Abstract
Further advances in the prevention, diagnosis and treatment of cancer requires a more complete knowledge of the molecular mechanisms that program the malignant state. Until recently, identifying and validating genetic alterations in tumors that contribute to cancer involved painstaking efforts focused primarily on single mutations. However, the application of whole genome approaches to the study of cancer now makes it possible to contemplate performing systematic characterizations of the structural basis of cancer by identifying mutations associated with each cancer type. In parallel, recent technological advances also make it possible to methodically characterize the function of putative oncogenes and tumor suppressor genes. The integration of these approaches now provides the means to not only derive a complete molecular description of cancer but will also provide well-validated targets for the development of therapeutic agents.
Keywords: Cancer, genomics, mutation, integration
1. Introduction
Cancer develops from normal tissues through a stepwise accumulation of genetic mutations. These mutations induce changes in both incipient tumor cells and surrounding cells that program initial neoplastic growth and full malignant transformation. Work in many laboratories over the past four decades has revealed and validated the role of some of the key genetic alterations that are responsible for oncogenesis in particular tissues. Indeed, the identification of oncogenic mutations such as BCR-ABL and activating mutations of EGFR has led to the development of molecularly targeted therapies now employed in the clinic. Despite these advances, it is clear that we continue to lack a full understanding of the mutations that drive cancer development for the majority of human cancer types.
In part, this situation exists because most human tumors, particularly those derived from epithelial cancers, exhibit global genomic alterations that make it difficult to identify mutations critical for cell transformation and to define the consequences of specific cancer-associated mutations. Moreover, since multiple mutations are required to program the transformed phenotype, dissecting how these mutations interact remains a critical step in not only understanding the molecular basis of cancer but also in applying this knowledge to develop effective anti-neoplastic therapeutics.
However, recent advances in sequencing technologies and comprehensive methods to map cancer-associated amplicons and deletions now make it possible to envision enumerating all of the genetic alterations harbored by a particular tumor. For example, advances in DNA sequencing technology have dramatically decreased the cost of sequencing while increasing throughput, and array-based technologies enable the systematic identification of recurrent regions of chromosomal amplification and deletion. Widespread implementation of these and related technologies has already begun to clarify the number and types of genetic alterations that occur in cancer genomes [1].
However, despite these advances in annotating structural alterations in cancer genomes, identifying the genes targeted by specific mutational, amplification or deletion events and deciphering the function of targeted gene mutations remains a major challenge. Thus, the parallel development of efficient methods to annotate the function of cancer-associated genes is necessary to distill validated cancer targets from this structural description of cancer genomes. The discovery that RNA interference (RNAi) operates in mammalian cells [2] and the collection and annotation of large collections of human cDNAs [3, 4] now provide a complementary systematic approach to interrogate the function of genes involved in cancer.
In this review we focus on recent advances in the application of genomics to the study of cancer. The integration of these approaches promises both to provide a comprehensive understanding of the molecular events that lead to cancer but will also provide the means to design novel therapeutic strategies.
2. Structural alterations in cancer genomes
2.1 Mutations and cancer genome sequencing
The RAS proto-oncogene was one of the first human oncogenes discovered [5]. Although changes in gene copy number or protein expression are found in some tumors [6], the early application of DNA sequencing revealed that a single nucleotide change in the proto-oncogene led to an amino acid substitution at an position that rendered the mutant protein constitutively active [7]. Subsequent studies have shown that a number of proto-oncogenes are converted into oncogenes through the acquisition of somatic mutations [8].
Based on these observations, several groups have applied capillary-based DNA sequencing technologies to analyze putative oncogenes that had been implicated in signaling pathways important for tumor cell proliferation and survival. For example, complete sequencing of RAS and the known RAS effector BRAF in 530 cancer cell lines and 378 tumor samples confirmed that RAS mutations occur in 15% of melanomas and 35% of colorectal cancers but also identified a high incidence of BRAF mutations (66%) [9]. Similarly, directed sequencing of genes that make up the phosphatidylinositol 3-kinase family (PI3K) of lipid kinases identified activating mutations in PIK3CA in colorectal, breast, brain and gastric cancers [10].
Directed sequencing of EGFR and kinases in lung cancer also led to the identification of recurrent mutations in EGFR in a significant subset of lung tumors [11-13]. Such tumors were more likely to have been derived from non-smoking women of Asian ancestry. Indeed, prior clinical trials had suggested that this subgroup of patients were more likely to respond to treatment with inhibitors of EGFR. Similarly, sequencing of kinases in neuroblastoma revealed activating mutations in ALK in 8% of primary tumors [14], and tumors harboring such mutations show increased sensitivity to ALK kinase inhibitors.
Based on these experiences, several groups have initiated efforts to sequence larger panels of genes in a range of cancer types. These efforts have confirmed the frequency of many of the known oncogenic mutations in kinases [15] and other classes of tumor suppressors and oncogenes [16-20]. In addition, deep sequencing of genes implicated in cancer development in specific cancer types such as glioblastomas has begin to reveal that even if mutations of a particular gene are present in only a fraction of tumors, that mutations in other members of the signaling pathway are often found in other cancers, suggesting that perturbation of certain pathways such as the retinoblastoma pathway are essential for cancer development [20]. Advances in sequencing technologies now make it possible to contemplate the complete sequencing of entire cancer genomes; indeed, the complete sequencing of the genome of tumor and skin cells derived from a patient with M1 acute myeloid leukemia identified 10 mutations, 8 of which were in genes not previously recognized as cancer genes [21].
However, these initial studies also identified a large number of mutations for which it remains unclear whether such mutations contribute to cancer development either because they occur at low frequency or have yet to be functionally characterized. Thus these initial studies confirm that a full characterization of cancer genomes will require a substantial increase in both the number of genes and tumors analyzed to achieve statistically meaningful results. The development of highly parallel approaches for DNA sequencing [22, 23] will certainly facilitate such studies but access to well-annotated cancer specimens remains a significant challenge.
Clinically, the observation that mutations in specific oncogenes such as EGFR or ALK predict response to targeted agents has led to the planning of clinical studies to test whether such testing will permit the selection of patients who are likely to respond to therapy. At the same time, recent results suggest that the presence of particular mutations, such as oncogenic mutations of KRAS, identify tumors that show little or no response to targeted agents [24]. The recent demonstration that mass spectrometric methods to identify sequence variations can be adapted for use in high throughput identification of known oncogenic mutations [25] will facilitate the identification of such predictive mutations until the means to perform full genome sequencing becomes possible.
2.2 Whole genome characterization of copy number alterations
In addition to sequence alterations, it has long been recognized that cancer genomes harbor numerous regions of copy number gain and loss. Indeed, many known oncogenes such as MYC and HER2 exhibit copy number gain more frequently than mutations and tumor suppressor genes are classically defined by loss of heterozygosity. Several whole genome techniques, comparative genome hybridization (CGH), high-density single nucleotide polymorphism (SNP) arrays and representational oligonucleotide microarray analysis (ROMA), permit the detection of chromosomal regions exhibiting decreased or increased copy number at high resolution [26-28]. These technologies have recently been used to identify genes and regions commonly altered in a wide range of cancers such as lung cancer [29, 30], breast cancer [31] and melanoma [32]. These studies have confirmed that some well defined regions of amplification or deletion target known oncogenes and tumor suppressor genes. In addition, the development of analytical tools together with the analysis of large numbers of samples has led to the identification of new oncogenes such as NKX2.1 [30] and MITF [32] in lung cancer and melanoma, respectively.
In addition, beyond the identification of specific oncogenes and tumor suppressor genes, several investigators have used these approaches to determine whether the pattern of recurrent alterations predicts the response to treatment. Indeed both whole genome copy number analyses and transcriptional profiles have been used to predict the response of cancer cell lines to specific chemotherapeutic agents [33, 34]. Moreover, recent work has shown that these methods can be used to demonstrate correlation between the presence of recurrent alterations and patient prognosis for breast cancer [35-38].
Although current analytical methods now permit identification of statistically significant regions of recurrent amplification and deletion [39], many of the regions identified by these approaches harbor dozens of candidate genes. Thus, the application of these methods to larger sample sets will provide a better definition of driver genes within these regions of copy number change. In addition, recent work in which the information obtained using these approaches have been combined with other types of analyses indicate that integrated methods will facilitate the identification and validation of oncogenes and tumor suppressor genes. These approaches will be discussed in Section 4.
2.3 Translocations in cancer
In many hematopoietic and pediatric malignancies, detailed analyses of chromosome structure have facilitated the identification of genetic alterations due to the translocation of sequences from one chromosome to another. Work from many laboratories has identified and characterized these chromosomal breakpoints. In some cases, such as in chronic myelogenous leukemia (CML), these translocations results in the creation of a fusion protein (BCR-ABL) that is constitutively active. In many other cancers, these translocations involve transcription factors, such as AML-ETO [40] and E2A-PBX1 [41] in subsets of leukemia. Although the pathways perturbed by these fusion proteins is known in some cases, identifying the functional consequences of these translocations continues to be an area of active research.
Until recently, due to the bewildering complexity of chromosomal aberrations found in epithelial cancers, most investigators have focused on the analysis of chromosomal translocations found in hematopoietic and pediatric cancers. However, recent work has identified similar types of translocations in lung and prostate cancers. Specifically, Soda et al. identified a cDNA from a lung cancer patient that encoded for an EML4–ALK fusion gene [42]. When expressed in the BaF3 experimental model, this fusion gene, which was found in approximately 6% of Japanese patients with lung cancer, conferred cytokine independence as well as sensitivity to inhibitor with ALK inhibitors.
Although successful, the identification of fusion proteins from expression libraries is technically difficult, in part due to the low levels of expression of such fusion genes. Recent work involving the application of analytical tools to study gene expression profiles derived from tumors has facilitated the identification of translocations in prostate and lung cancer. Specifically, Chinnaiyan and colleagues reasoned that evaluating variance in a data set using the median instead of the mean would maintain the peaks of outliers in expression profiling datasets [43]. Using this analytical method, called cancer outlier profile analysis (COPA), they identified recurrent rearrangements involving the 5′ regulatory sequences of the androgen regulated gene TMPRRS2 and members of the ETS family of transcription factors (ERG or ETV1) [44]. Subsequent work showed that these rearrangements are found in a large fraction of human prostate cancers. Taken together, these observations suggest that other such translocations also occur in other epithelial cancers, and that the application of COPA and paired-end high throughput sequencing will make it possible to discover translocations.
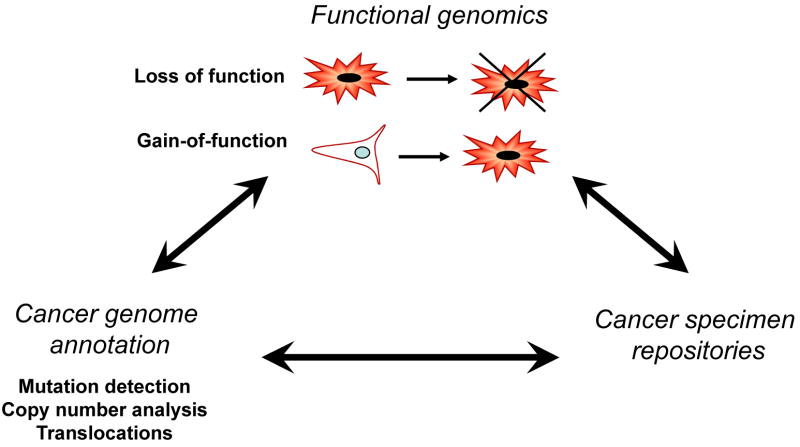
3. Functional approaches to deciphering cancer genomes
As described in the previous sections, technological advances will enable the construction of comprehensive views of genetic alterations in cancer genomes, and will, in many cases, identify specific genes that drive cancer development or progression. However, since most tumors exhibit hundreds of genetic alterations, deciphering the specific alterations necessary to program malignant transformation remains a significant challenge. In recent years, several laboratories have developed complementary high throughput approaches to manipulate gene expression in experimental models. The basic tenet of these functional genomics studies is that by perturbing the activity of a gene, one can gain insight into its biological functions by assessment in phenotypic assays.
3.1 Gain of function approaches
One approach to investigate gene function is to overexpress genes and determine the consequences in specific assays. Although cDNA libraries have been used for many years [45-47], early efforts relied on DNA transfection and thus required strategies that employed many rounds of selection to enrich for the gene of interest. Over the past several years, several groups have created expression libraries in retroviral vectors, which permit higher efficiency gene transduction [48].
In particular, such retroviral libraries have been used to identify genes that when overexpressed bypass the proliferative arrest induced by various stimuli. For example, this approach has been used to identify TBX2 as a gene amplified in breast cancer that permits cells to proliferate in Bmi-deficient fibroblasts [49], DRIL1 as a gene that bypasses RAS induced senescence [50], and BCL6 as a gene that permits proliferation in the presence of active p19ARF/p53 signaling [51]. Using a different library, Huang et al. identified a network of genes that regulate p53 function [52]. In each of these cases, the investigators employed a positive selection strategy. Interestingly, further mechanistic studies link all of these genes to the regulation of the retinoblastoma and p53 pathways, corroborating the several lines of evidence that implicate these pathways as central regulators of proliferation.
More recently, several groups have used similar approaches to identify genes involved in other aspects of cell transformation beyond proliferation. For example, Brugge and her colleagues used a cDNA expression library derived from MCF7 breast cancer cells to identify the prostate derived Ets factor PDEF as a gene that permits immortalized mammary epithelial cells to invade and migrate [53]. This gene, which is overexpressed in breast and prostate cancers, cooperates with receptor tyrosine kinases such as HER2 and CSF-1 to induce cell transformation. Similarly, by expressing an expression library derived from a cancer cell line capable of metastasis in non-metastatic cells, Martin et al. identified BCL-XL as a gene that permits metastatic growth [54].
In each of these examples, the cDNA libraries used by these investigators were derived from cell lines by reverse transcription of mRNA. Although this approach has been used successfully, two limitations of this methodology are that each gene is not represented at equal frequency in the library and longer cDNAs are under represented. With the development of large collections of open reading frames (ORFs) [3, 4], it is now possible to create expression libraries in which there is equal representation. For example, Boehm et al. used a relatively small cDNA library targeting 353 kinases in an assay to identify IKBKE as a breast cancer oncogene that substitutes for AKT to permit cell transformation [55].
Another type of gain of function approach utilizes microRNA (miRNA) expression libraries to screen using phenotypic assays. MiRNAs are endogenous small RNAs that function by downregulating expression of their target genes, either through induction of transcript degradation or translational inhibition. Nearly 500 annotated human miRNAs have been described, most of which do not have identified targets or functions [56]. miRNAs implicated in cancer include let-7, a negative regulator of RAS found upregulated in lung cancers [57], the miR-17-92 cluster, which is upregulated in lymphomas and can promote lymphomagenesis [58] and miR-15 and miR-16, negative regulators of BCL2, that are downregulated in chronic lymphoctic leukemia [59]. While it is clear that hese miRNAs play a key role in the pathogenesis of these tumors, recent work suggests that miRNAs may play a key role in cancer development as mice lacking Dicer, the endoribonuclease that is required for miRNA processing, show an increased susceptibility to cancer [60]. Indeed, using a retroviral expression library of miRNAs, Voorhoeve et al. identified miR-372 and miR-373 in a Ras-induced senescence bypass screen [61]. In aggregate, these observations provide strong evidence that gain of function approaches, particularly as new ORF and miRNA libraries become available, will continue to permit the identification of genes involved in cancer.
3.2 Loss of function approaches
Determining the consequences of gene loss-of-function is a classic means of elucidating gene function. The finding that RNAi operates in mammalian cells now permits researchers to generate loss of function phenotypes in mammalian cells that previously was possible in model organisms. Similar to the cDNA or ORF libraries used for gain-of-function approaches, RNAi libraries can be introduced into cells either stably or transiently. In mammalian cells, RNAi-mediated gene suppression can be induced by the introduction of chemically synthesized siRNAs, or plasmids expressing RNA hairpins, known as shRNAs, which get processed to siRNAs by Dicer [62]. In either case, the siRNA becomes incorporated into the RNA-induced silencing complex (RISC) and directs sequence-specific mediated degradation or translational suppression of the target mRNA, resulting in decreased protein expression [63]. Although siRNAs are easily synthesized and highly effective in inducing gene knockdown, such oligonucleotide reagents are relatively expensive and can only be used for transient loss of function experiments. Vector based systems provide stable expression of the RNAi construct, are renewable resource through propagation in E. coli, and can be used to create retroviruses expand the range and type of cells into which such constructs can be introduced [64].
Both siRNA and shRNA libraries have been used successfully in transfection-based arrayed screens looking at phenotypes that develop shortly after gene suppression, such as apoptosis, cell signaling events or cell cycle distribution [65-68]. For many other cancer-related phenotypic assays, such as anchorage independent colony formation, bypass of senescence or tumor xenografts, long-term gene suppression is essential, requiring stable integration and expression of the RNAi vector. Recent work from several laboratories has shown that these approaches are tractable in human cells. For example, PITX1 was found as a negative regulator of RAS signaling [69], REST1 has been identified a negative regulator of PI3K signaling [70], and CDK8 has been identified as a regulator of β-catenin signaling in colon cancer [71]. Although such arrayed format screens require assays that are amenable to well-based miniaturation, this experimental design permits the use of high content imaging to identify subtle or complex phenotypes [72, 73].
In addition, it is possible to use these vector-based shRNA libraries in pooled formats. The advantages of this approach are that such pooled screens permit the study of a larger number of genes with decreased cost and provide the possibility of using loss of function genetics in assays that cannot be performed in vitro. Several large-scale screens using pooled libraries have been performed [74-77], demonstrating that both positive and negative selection screens are possible using these formats. To facilitate the deconvolution of genes targeted by shRNAs in these screens, each of these groups has developed strategies to quantify the abundance of each shRNA at the beginning and end of each screen by using the sequence of the shRNA or another unique sequence in the shRNA vector.
Taken together, these observations indicate that both gain of function and loss of function approaches provide a complementary path to discovering and validating genes involved in cancer. Although the tools used for these studies continue to evolve, further studies using these methods will help identify both oncogenes and tumor suppressor genes.
4. Integrated approaches to discover cancer drivers
The development of technologies that permit the unbiased investigation of structural alterations in cancer genomes as well as those genes essential for cancer phenotypes now provide the mans to generate comprehensive molecular views of cancer. Although each of the approaches described in the prior sections is powerful, the integration of these approaches will provide a much more efficient and effective means to identify and validate novel cancer genes.
Indeed, for several of the genes identified by the gain of function and loss of function approaches described above, a key element that supported a role for the gene in cancer was the finding that the expression of the specific genes was altered in particular types of cancer. For example, PITX1 not only replaced RAS in in vitro transformation assays but was also found to be downregulated in prostate, bladder and colon cancers [69]. Similarly, REST emerged from an RNAi screen for suppressors of epithelial cell transformation and was found to lie within a chromosomal region commonly deleted in colon cancers, suggesting tumor suppressor function for this gene as well [70]. Moreover, the two miRNA identified in the screen to bypass senescence, miRNA-372 and miRNA-373, were also found to be overexpressed in testicular germ cell tumors, with concomitant loss of expression of the putative miRNA target LATS2, supporting a positive role for these miRNAs in cell transformation [61]. Similarly, functional studies focused on Nkx2.1, ALK and MITF provided strong evidence that genes mutated in specific types of cancer contribute directly to the transformed phenotype [14, 30, 32, 42].
In these examples, functional or expression studies were performed to validate initial findings. The increasing availability of large-scale datasets now provides the opportunity to combine these approaches. For example, the breast cancer oncogene IKBKE was identified by combining two functional screens with comprehensive analyses of copy number alterations in primary breast cancer cell lines and tumors. Specifically, IKBKE scored in a gain of function screen for the ability to promote transformation in vitro and a loss of function screen for genes essential for viability of cancer cells, two hallmark characteristics of typical oncogenes [55], and IKBKE resides within a region commonly amplified in primary breast tumors and breast cancer cell lines. Similarly, the CDK8 oncogene was found by combing two RNAi screens, one to identify genes that regulate β-catenin/TCF transcriptional activity and a second to uncover genes essential for colon cancer cell line proliferation, with a whole genome analysis of copy number alterations in colon cancer [71]. More recently, Zender et al. identified regions recurrently deleted in heptatocellular carcinomas by ROMA and then performed a loss of function screen in an experimental model of murine hepatocellular cancer to identify several putative tumor suppressor genes among the genes harbored by these deleted regions including XPO4, a protein that regulates nuclear export [78].
Taken together, these experiments provide strong proof of principle evidence that the integration of both structural and functional genomic approaches will accelerate the discovery and validation of new oncogenes and tumor suppressor genes. One clear advantage of these combined approaches is that the limitations of any specific methodology are offset by information derived from a complementary but distinct technology. Moreover, the evidence supporting the role for genes that emerge from these approaches in cancer is certainly stronger.
5. Future directions
Although cancer genomes exhibit a staggering complexity, recent advances in genome technologies now provide multiple ways to identify genes involved in cancer initiation or progression. Integrating these different approaches will enable rapid and efficient triangulation of bona fide cancer genes and help validate new targets that show promise for therapeutic targeting.
However, it is also clear that further developments in technology are necessary to truly apply these approaches in a comprehensive manner. It is already clear that new generation sequencing technologies will provide the means to perform more detailed analyses of genetic alterations and may make it possible to sequence entire cancer genomes for specific tumors [21]. Moreover, the prospective implementation of mass spectrometric [25] and eventually sequencing in patient samples will not only facilitate the discovery of new mutations but will also provide the means to select treatments tailored to the mutations harbored by particular tumors.
In parallel, several groups continue to expand and to improve the coverage and efficiency of reagent libraries that are necessary for gain and loss of function approaches. The use of these tools in both cell and animal studies in a range of assays will provide comprehensive views of genes and pathways involved in specific cancer phenotypes. In addition, initial efforts to provide tools that allow investigators to compare data derived from different sources (e.g. http://www.broad.mit.edu/igv/) now provide the means to perform such integrated analyses. However, the rapid expansion of the types and size of datasets derived from both structural and functional interrogation of cancer genomes will require a new generation of analytical tools to identify genes casually involved in cancer development from the large number of random mutations that occur during cancer development. Indeed, without the further development of statistically rigorous methods, deciphering useful information from these high throughput approaches will not occur.
Moreover, further advances in our understanding of cancer will require both new approaches beyond these described here. For example, new technologies that permit the investigation of epigenetic alterations in cancer genomes [79] will provide complementary datasets to identify other genes involved in cancer, while new assays that interrogate cancer phenotypes such as invasion and metastasis will likely open new avenues in cancer biology. In addition, a concerted effort dedicated to collection and annotation of patient-derived primary and metastatic tumors is necessary in order to maximize the generality of these approaches. Although significant challenges remain, the pace and scale of progress in applying genomics to the study of cancer should continue to accelerate and provide the means to elucidate a comprehensive view of the key cooperative interactions that drive cancer.
Figure 1.
Acknowledgments
We thank the members of the Hahn laboratory, DFCI Center for Cancer Genome Discovery and the Broad Institute Cancer Program for support and encouragement. This work was supported in part by grants from the NIH (R33 CA128625, U54 CA112962, P01 CA095616), the DoD (W81XWH-07-1-0408), the Prostate Cancer Foundation and the Starr Cancer Consortium.
This work was supported in part by the NIH (R33 CA128625), the Starr Cancer Consortium (I1-A11) and the Prostate Cancer Foundation.
Biography

William C. Hahn, M.D., Ph.D.
Associate Professor of Medicine
Harvard Medical School
Department of Medical Oncology, Dana-Farber Cancer Institute
Senior Associate Member, Broad Institute of MIT and Harvard
Dr. William C. Hahn is a medical oncologist and Associate Professor in the Department of Medical Oncology at the Dana-Farber Cancer Institute and a Senior Associate Member of the Broad Institute of MIT and Harvard. He directs the Center for Cancer Genome Discovery at the Dana-Farber Cancer Institute.
Dr. Hahn and his colleagues helped demonstrate that activation of the reverse transcriptase telomerase plays an essential role in malignant transformation. His current work focuses on the understanding the cooperative genetic interactions that lead to malignant transformation and the creation of novel experimental model systems for the study of normal and malignant epithelial biology. In addition, he is a founding member of The RNAi Consortium, Broad Institute-based effort to develop genome scale RNA interference reagents and the technologies for their use. His laboratory has pioneered the use of integrated functional genomic approaches to identify and validate cancer targets. Clinically, he is a member of the Lank Center for Genitourinary Oncology and is devoted to the development of new therapeutic strategies for the treatment of prostate and other cancers.
Dr. Hahn has been the recipient of many honors and awards including a Harvard National Scholarship, a Damon Runyon-Walter Winchell Cancer Research Fund Fellowship, Howard Hughes Medical Institute Pre- and Postdoctoral Fellowships, a Doris Duke Charitable Foundation Clinical Scientist Development Award, the 2000 Wilson S. Stone Award from M.D. Anderson Cancer Center for outstanding research in cancer, a Kimmel Scholar Award, and the Howard Temin Award from the National Cancer Institute. In 2005, Dr. Hahn was elected to the American Society of Clinical Investigation.
Dr. Hahn received his A.B. from Harvard University in Biochemical Sciences summa cum laude in 1987 and his M.D and Ph.D. from Harvard Medical School in 1994. He then completed clinical training in Internal Medicine at the Massachusetts General Hospital and Medical Oncology at the Dana-Farber Cancer Institute. He conducted his postdoctoral studies with Dr. Robert Weinberg at the Whitehead Institute for Biomedical Research and joined the faculty of DFCI and Harvard Medical School in 2001.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 3.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, Rogers J, Lawlor S, McLaren S, Dricot A, Borick H, Cusick ME, Vandenhaute J, Dunham I, Hill DE, Vidal M. Human ORFeome 3.1: A Resource of Human Open Reading Frames Covering over 10,000 Human Genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolfs A, Montor WR, Yoon SS, Hu Y, Bhullar B, Kelley F, McCarron S, Jepson DA, Shen B, Taycher E, Mohr SE, Zuo D, Williamson J, Mekalanos J, Labaer J. Production and sequence validation of a complete full length ORF collection for the pathogenic bacterium Vibrio cholerae. Proc Natl Acad Sci U S A. 2008;105:4364–4369. doi: 10.1073/pnas.0712049105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taya Y, Hosogai K, Hirohashi S, Shimosato Y, Tsuchiya R, Tsuchida N, Fushimi M, Sekiya T, Nishimura S. A novel combination of K-ras and myc amplification accompanied by point mutational activation of K-ras in a human lung cancer. EMBO J. 1984;3:2943–2946. doi: 10.1002/j.1460-2075.1984.tb02236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downward J, Riehl R, Wu L, Weinberg RA. Identification of a nucleotide exchange-promoting activity for p21ras. Proc Natl Acad Sci U S A. 1990;87:5998–6002. doi: 10.1073/pnas.87.15.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 10.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 11.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 12.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George RE, Sanda T, Hanna M, Frohling S, Luther W, 2nd, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, Xue L, Zozulya S, Gregor VE, Webb TR, Gray NS, Gilliland DG, Diller L, Greulich H, Morris SW, Meyerson M, Look AT. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008 doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, Jones S, Sjoblom T, Park BH, Parsons R, Willis J, Dawson D, Willson JK, Nikolskaya T, Nikolsky Y, Kopelovich L, Papadopoulos N, Pennacchio LA, Wang TL, Markowitz SD, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008 doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 20.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A, Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC, Graubert TA, DiPersio JF, Wilson RK. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, O'Neill K, Sasaki H, Lindeman N, Wong KK, Borras AM, Gutmann EJ, Dragnev KH, Debiasi RM, Chen TH, Glatt KA, Greulich H, Desany B, Lubenski CK, Brockman W, Alvarez P, Hutchison SK, Leamon JH, Ronan MT, Turenchalk GS, Egholm M, Sellers WR, Rothberg JM, Meyerson M. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nature Medicine. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 24.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, Klein P, Miller VA, Ostland MA, Ramies DA, Sebisanovic D, Stinson JA, Zhang YR, Seshagiri S, Hillan KJ. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 25.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 26.Weir B, Zhao X, Meyerson M. Somatic alterations in the human cancer genome. Cancer Cell. 2004;6:433–438. doi: 10.1016/j.ccr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, West JA, Rostan S, Nguyen KC, Powers S, Ye KQ, Olshen A, Venkatraman E, Norton L, Wigler M. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, Sugarbaker D, Chen F, Rubin MA, Janne PA, Girard L, Minna J, Christiani D, Li C, Sellers WR, Meyerson M. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 30.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, Brennan C, Polyak K. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–4078. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- 32.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 33.Bussey KJ, Chin K, Lababidi S, Reimers M, Reinhold WC, Kuo WL, Gwadry F, Ajay, Kouros-Mehr H, Fridlyand J, Jain A, Collins C, Nishizuka S, Tonon G, Roschke A, Gehlhaus K, Kirsch I, Scudiero DA, Gray JW, Weinstein JN. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5:853–867. doi: 10.1158/1535-7163.MCT-05-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, Spichtin H, Maurer R, Mirlacher M, Kochli O, Zuber M, Dieterich H, Mross F, Wilber K, Simon R, Sauter G. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534–8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- 36.Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 37.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, Yoon S, Wigler M, Ye K, Borresen-Dale AL, Naume B, Schlicting E, Norton L, Hagerstrom T, Skoog L, Auer G, Maner S, Lundin P, Zetterberg A. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nucifora G, Birn DJ, Erickson P, Gao J, LeBeau MM, Drabkin HA, Rowley JD. Detection of DNA rearrangements in the AML1 and ETO loci and of an AML1/ETO fusion mRNA in patients with t(8;21) acute myeloid leukemia. Blood. 1993;81:883–888. [PubMed] [Google Scholar]
- 41.Hunger SP, Galili N, Carroll AJ, Crist WM, Link MP, Cleary ML. The t(1;19)(q23;p13) results in consistent fusion of E2A and PBX1 coding sequences in acute lymphoblastic leukemias. Blood. 1991;77:687–693. [PubMed] [Google Scholar]
- 42.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 43.Hanauer DA, Rhodes DR, Sinha-Kumar C, Chinnaiyan AM. Bioinformatics approaches in the study of cancer. Curr Mol Med. 2007;7:133–141. doi: 10.2174/156652407779940431. [DOI] [PubMed] [Google Scholar]
- 44.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 45.Lin HY, Harris TL, Flannery MS, Aruffo A, Kaji EH, Gorn A, Kolakowski LF, Jr, Lodish HF, Goldring SR. Expression cloning of an adenylate cyclase-coupled calcitonin receptor. Science. 1991;254:1022–1024. doi: 10.1126/science.1658940. [DOI] [PubMed] [Google Scholar]
- 46.Seed B, Aruffo A. Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc Natl Acad Sci U S A. 1987;84:3365–3369. doi: 10.1073/pnas.84.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang XF, Lin HY, Ng-Eaton E, Downward J, Lodish HF, Weinberg RA. Expression cloning and characterization of the TGF-beta type III receptor. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 48.Koh EY, Chen T, Daley GQ. Novel retroviral vectors to facilitate expression screens in mammalian cells. Nucleic Acids Res. 2002;30:e142. doi: 10.1093/nar/gnf142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 50.Peeper DS, Shvarts A, Brummelkamp T, Douma S, Koh EY, Daley GQ, Bernards R. A functional screen identifies hDRIL1 as an oncogene that rescues RAS-induced senescence. Nat Cell Biol. 2002;4:148–153. doi: 10.1038/ncb742. [DOI] [PubMed] [Google Scholar]
- 51.Shvarts A, Brummelkamp TR, Scheeren F, Koh E, Daley GQ, Spits H, Bernards R. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 2002;16:681–686. doi: 10.1101/gad.929302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Q, Raya A, DeJesus P, Chao SH, Quon KC, Caldwell JS, Chanda SK, Izpisua-Belmonte JC, Schultz PG. Identification of p53 regulators by genome-wide functional analysis. Proc Natl Acad Sci U S A. 2004;101:3456–3461. doi: 10.1073/pnas.0308562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gunawardane RN, Sgroi DC, Wrobel CN, Koh E, Daley GQ, Brugge JS. Novel role for PDEF in epithelial cell migration and invasion. Cancer Res. 2005;65:11572–11580. doi: 10.1158/0008-5472.CAN-05-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin SS, Ridgeway AG, Pinkas J, Lu Y, Reginato MJ, Koh EY, Michelman M, Daley GQ, Brugge JS, Leder P. A cytoskeleton-based functional genetic screen identifies Bcl-xL as an enhancer of metastasis, but not primary tumor growth. Oncogene. 2004;23:4641–4645. doi: 10.1038/sj.onc.1207595. [DOI] [PubMed] [Google Scholar]
- 55.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson A, Greulich H, Stewart CJ, Mulvey LA, Shen RR, Ambrogio L, Hirozane-Kishikawa T, Hill DE, Vidal M, Meyerson M, Grenier JK, Hinkle G, Root DE, Roberts TM, Lander ES, Polyak K, Hahn WC. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 56.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 57.Johnson BE, Janne PA. Epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Cancer Res. 2005;65:7525–7529. doi: 10.1158/0008-5472.CAN-05-1257. [DOI] [PubMed] [Google Scholar]
- 58.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 61.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 62.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 63.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 64.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 65.Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell. 2003;12:627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- 66.Kittler R, Buchholz F. Functional genomic analysis of cell division by endoribonuclease-prepared siRNAs. Cell Cycle. 2005;4:564–567. [PubMed] [Google Scholar]
- 67.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 68.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 69.Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM, Agami R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 70.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, Depinho RA, Chin L, Elledge SJ. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 71.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG, Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, Bojarski E, Mermel C, Barretina J, Chan JA, Baselga J, Tabernero J, Root DE, Fuchs CS, Loda M, Shivdasani RA, Meyerson M, Hahn WC. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 74.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 75.Schrock E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 76.Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 77.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, McCombie WR, Wigler M, Hicks J, Hannon GJ, Powers S, Lowe SW. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plass C. Cancer epigenomics. Hum Mol Genet. 2002;11:2479–2488. doi: 10.1093/hmg/11.20.2479. [DOI] [PubMed] [Google Scholar]