, Shematorova E, Van Driessche B, Vandenhaute J, Thuriaux P, Van Mullem V. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. Embo J. 2004;23:4232–4242. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davie JK, Kane CM. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Mol Cell Biol. 2000;20:5960–5973. doi: 10.1128/mcb.20.16.5960-5973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malagon F, Tong AH, Shafer BK, Strathern JN. Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation-elongation transition. Genetics. 2004;166:1215–1227. doi: 10.1534/genetics.166.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prather DM, Larschan E, Winston F. Evidence that the elongation factor TFIIS plays a role in transcription initiation at GAL1 in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:2650–2659. doi: 10.1128/MCB.25.7.2650-2659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 55•.Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci U S A. 2007;104:16062–16067. doi: 10.1073/pnas.0704534104. This paper demonstrates a role for TFIIS in stimulation of PIC assembly at several active promoters in vivo. This function is particularly important in strains containing mutations of Mediator subunits, thereby explaining previous genetic interactions between these two factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ito T, Saso K, Arimitsu N, Sekimizu K. Defective FESTA/EAF2-mediated transcriptional activation in S-II-deficient embryonic stem cells. Biochem Biophys Res Commun. 2007;363:603–609. doi: 10.1016/j.bbrc.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 57••.Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. This work provides a genomewide occupancy map of TFIIS in S. cerevisiae, and finds that it localizes to most RNA Polymerase III genes. In vitro and in vivo data suggest TFIIS is important for initiation for these genes, possibly by aiding in recruitment of TFIIIB and RNApIII to the promoters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pereira LA, Klejman MP, Timmers HT. Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription. Gene. 2003;315:1–13. doi: 10.1016/s0378-1119(03)00714-5. [DOI] [PubMed] [Google Scholar]
- 60.Auble DT. The dynamic personality of TATA-binding protein. Trends Biochem Sci. 2008 doi: 10.1016/j.tibs.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geisberg JV, Holstege FC, Young RA, Struhl K. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci U S A. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geisberg JV, Moqtaderi Z, Kuras L, Struhl K. Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:8122–8134. doi: 10.1128/MCB.22.23.8122-8134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geisberg JV, Struhl K. Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Mol Cell. 2004;14:479–489. doi: 10.1016/j.molcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 65••.van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008;22:2359–2369. doi: 10.1101/gad.1682308. A genomewide occupancy map of the NC2 complex and Mot1 demonstrates strong co-localization of NC2, Mot1 and TBP at many active promoters. A complex composed of NC2, Mot1, TBP and DNA was isolated from chromatin extracts suggesting these complexes function together to regulate promoter bound TBP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Albert TK, Grote K, Boeing S, Stelzer G, Schepers A, Meisterernst M. Global distribution of negative cofactor 2 subunit-alpha on human promoters. Proc Natl Acad Sci U S A. 2007;104:10000–10005. doi: 10.1073/pnas.0703490104. A genomewide analysis of NC2 localization at promoters in mammalian cells shows that NC2 promoter binding correlates positively with gene expression levels, and that NC2 occupancy is negatively correlated with the presence of TFIIB recognition elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klejman MP, Pereira LA, van Zeeburg HJ, Gilfillan S, Meisterernst M, Timmers HT. NC2alpha interacts with BTAF1 and stimulates its ATP-dependent association with TATA-binding protein. Mol Cell Biol. 2004;24:10072–10082. doi: 10.1128/MCB.24.22.10072-10082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li XY, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 69•.Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol. 2007;14:1196–1201. doi: 10.1038/nsmb1328. Using a variety of biophysical methods, the authors find that NC2 induces rapid dynamic changes in the conformation of the TBP–DNA complex, and can mobilize TBP away from TATA regions of promoters. The data suggest that NC2 not only binds TBP to inhibit PIC formation, but also to mobilize TBP on the DNA. [DOI] [PubMed] [Google Scholar]
- 70•.Sprouse RO, Wells MN, Auble DT. TATA-binding protein variants that bypass the requirement for Mot1 In Vivo. J Biol Chem. 2008 doi: 10.1074/jbc.M808951200. A screen for TBP mutants in yeast finds that strains with defects in PIC assembly no longer require Mot1. The authors suggest that Mot1 may help to maintain appropriate levels of PIC instability which is critical for the regulation of transcription in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, Auble DT. Regulation of TATA-binding protein dynamics in living yeast cells. Proc Natl Acad Sci U S A. 2008;105:13304–13308. doi: 10.1073/pnas.0801901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc Natl Acad Sci U S A. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Huisinga KL, Pugh BF. A TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol. 2007;8:R46. doi: 10.1186/gb-2007-8-4-r46. The authors use a computational approach to model PIC assembly regulation in terms of defined biochemical interactions that regulate the function of TBP. Perturbations to the TBP regulatory network are simulated and tested experimentally via genetic mutations. The work shows that pathways to PIC assembly can be modeled with biochemically defined regulatory pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Malecova B, Gross P, Boyer-Guittaut M, Yavuz S, Oelgeschlager T. The initiator core promoter element antagonizes repression of TATA-directed transcription by negative cofactor NC2. J Biol Chem. 2007;282:24767–24776. doi: 10.1074/jbc.M702776200. This work shows that presence of INR elements on TATA containing promoters (but not TATA-less promoters) confers resistance of these promoters to NC2 mediated inhibition in vitro. This is an example of how a specific set of promoter elements restrict the binding of a transcriptional regulatory factor. [DOI] [PubMed] [Google Scholar]
- 75•.Deng W, Malecova B, Oelgeschlager T, Roberts SG. TFIIB recognition elements control the TFIIA-NC2 axis in transcriptional regulation. Mol Cell Biol. 2008 doi: 10.1128/MCB.01346-08. The authors find that TFIIA preferentially associates with BRE-containing promoters while NC2 is recruited to promoters that lack consensus BREs. Moreover, TFIIA assembly at BRE-containing promoters results in reduced transcriptional activity, while NC2 acts as a positive factor at promoters that lack functional BREs, suggesting a model where by promoter sequence elements can direct the positive or negative functions of initiation factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 77••.Hsu JY, Juven-Gershon T, Marr MT, 2nd, Wright KJ, Tjian R, Kadonaga JT. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. The work shows that TBP overexpression inhibits DPE-dependent transcription Depletion of Mot1 and NC2 inhibited transcription more strongly from DPE-dependent promoters compared to TATA-dependent promoters. The authors suggest a model whereby NC2 and Mot1 stimulate DPE-dependent transcription by removing TBP from these promoters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dasgupta A, Juedes SA, Sprouse RO, Auble DT. Mot1-mediated control of transcription complex assembly and activity. Embo J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Masson P, Leimgruber E, Creton S, Collart MA. The dual control of TFIIB recruitment by NC2 is gene specific. Nucleic Acids Res. 2008;36:539–549. doi: 10.1093/nar/gkm1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80•.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81•.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82•.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 83•.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. This group of papers describes a new class of transcripts that initiate near the expected transcription start sites of protein-encoding genes. These RNAs are short, present at low abundance, and often are transcribed in the opposite direction of the protein-encoding mRNA. This challenges our models of how promoter sequence directs initiation of transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84•.Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. Embo J. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. The authors provide an analysis of TBP binding sites in human cells, and then obtain binding profiles for 26 transcription factors at these locations, including several subunits of the basal initiation machinery. The authors find many TBP binding sites outside of canonical promoter regions, including in introns, and provide distinct profiles for basal factors at promoters in CpG and non-CpG islands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85•.Sprouse RO, Shcherbakova I, Cheng H, Jamison E, Brenowitz M, Auble DT. Function and structural organization of Mot1 bound to a natural target promoter. J Biol Chem. 2008;283:24935–24948. doi: 10.1074/jbc.M803749200. The authors show that TBP can bind in the wrong orientation at some promoters in vitro, which inhibits PIC formation. Moreover, Mot1 functions positively at these promoters in part by facilitating redistribution of TBP binding orientation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Topalidou I, Papamichos-Chronakis M, Thireos G, Tzamarias D. Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. Embo J. 2004;23:1943–1948. doi: 10.1038/sj.emboj.7600199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arnett DR, Jennings JL, Tabb DL, Link AJ, Weil PA. A proteomics analysis of yeast Mot1p protein-protein associations: insights into mechanism. Mol Cell Proteomics. 2008;7:2090–2106. doi: 10.1074/mcp.M800221-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]