Abstract
Apolipoprotein E4 (apoE4) is a risk factor for Alzheimer’s disease and has been associated with a variety of neuropathological processes. ApoE4 C-terminally truncated forms have been found in brains of Alzheimer’s disease patients. Structural rearrangements in apoE4 are known to be key to its physiological functions. To understand the effect of C-terminal truncations on apoE4 lipid-free structure, we produced a series of recombinant apoE4 forms with progressive C-terminal deletions between residues 166 and 299. Circular dichroism measurements show a dramatic loss in helicity upon removal of the last 40 C-terminal residues, whereas further truncations of residues 203–259 lead to recovery of helical content. Further deletion of residues 186–202 leads to a small increase in helical content. Thermal denaturation indicated that removal of residues 260–299 leads to an increase in melting temperature but truncations down to residue 186 did not further affect the melting temperature. The progressive C-terminal truncations, however, gradually increased the cooperativity of thermal unfolding. Chemical denaturation of the apoE4 forms revealed a two-step process with a clear intermediate stage that is progressively lost as the C-terminus is truncated down to residue 230. Hydrophobic fluorescent probe binding suggested that regions 260–299 and 186–202 contain hydrophobic sites, the former being solvent accessible in the wild-type molecule and the latter being accessible only upon truncation. Taken together, our results show an important but complex role of apoE4 C-terminal segments in secondary structure stability and unfolding and suggest that interactions mediated by the C-terminal segments are important for the structural integrity and conformational changes of apoE4.
Apolipoprotein E (apoE)1 is an important protein of the lipid transport system that has an indisputable role in atherosclerosis, dyslipidemia, and Alzheimer’s disease (AD) (1, 2). ApoE, expressed in liver, brain, and other tissues, has three common isoforms (apoE2, apoE3, apoE4) in the general population, each differing in the amino acid positions 112 and 158 (3, 4). ApoE3, the most common form, contains cysteine and arginine, respectively, whereas apoE2 has two cysteine residues and apoE4 has two arginine residues at these positions. ApoE4 has been associated with a variety of neuropathological processes, including AD (2). ApoE4 is a major genetic risk factor for AD since 40% of all patients have at least one ε4 allele (5). Being homozygous for the ε4 allele increases the risk of AD 4-fold and lowers the age of onset of late-onset AD (5). Recent studies have shown that apoE4 is also associated with carotid atherosclerosis and is a significant risk factor for coronary heart disease (6, 7).
ApoE contains 299 residues and in the lipid-free state is folded into two independent structural domains. Digestion with thrombin produces a 22 kD N-terminal fragment (residues 1–191) and a 10-kD C-terminal fragment (residues 216–299) (8, 9). X-ray crystallography studies showed that the N-terminal domain is folded into a four-helix bundle of amphipathic α-helices spanning residues 24–164 (10–12). The C-terminal domain is highly α-helical, as determined by computer modeling and circular dichroism spectroscopy (8, 13, 14), but its exact structure is unknown. A recent NMR study showed that the C-terminal domain becomes more structured in full length apoE3, probably because of inter-domain interactions (15).
In apoE4, the N- and C-terminal domains interact differently than in the other isoforms, since Arg-112 results in the orientation of the Arg-61 side chain in the N-terminal domain of apoE4 away from the four-helix bundle. This orientation allows Arg-61 to form a salt bridge with Glu-255 in the C-terminal domain (11, 16). The interaction between the N- and C-terminal domains in apoE4 has been proposed to be responsible for the conformational lability of apoE4 (17). Upon binding to phospholipids, apoE undergoes large conformational changes and the presence of partially folded states could facilitate such changes (18). Previous studies suggested that the C-terminal domain initiates the binding of apoE to phospholipids and has a higher avidity for lipid than the N-terminal domain (11, 19). It has been proposed that compared with the folded conformation, partially unfolded conformations of apoE can bind to lipid particles, such as lipoproteins or membranes, with higher affinities or on rates (20, 21). However, partially unfolded conformations are more sensitive to proteolysis and could therefore be more vulnerable to degradation pathways (22).
ApoE4 has been found to be much more susceptible to proteolysis than apoE3, creating bioactive carboxy-terminal truncated fragments (29–30 and 14–20 kDa) in brains of AD patients and apoE transgenic mice (23–25). The proteolytic cleavage of apoE4 occurs in neurons but not in astrocytes (25). In one study it was shown that the apoE cleaving enzyme is a chymotrypsin-like serine protease that cuts apoE initially at Met-272 and/or Leu-268 (24). In another study it was proposed that the proteolytic cleavage site is close to residue 187 of apoE (26). The C-terminal truncated apoE4 fragments and especially apoE4Δ(273–299), which was shown to lead in a less-ordered organization in the C-terminal domain (27), are associated with increased phosphorylation of tau, mitochondrial dysfunction, and neurotoxicity in cultured neuronal cells and transgenic mice and may play a key role in the development of neuronal degeneration observed in AD (23–25, 28). Furthermore, the C-terminal region of apoE has been found to be implicated in triglyceride homeostasis because C-terminal deletion mutants (within the region 185–299) are able to correct the high cholesterol levels of apoE knock out mice without induction of hypertriglyceridemia (29).
To gain insight to the role of the C-terminal domain of apoE4 in the molecule’s structure and stability, we examined the effect of progressive truncations of the C-terminal domain (Δ260–299, Δ230–299, Δ203–299, Δ186–299, and Δ166–299, with respective molecular weights of 29.5–18.5 kDa) in lipid-free apoE4. The truncations were designed at about every 30 amino acids and in-between predicted helical regions of the protein (13). We produced all mutants in an adenovirus-driven mammalian expression system and used the purified recombinant proteins in experiments to measure their secondary structure by circular dichroism, their hydrophobic binding properties by ANS fluorescence, and their stability versus chemical denaturants and temperature. To the best of our knowledge, this is the most systematic study of apoE4 C-terminal truncations to date. Our results, taken as a whole, provide evidence that different C-terminal segments of apoE4 have strong yet discrete effects upon the molecule’s structural integrity and unfolding properties. These findings combined with previous functional characterization of the C-terminal moiety suggest a crucial role of the C-terminus in the biological function of apoE and provide the basis for the explanation of altered functionality of truncated apoE4 forms.
MATERIALS AND METHODS
Materials
Leibovitz’s L-15 medium, fetal bovine serum, trypsin/EDTA, and penicillin/streptomycin were purchased from Invitrogen Corp. (Carlsbad, CA). Dextran sulfate and epoxy-activated Sepharose 6B were obtained from Amersham Biosciences (Piscataway, NJ); dialysis tubing was obtained from Spectrum Medical Industries, Inc. (Los Angeles, CA) and Sigma Aldrich Corp. (St. Louis, MO), and the Dc protein assay kit was obtained from Bio-Rad (Hercules, CA). All other reagents were purchased from standard commercial sources described previously (30, 31).
Production and Purification of apoE Using the Adenovirus System
We chose a mammalian system for protein production to ensure that the produced protein resembles the most the naturally occurring human protein in terms of correct folding and post-translational modifications such as glycosylation (32, 33). All plasmids and recombinant adenoviruses containing the wild-type and mutated human apoE4 genes were constructed as described previously (30). Human HTB13 cells (SW 1783, human astrocytoma) grown to 80% confluence in Leibovitz’s L-15 medium containing 10% (v/v) fetal bovine serum (FBS) in roller bottles were infected with adenoviruses expressing wild-type or mutant apoE4 forms at a multiplicity of infection of 20. After 24 h of infection, cells were washed twice with serum-free medium and preincubated in serum-free medium for 30 min; fresh serum-free medium was then added. After 24 h, the medium was harvested and fresh serum-free medium was added to the cells. The harvests were repeated ~8–10 times. ApoE was purified from the culture medium of adenovirus-infected HTB-13 cells as described previously (34). All preparations were at least 95% pure as estimated by SDS-PAGE. Proteins were stored in −80 °C in lyophilized form until use.
Protein Refolding
Before all measurements, lyophilized stocks of wild-type apoE4 or C-terminal deletion mutants were treated with a refolding step that was necessary to get rid of small amounts of aggregation products that accumulated during storage or reconstitution of the lyophilized protein. In all cases, the protein was reconstituted to about 0.1 mg/mL in 20 mM Hepes, pH 7.2, 50 mM NaCl, 8 M guanidine hydrochloride, and incubated for 30 min at room temperature to completely denature the protein. Following incubation, the protein samples were dialyzed at 4 °C versus 2lt of 20 mM Hepes pH 7.2, 50 mM NaCl, overnight. The dialysis buffer was changed twice over the period of the following day, and the dialysis was allowed to proceed again overnight to ensure the complete removal of guanidine. The next day the samples were recovered from dialysis and centrifuged at 15 000 g for 15 min to remove any precipitated protein. The supernatant was recovered and the freshly refolded proteins were quantitated by measuring the absorbance at 280 nm using calculated extinction coefficients as follows: apoE4 = 1.34 mg−1 mL cm−1, apoE4(Δ166–299) = 1.45 mg−1 mL cm−1, apoE4(Δ186–299) = 1.31 mg−1 mL cm−1, apoE4(Δ203–299) = 1.21 mg−1 mL cm−1, apoE4(Δ230–299) = 1.28 mg−1 mL cm−1, apoE4(Δ260–299) = 1.13 mg−1 mL cm−1. Proteins were kept on ice and immediately used for measurements. Protein samples were kept at low concentrations (<0.1 mg/mL) to avoid aggregation.
Circular Dichroism (CD) Measurements
All CD spectroscopy was performed on a Jasco-715 circular dichroism spectrometer using quartz cuvettes with an optical path length of 1 mm. Scan range was from 190nm to 260nm with a bandwidth of 1nm, 8 s response time, 0.2nm step size and 50nm/min scan speed. Each spectrum is the average of 5 accumulations. The sample volume typically used was 140 µL and spectra were acquired either at 15 or 25 °C. For thermal denaturation experiments, the CD signal at 222 nm was followed as the sample chamber temperature was increased from 20 to 80 °C using a step size of 0.2 °C and at a rate of 1 °C/min. For calculation of the first-order derivative of the melting curves, the data were fit to a ninth degree polynomial. The melting temperature was calculated as the temperature at the maximum of the derivative curve. Helical content based on the CD signal at 208 nm was calculated according to Greenfield N. et al. (35) using the equation:
| (1) |
Helical content was also calculated on the basis of the CD signal at 222 nm according to Morrisett et al. (36) using the equation
| (2) |
The cooperativity index, n, was calculated as previously described (27), using the Hill equation n = (log 81)/log(T0.9/T0.1), where T0.9 and T0.1 are the temperatures where the unfolding transition has reached a fractional completion of 0.9 and 0.1.
ANS Fluorescence
1,8 ANS (1-anilinonaphthalene-8-sulfonic acid, Sigma-Aldrich) was dissolved into dimethylsulfoxide (DMSO) to a final concentration of 50 mM (ANS stock solution) and stored at −80 °C. Freshly refolded wild-type apoE4 or apoE4 C-terminal deletion mutants in 20 mM Hepes pH 7.2, 50 mM NaCl were put in a fluorescence cuvette after adjusting their concentration to be at same level for all mutants. Fluorescence measurements were preformed in a Quantamaster 4 fluorescence spectrometer (Photon Technology International, New Jersey) in a 1.5 mL plastic fluorimeter cuvette (Roth, Germany), using a 2 nm excitation slit width and 4 nm emission slit width. The scan rate was 1 nm/s with excitation at 395 nm and emission range from 410 to 550 nm. After measuring the background protein fluorescence, we added 5 µL of ANS stock solution and mixed the solution. The final ANS concentration was 250 µM. Final protein concentration during measurements was typically 0.04 to 0.1 mg/mL.
Chemical Denaturation Experiments
About 0.05 mg/mL of either wild-type apoE4 or C-terminal deletion mutants were added in a 2 mL quartz fluorimeter cuvette, and the intrinsic tryptophan protein fluorescence was measured using the following parameters: excitation 295 nm, emission 340nm, excitation slit width 1 nm, emission slit width 4 nm, 20 s integration. Small amounts of an 8 M guanidine hydrochloride solution were gradually added in the cuvette; the mixture was stirred using a small magnetic stir bar for 10 s and incubated in the dark for 2 min, at which point the fluorescence signal of the sample was measured as described above. The titration was carried on until about a 5 M final concentration of guanidine hydrochloride was reached inside the cuvette. Mock titrations with the addition of buffer instead of guanidine hydrochloride were used to correct the signal for dilution during the titration. In all cases and after this correction, a stable plateau was observed at high guanidine concentrations, and this plateau was used to normalize the signal to total fraction of the protein unfolded.
RESULTS
CD Spectroscopy
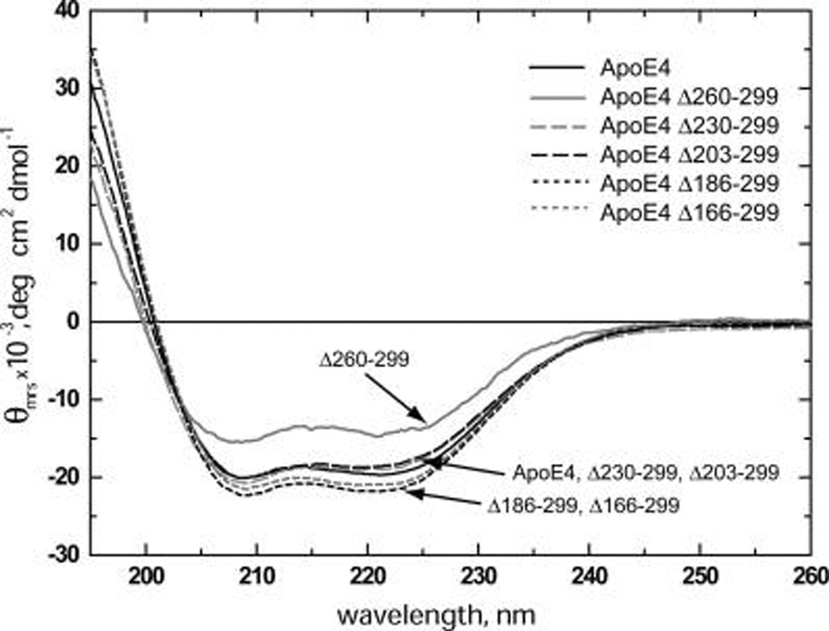
The far-UV CD spectra of freshly dialyzed wild-type apoE4 and deletion mutants were recorded and the normalized spectra were used to estimate the α-helical content of each mutant (spectra in Figure 1 and parameters in Table 1). The CD spectrum of wild-type apoE4 has a characteristic shape identical to previously reported spectra for that molecule (27, 37–39), resulting in a calculated α-helical content, on the basis of the CD signal at 222 nm, of 57.9 ± 1.5%. Deletion of the last 40 C-terminal amino acids (Δ260–299) resulted in a marked decrease on the CD spectrum intensity and a calculated α-helical content that was 23% smaller compared to wild-type apoE4. Surprisingly further deletions (Δ230–299 and Δ203–299) resulted in CD spectra that resemble the wild-type with a similar α-helical content. More drastic deletions (Δ186–299 and Δ166–299) resulted in CD spectra of slightly higher intensity than that of wild-type apoE4 and a respective statistically significant increase in α-helical content by 9 and 6% (Table 1). Similar results were observed when the α-helical content was measured on the basis of the CD signal at 208 nm (Table 1).
FIGURE 1.
Circular dichroism spectra of wild-type apoE4 and 5 C-terminal deletion mutants. Spectra shown are averages of 3–5 separate experiments. Solid black line, wild-type apoE4; solid gray line, apoE4 Δ260–299; long-dash gray line, apoE4 Δ230–299; long-dash black line, apoE4 Δ203–299; short-dash black line, apoE4 Δ186–299; short-dash gray line, apoE4 Δ166–299. Both wild-type apoE4 and Δ230–299 and Δ203–299 deletion mutants have very similar CD spectra with some small differences in shape, especially near the 220 nm area. ApoE4 deletions Δ186–299 and Δ166–299 consistently showed slightly higher CD signal compared to wild-type apoE4. Deletion Δ260–299 exhibited significantly lower CD signal in both the 208 and 220 nm regions.
Table 1.
α-Helical Content and Melting Temperatures Calculated from Circular Dichroism Experiments for Wild-Type ApoE4 and C-Terminal Deletion Mutants
| % α-helix208nma | %α-helix222nma | Tm (°C)b | cooperativity index, n | |
|---|---|---|---|---|
| WT | 54.8 ± 1.1 | 57.9 ± 1.5 | 50.2 | 5.18 |
| Δ166–299 | 58.9 ± 0.8c | 61.2 ± 0.4c | 51.1 | 7.10 |
| Δ186–299 | 61.8 ± 1.1c | 63.1 ± 1.3c | 56.9 | 6.58 |
| Δ203–299 | 54.5 ± 1.1 | 55.0 ± 0.9 | 56.5 | 6.54 |
| Δ230–299 | 56.8 ± 2.8 | 56.0 ± 1.5 | 55.5 | 5.71 |
| Δ260–299 | 39.5 ± 4.9d | 44.4 ± 1.0e | 55.6 | 5.28 |
Means ± SD from 3–5 independent experiments.
The reproducibility in Tm is ±1.2 °C.
Significance of difference from the value for WT: p < 0.05.
Significance of difference from the value for WT: p < 0.005.
Significance of difference from the value for WT: p < 0.0005.
Thermal Unfolding of apoE4
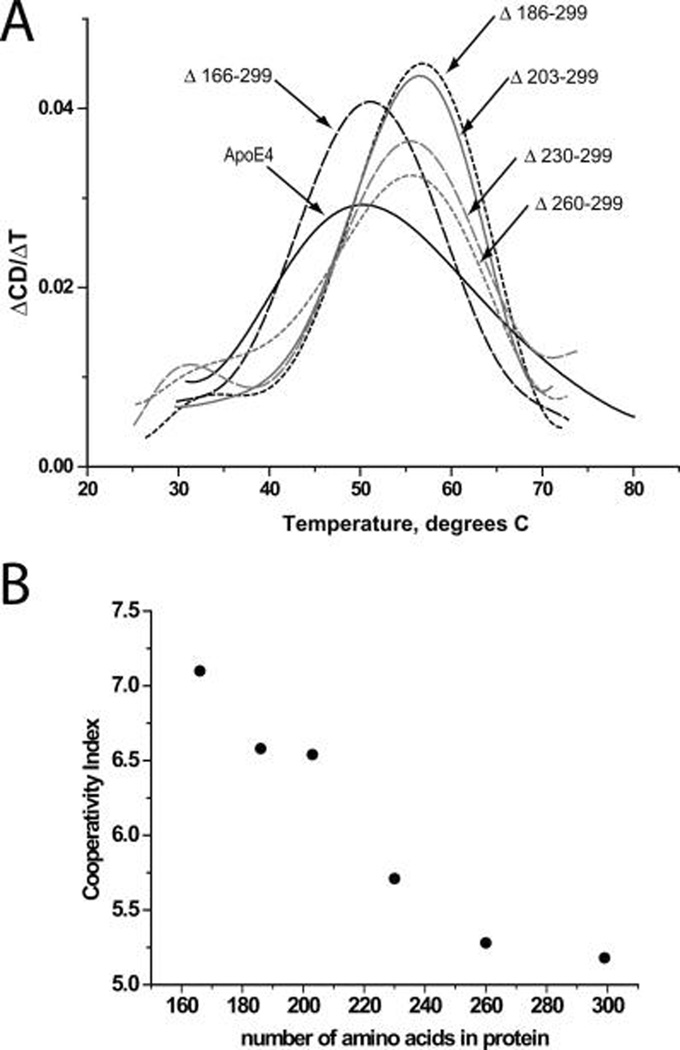
To gain insight on the role of the C-terminus moiety of apoE4 in the secondary structure changes the molecule undergoes during heat-induced denaturation, we measured the CD signal at 222 nm while the sample was gradually heated to 90 °C. Figure 2A shows the first derivative of the CD signal during the thermal unfolding of wild-type apoE4 and deletion mutants. Table 1, shows the melting temperatures (Tm) calculated from the maximum of each curve in Figure 2A. Deletions up to amino acid 185 (Δ260–299, Δ230–299, Δ203–299 and Δ186–299) resulted in an increase in Tm of about 5 °C, indicating that even the smallest truncation (Δ260–299) is sufficient to modify the thermal unfolding parameters of the molecule. This suggests that the C-terminus of the apoE4 plays a crucial role in the structural integrity of the protein during thermal unfolding. Further truncation to amino acid 165 (Δ166–299) reduced the Tm back to similar to wild-type value possibly due to destabilization of the N-terminal folding domain. All the C-terminal deletions resulted in apoE4 molecules that underwent thermal denaturation in a more cooperative manner (Table 1). Interestingly, the cooperativity index appears to increase in an almost linear fashion as the C-terminus of apoE4 is truncated (Figure 2B). This finding is consistent with the idea that the full length of the C-terminal domain of apoE4 is less-organized in structure than the N-terminal domain.
FIGURE 2.
(A) First-order derivative of thermal denaturation traces of wild-type apoE4 and C-terminal deletion mutants. Solid black line, wild-type apoE4; short-dash gray line, apoE4 Δ260–299; long-dash gray line, apoE4 Δ230–299; solid gray line, apoE4 Δ203–299; short-dash black line, apoE4 Δ186–299; long-dash black line, apoE4 Δ166–299. Wild-type apoE4 had a Tm of 50.2 °C and all deletion mutants with the exception of Δ166–299 exhibited elevated Tm values. (B) Plot of cooperativity index for thermal unfolding for wild-type and apoE4 mutants versus total number of amino acids in the mutant protein. Notice how the cooperativity index increases in almost a linear fashion as the C-terminus of apoE4 is truncated.
Chemical Unfolding of apoE4
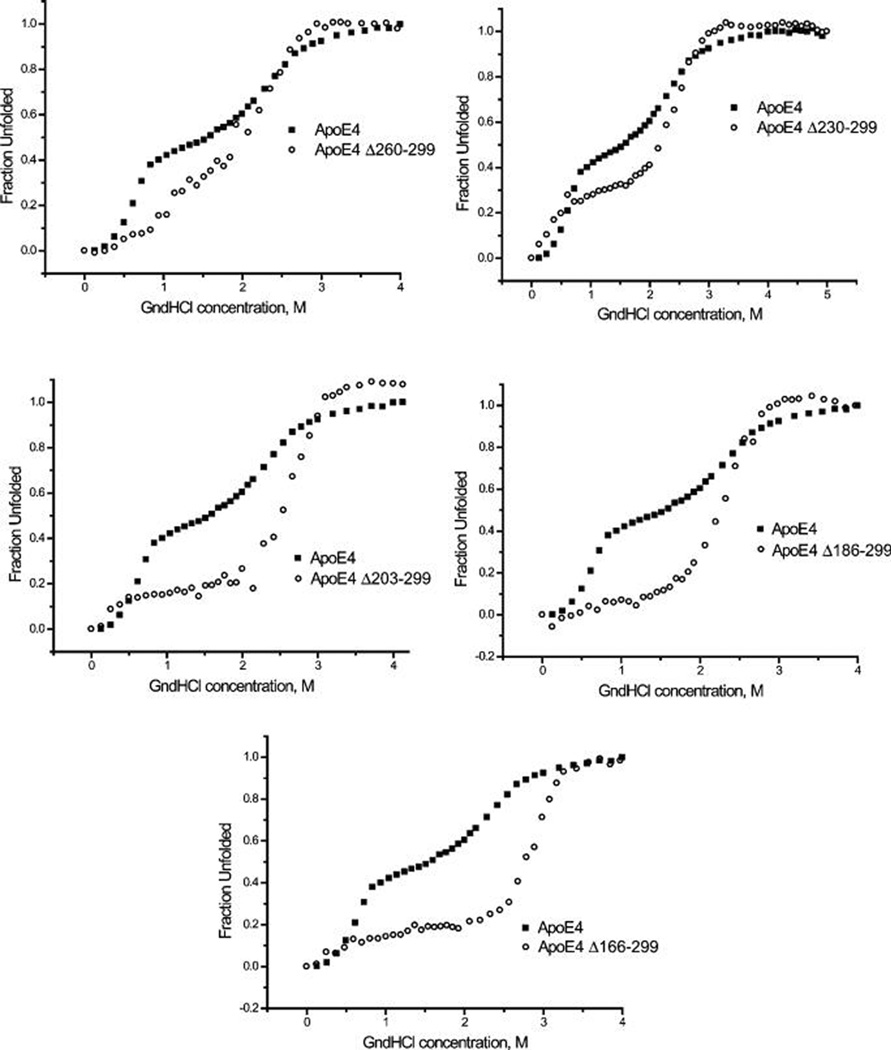
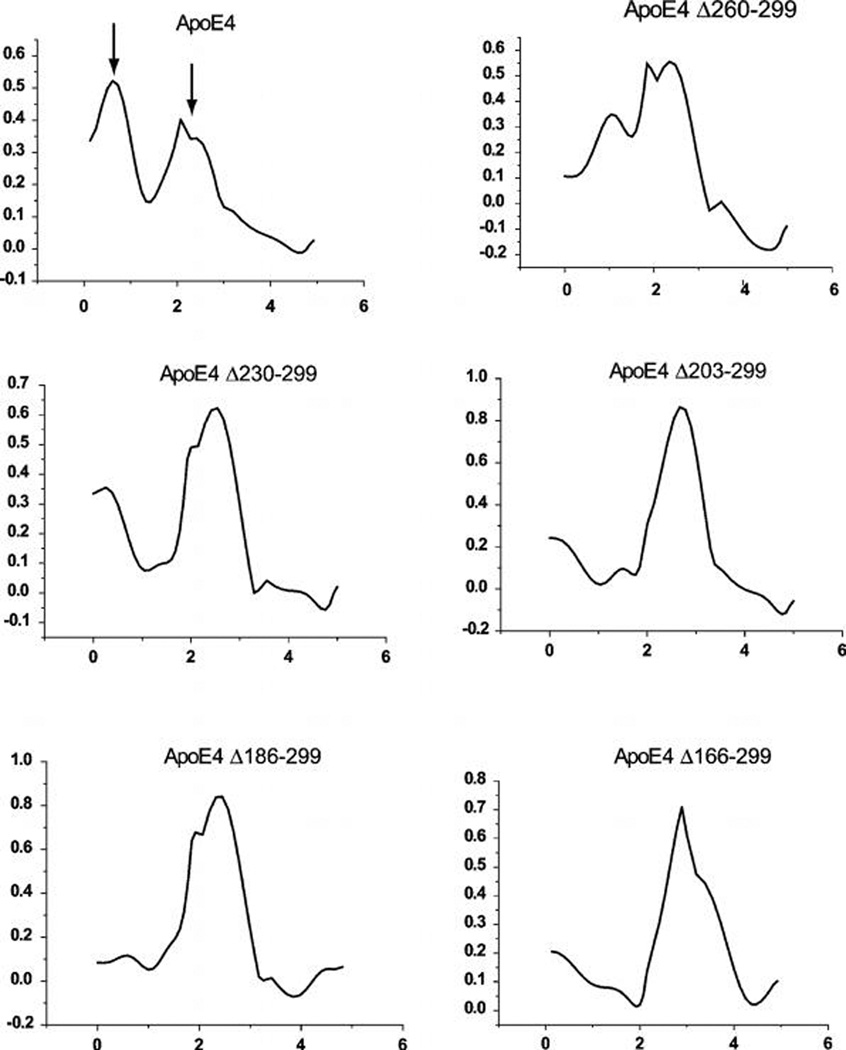
The intrinsic tryptophan fluorescence of the apoE4 variants was followed upon the chemical denaturation of the protein by titration of guanidine hydrochloride. In Figure 3, the denaturation traces of each apoE4 deletion variant is depicted in pair wise comparison to the denaturation trace of wild type apoE4. As previously shown (27, 40), the chemical denaturation profile of wild-type apoE4 consists of two transitions with a clearly discernible intermediate between 1 and 1.5 M guanidine hydrochloride. This-two step denaturation profile of wild-type apoE4 suggest the existence of a relatively labile structural element in the apoE4 molecule that is more sensitive to the chemical denaturant than the rest of the apoE4 molecule. The presence of a chemical melting intermediate is consistent with the relatively low cooperativity seen in the thermal melting of apoE4. As can be seen in Figure 3, in each of the deletion variants, the presence of this intermediate is reduced or completely abolished, leading to denaturation traces that are shifted to higher guanidine hydrochloride concentrations. This is clearer when the first derivative of the chemical denaturation profile is analyzed (Figure 4). In the first derivative of the chemical denaturation, distinct transitions appear as peaks, indicated by arrows in the wild-type apoE4 trace. As progressive C-terminal deletion variants are analyzed, the first transition peak is greatly diminished in apoE4 variant Δ260–299 and is essentially absent in the apoE4 variants Δ230–299, Δ203–299 and Δ186–299 and Δ166–299. Abrogation of this chemical melting intermediate step appears to correspond to the increased thermal melting cooperativity seen upon C-terminal truncations. These results suggest that the C-terminal region is critical for the destabilization of apoE4 into an intermediate partially unfolded state.
FIGURE 3.
Chemical denaturation traces for wild-type apoE4 and C-terminal deletion mutants. Changes in tryptophan fluorescence (excitation 295, emission 340 nm) of apoE4 was measured in solution upon titration with increasing amounts of the chemical denaturant guanidine hydrochloride (GndHCl). All data were corrected for dilution and normalized. Each panel shows the denaturation trace for wild-type apoE4 and one of each of the C-terminal deletion mutants. Note that the trace is shifted to higher guanidine concentrations for all mutations. ApoE4 deletions also gradually alter the shape of the denaturation curve, eventually abrogating the presence of an intermediate.
FIGURE 4.
First-order derivative of chemical denaturation traces shown in Figure 3 for wild-type apoE4 and C-terminal deletion mutants. X-axis is the guanidine hydrochloride molar concentration. Data from Figure 3 were smoothed using a moving-average algorithm to reduce noise spikes in the first derivative traces. Peaks indicate transition points during the chemical denaturation. Note the two distinct peaks for wild-type apoE4 (marked by arrows). The first peak that appears in lower guanidine concentration is a major transition for wild-type apoE4 but rapidly disappears with progressive C-terminal deletions.
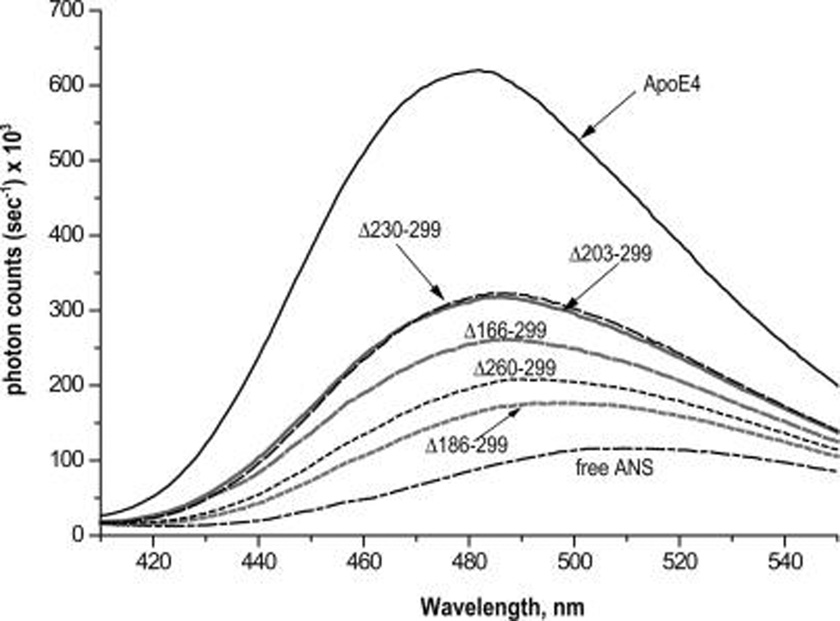
ANS Binding
The fluorescence of the ANS in the presence of wild type apoE4 and apoE4 deletion mutants was measured to determine how the deletions affect the exposure of the hydrophobic surfaces of apoE4 (Figure 5). The fluorescence signal was significantly reduced for all C-terminal deletion mutants compared to wild-type apoE4, indicating either loss of exposure of a hydrophobic surface in the mutants or possible complete abrogation of that surface because of truncation. Because the minimum deletion of Δ260–299 was enough to significantly reduce the ANS fluorescence signal, these data suggest that the region between amino acids 260 and 299 is either a component of an ANS binding hydrophobic surface or critical to its exposure to the solvent. Interestingly, further truncation down to residue 202 (Δ230–299 and Δ203–299) resulted in ANS fluorescence signal increase, suggesting the presence of a second ANS binding site that is exposed with truncation. The fluorescence signal is reduced upon further truncation to amino acid 185 (Δ186–299) indicating that the region 186–202 may be a component of a hydrophobic surface. Further trimming to amino acid 165 resulted in an increase of ANS signal. This is probably due to the destabilization of the N-terminal apoE4 folding domain (8, 9, 11) and partial exposure of its hydrophobic core.
FIGURE 5.
ANS fluorescence spectra in the presence of wild-type apoE4 and apoE4 C-terminal deletion mutants; 250 µM of ANS were incubated with 0.044 mg/mL of protein (wild-type apoE4 or deletion mutants) and the fluorescence spectrum was recorded by exciting at 395 nm. Solid black line, wild-type apoE4; long-dash black line, apoE4 Δ230–299; solid gray line, apoE4 Δ203–299; long-dash gray line, apoE4 Δ166–299; short-dash black line, apoE4 Δ260–299; short-dash gray line, apoE4 Δ186–299; dash-dot black line, free ANS.
DISCUSSION
Although the importance of the C-terminal moiety of apoE in its physiological functions has been demonstrated (23–25, 28, 29, 41, 42) the mechanism by which the C-terminus of apoE4 affects protein function is still poorly understood. ApoE is known to undergo major conformational changes upon lipid binding that may involve the protein’s C-terminal region (18, 43, 44), although the exact role of the C-terminus of apoE in the protein’s structure is still vague. The finding of C-terminally truncated apoE4 variants in the brain of Alzheimer’s disease patients in conjunction with apoE4’s susceptibility to proteolysis further necessitate the understanding of the exact role of apoE4’s C-terminus in the protein’s structure, stability and function. Previous studies have shown that the C-terminus of apoE4 is largely helical and participates in weak interactions with the N-terminus (8, 13–15, 39). Tanaka et al. demonstrated that removal of residues 273–299 or 261–299 leads to a less-organized C-terminal structure and affects the stability of the whole apoE4 molecule (27). Sakamoto et al. examined the effect of removing residues 273–299 in apoE3 as compared to apoE4 and suggested that this region is crucial for the different lipid binding properties of apoE4 (44). In this study, we have examined the effect of further C-terminal truncations in apoE4 in an effort to clarify the role of apoE4’s C-terminus in the molecule’s structure and stability. To that end, we characterized 5 progressive C-terminal deletions (Δ260–299, Δ230–299, Δ203–299, Δ186–299, and Δ166–299) of apoE4. Because of the lack of any direct structural information on the C-terminal moiety of apoE4, these deletions were designed largely on the basis of previous molecular modeling studies (13). To generate the mutations, we introduced stop codons at the turns that connect the various postulated C-terminal helices (13). We measured the helical content of the apoE4 mutants by circular dichroism spectroscopy to gain insight on the secondary structure of the C-terminal moiety of apoE4. The hydrophobic site exposure in those regions was evaluated by ANS probe binding. Finally, the thermal and chemical melting profiles of the progressive truncations were used to evaluate unfolding structural transitions that may parallel lipid binding structural transitions. Analysis of the physicochemical properties of full-length and truncated apoE4 forms in the C-terminal moiety of the protein (residues 166–299) offers the opportunity to extract information regarding interactions between different segments of the protein by pairwise comparisons. However, it should be noted that in a study of deletion mutants like this, the effect of the deleted segment on the function of the full-length protein can be inferred only indirectly, because only the properties of the remaining molecule, and not of the deleted segments, are analyzed. Still, useful insights can be drawn that can at the very least guide future efforts.
Consistent with previous studies we find wild-type apoE4 to be highly helical based on CD spectroscopy and to be able to bind ANS probe efficiently, enhancing its fluorescence, indicating the presence of solvent-exposed hydrophobic regions in the molecule (17, 27, 37–40). Wild-type apoE4 can be thermally denatured with a Tm of 50.2 °C, exhibiting low degree of cooperativity as evidenced by the broad thermal transition. Chemical denaturation by guanidine hydrochloride indicated a two-step process with a weak but clear intermediate stage. Both the low cooperativity of the thermal melting and the two-step process of the chemical melting suggest a structurally labile molecule and the possible coexistence of multiple conformational states in the ensemble.
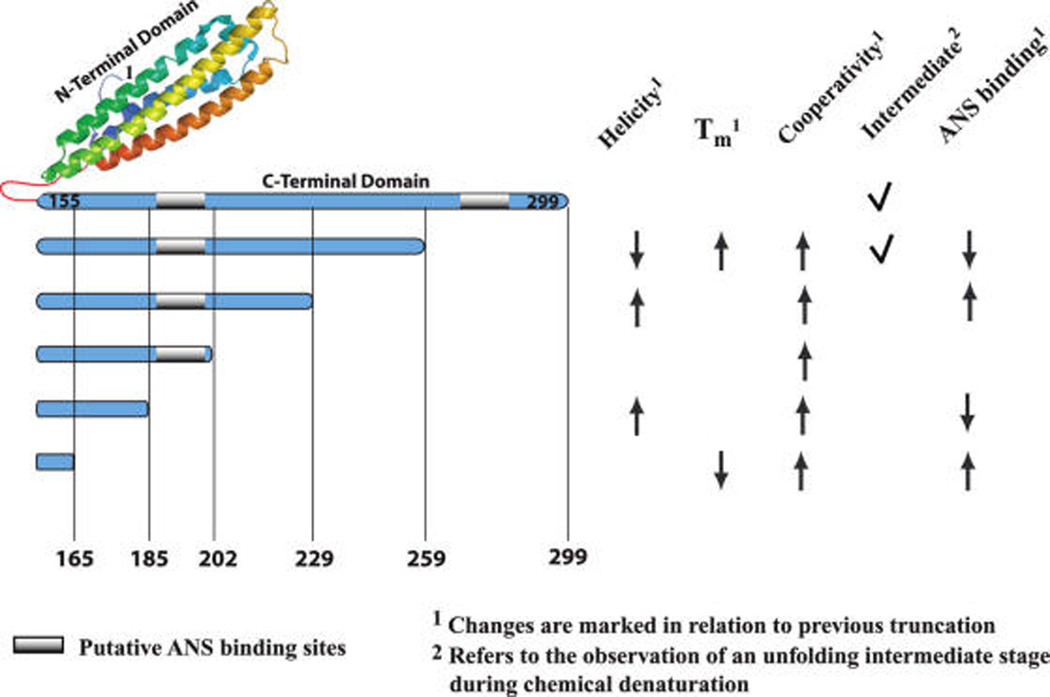
Figure 6 summarizes the effects of C-terminal truncations to the biophysical properties of the remaining molecule. The effects of the progressive deletion of C-terminal segments are discussed below.
FIGURE 6.
Summary of the observed changes in biophysical properties of apoE4 upon C-terminal truncations. ApoE4 N-terminal moiety (residues 1–155, PDB code: 1YA9) is depicted in cartoon format (made by PYMOL, http://www.pymol.org). The C-terminal moiety of apoE4 is depicted as a single rod structure to allow easy visualization of the relative extent of the truncations. Gray regions denote putative ANS binding sites. All changes in biophysical properties (helicity, melting temperature, thermal melting cooperativity, existence of a chemical melting intermediate form, and extent of ANS probe binding) are noted on the right part of the figure. Changes are shown in relation to the previous truncation only; an upward-pointing arrow indicates that the property has increased and a downward-pointing arrow indicates that the property has decreased. A tick mark indicates the presence of a chemical unfolding intermediate stage.
Deletion of Residues 260–299
Deletion of the last 40 C-terminal residues of apoE4 led to a significant loss of helicity as measured by CD spectroscopy. This is consistent with an earlier study of apoE4(Δ261–299) (27) where the authors concluded that region 261–299 is highly helical. However, the absence of any knowledge about the interactions of residues 261–299 with other parts of apoE4 makes direct conclusions regarding the structure of the deleted region uncertain. An alternate explanation is that region 260–299 is important for stabilizing helical content of apoE4. Region 260–299 appears critical to the unfolding of apoE4 since deletion of this region leads to an apoE4 form that has higher Tm and melting cooperativity. Thus, it appears that region 260–299 is important for the structural plasticity of wild-type apoE4, contributing to a lower melting temperature and a higher degree of conformational heterogeneity. This is further evidenced by chemical denaturation experiments that show a partial loss of an unfolding intermediate upon truncation of region 260–299. Finally, truncation leads to a marked reduction of exposed hydrophobic sites as evidenced by ANS titration, indicating the existence of a solvent-exposed hydrophobic site in region 260–299 in wild-type apoE4, a site that may be important in the first steps of interactions between apoE4 and lipids. This idea is further supported by a recent study by Sakamoto et al. in which the authors compared a C-terminal truncation (273–299) in apoE3 and apoE4 concluding that the C-terminal domain of apoE4 is crucial for the protein’s lipid binding properties (44).
Deletion of Residues 230–259
Further deletion of residues 230–259 of apoE4 resulted in the complete recovery of the initial CD signal of wild-type apoE4. This striking result suggests that region 230–259 is either partially nonhelical in apoE4(Δ260–299) and/or that it destabilizes the helical structure of the remaining apoE4 molecule through atomic interactions. The region 230–259 does not seem to affect greatly the thermal unfolding of apoE4 but contributes in the two-step process of chemical unfolding, because its deletion further abrogates the unfolding intermediate. Tanaka et al. have previously suggested that residues 192–272 do not have a significant impact on the stability of apoE4. However, in that study, the authors did not examine intermediate deletions in the region 261 and 192 as we did in this study and may have missed the effect of the contribution of the 230–259 region. Deletion of the region 230–259 also enhances binding of the ANS probe, suggesting that this region buries a hydrophobic site that is not available in full-length apoE4, indicating long-distance interactions between this region and other parts of the molecule.
Deletion of Residues 203–229
Further deletion of residues 203–229 does not produce discernible change in the CD signal indicating that this region does not affect the helical stability of the molecule. Thermal and chemical melting profiles did not change by this deletion, suggesting that there is no direct and separate involvement of this region in the unfolding pathway of apoE4. No further exposure of hydrophobic sites is evident upon deletion of this region. Overall, it appears that residues 203–229 are helical and do not clearly participate in interactions affecting the stability of the molecule or its conformational transitions.
Deletion of Residues 186–202
We observed a small but reproducible gain in helicity upon truncation of this region, suggesting that, similarly to region 230–259 (but to a lesser degree), residues 186–202 are either nonhelical in apoE4(Δ203–299) or can destabilize the helical structure of the remaining apoE4 molecule. Thermal and chemical melting profiles were not affected by this progressive truncation. Loss in ANS binding indicates that this region contains a hydrophobic site that is exposed by previous truncations (particularly of region 230–259). In conclusion, residues 186–202 appear to have the capacity to affect the secondary structure of apoE4 and to contain a buried hydrophobic site that can be exposed by truncation of region 230–259. It is conceivable that exposure of this hydrophobic site is part of the conformational rearrangements that apoE4 undergoes upon interactions with lipids. This deletion contains part of the previously proposed hinge region of apoE4 (residues 192–215) that may mediate interdomain interactions (15).
Deletion of Residues 166–185
Further deletion of the 166–185 region does not lead to a change in overall helicity, suggesting that this region does not affect the helical stability of the molecule. Truncation of this region resulted in reduction of Tm, possibly due to destabilization of the N-terminal apoE 22 kDa folding domain (residues 1–191). Increase in ANS binding suggests exposure of a hydrophobic region that is normally buried in full-length apoE4. Overall, residues 166–185 appear to be important for the structural stability of the N-terminal moiety of apoE4. X-ray analysis of the N-terminal moiety of apoE has demonstrated a four-helix bundle structure spanning residues 24–164 that has been shown to unfold in response to phospholipid binding (10, 12). Our results suggest that the neighboring residues 166–185 are important for the structural integrity and lability of this four-helix bundle.
Functional Properties of apoE4 C-Terminal Truncation Mutants
Dimyristoyl-l-α-phosphatidylcholine (DMPC) clearance assays for the apoE4 C-terminal truncation mutants used in this study have shown that progressive C-terminal truncations abrogate the ability of apoE4 to bind and solubilize suspensions of DMPC multilamellar vesicles. Deletion of residues beyond amino acid 259 resulted in a great decrease in the rate of solubilization of multilamellar DMPC vesicles (30). Furthermore, recent studies from the laboratory of Dr. Vassilis Zannis (V. Zannis, personal communication) showed that ATP-binding cassette transporter A1 (ABCA1) mediated cholesterol efflux (45, 46) from macrophages in the presence of apoE4(Δ260–299) was 70% of the wild type apoE4 while in the presence of all other C-terminal truncation mutants ABCA1-mediated cholesterol efflux was essentially abrogated. In this study we demonstrate that both wild-type apoE4 as well as apoE4 (Δ260–299) unfold through an intermediate stage that is lost upon further truncations (Figure 4). The ability of apoE4 mutants to interact with lipids and induce cholesterol efflux appears to correlate with the availability of this unfolding intermediate. This interesting correlation between structural stabilization and loss of function supports the concept that the C-terminus of apoE4 is responsible for providing conformational lability to allow for structural rearrangements necessary for functional interactions.
Critical Regions in the C-Terminus of apoE4
The progressive deletions used in this study, although designed on the basis of previous molecular modeling studies because of the lack of an experimentally determined three-dimensional structure of the C-terminal domain of apoE (13), provide interesting insights on the location of sequence segments in the C-terminus of apoE4 that affect the protein’s conformational stability and function. Region 260–299 appears to be important for maintaining apoE4 helicity, whereas regions 230–259 and 186–202 appear to be reducing apoE4 helical content. This is a novel finding that suggests that the secondary structure of apoE4 and in particular its C-terminus is labile, although it is not possible to say if those effects are direct or through long-range interactions. Region 230–299 appears to contain necessary segments for the formation of the intermediate stage during chemical unfolding but also for lipid binding and ABCA1 mediated cholesterol efflux from cells. Hydrophobic binding sites are probably contained within regions 260–299 and 186–202. Finally, region 260–299 appears to be critical for thermal destabilization of apoE4. It should be noted though that the deletions used here are coarse enough to preclude exact sequence segment definition and further mutational studies based on these results might be necessary to pinpoint the exact locations of sequences within apoE4 that are critical to its conformational stability and function. However, the progressive gain in thermal cooperativity upon progressive deletions, suggest that some of the properties of the C-terminus of apoE4 may be impossible to define in a short sequence segment and are a property of the C-terminus as a whole.
In summary, we present here a systematic characterization of the secondary structure and unfolding profiles of a series of progressive C-terminal truncations of human apoE4 spanning residues 166–299. Taken as a whole, our results demonstrate that segments of the C-terminal moiety of apoE4 have strong but yet discrete effects in the secondary structure and unfolding transitions of the molecule. The secondary structure of apoE4 can be greatly affected in a complex manner by C-terminal truncations, suggesting that secondary structure elements within apoE4 are labile and can be affected by interactions with neighboring regions. C-terminal truncations lead to significant stabilization of the remaining molecule suggesting that at least part of the C-terminus of apoE4 is responsible for the conformational lability of the protein. This lability may be mediated by intermediate conformations that are evident during chemical unfolding experiments. Our results also suggest a complex but labile tertiary structural organization within the C-terminal moiety of apoE4 that may be important for its physiological function. Alterations of the structure of apoE4 brought forth by truncations may be responsible for the abnormal apoE4 functionality observed in Alzheimer disease patients’ brain (23–25, 28).
ACKNOWLEDGMENT
The authors thank Ms. Evagelia Karatairi for assisting with the chemical denaturation experiments.
Funding for this work was provided by the sixth Framework Programme of the European Union (LSHM-CT-2006-037631 to A.C. and V.Z. and Marie Curie International Reintegration Grants 017157 to E.S. and 031070 to A.C.), by the General Secretariat of Research and Technology of Greece, and by the National Institutes of Health Grant (HL68216 to V.Z.).
Footnotes
Abbreviations: ABCA1, ATP-binding cassette transporter A1; ANS, 1-anilinonaphthalene-8-sulfonic acid; apoE, apolipoprotein E; CD, circular dichroism; GndHCl, guanidine hydrochloride; Tm, melting temperature; DMPC, dimyristoyl-l-α-phosphatidylcholine; WT, wild-type.
REFERENCES
- 1.Zannis VI, Kypreos KE, Chroni A, Kardassis D, Zanni EE. Lipoproteins and Atherogenesis. In: Loscalzo J, editor. Molecular Mechanisms of Atherosclerosis. London: Taylor & Francis Books; 2004. pp. 111–174. [Google Scholar]
- 2.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zannis VI, Just PW, Breslow JL. Human apolipoprotein E isoprotein subclasses are genetically determined. Am. J. Hum. Genet. 1981;33:11–24. [PMC free article] [PubMed] [Google Scholar]
- 4.Zannis VI, Breslow JL, Utermann G, Mahley RW, Weisgraber KH, Havel RJ, Goldstein JL, Brown MS, Schonfeld G, Hazzard WR, Blum C. Proposed nomenclature of apoE isoproteins, apoE genotypes, and phenotypes. J. Lipid Res. 1982;23:911–914. [PubMed] [Google Scholar]
- 5.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 6.Graner M, Kahri J, Varpula M, Salonen RM, Nyyssonen K, Jauhiainen M, Nieminen MS, Syvanne M, Taskinen MR. Apolipoprotein E polymorphism is associated with both carotid and coronary atherosclerosis in patients with coronary artery disease. Nutr., Metab. Cardiovasc. Dis. 18:271–277. doi: 10.1016/j.numecd.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Song Y, Stampfer MJ, Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann. Intern. Med. 2004;141:137–147. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]
- 8.Wetterau JR, Aggerbeck LP, Rall SC, Jr, Weisgraber KH. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J. Biol. Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 9.Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J. Biol. Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 10.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 11.Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 12.Wilson C, Mau T, Weisgraber KH, Wardell MR, Mahley RW, Agard DA. Salt bridge relay triggers defective LDL receptor binding by a mutant apolipoprotein. Structure. 1994;2:713–718. doi: 10.1016/s0969-2126(00)00072-1. [DOI] [PubMed] [Google Scholar]
- 13.Nolte RT, Atkinson D. Conformational analysis of apolipoprotein A-I and E-3 based on primary sequence and circular dichroism. Biophys. J. 1992;63:1221–1239. doi: 10.1016/S0006-3495(92)81698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 15.Zhang Y, Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 16.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 17.Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Phillips MC, Lund-Katz S. Effects of polymorphism on the lipid interaction of human apolipoprotein E. J. Biol. Chem. 2003;278:40723–40729. doi: 10.1074/jbc.M304814200. [DOI] [PubMed] [Google Scholar]
- 18.Peters-Libeu CA, Newhouse Y, Hatters DM, Weisgraber KH. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J. Biol. Chem. 2006;281:1073–1079. doi: 10.1074/jbc.M510851200. [DOI] [PubMed] [Google Scholar]
- 19.Segall ML, Dhanasekaran P, Baldwin F, Anantharamaiah GM, Weisgraber KH, Phillips MC, Lund-Katz S. Influence of apoE domain structure and polymorphism on the kinetics of phospholipid vesicle solubilization. J. Lipid Res. 2002;43:1688–1700. doi: 10.1194/jlr.m200157-jlr200. [DOI] [PubMed] [Google Scholar]
- 20.Weers PM, Narayanaswami V, Ryan RO. Modulation of the lipid binding properties of the N-terminal domain of human apolipoprotein E3. Eur. J. Biochem. 2001;268:3728–3735. doi: 10.1046/j.1432-1327.2001.02282.x. [DOI] [PubMed] [Google Scholar]
- 21.Morrow JA, Hatters DM, Lu B, Hochtl P, Oberg KA, Rupp B, Weisgraber KH. Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. J. Biol. Chem. 2002;277:50380–50385. doi: 10.1074/jbc.M204898200. [DOI] [PubMed] [Google Scholar]
- 22.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, Mahley RW. Apolipoprotein E fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8838–8843. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, Fish JD, Masliah E, Hopkins PC, Scearce-Levie K, Weisgraber KH, Mucke L, Mahley RW, Huang Y. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brecht WJ, Harris FM, Chang S, Tesseur I, Yu GQ, Xu Q, Dee FJ, Wyss-Coray T, Buttini M, Mucke L, Mahley RW, Huang Y. Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 2004;24:2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellnitz S, Friedlein A, Bonanni C, Anquez V, Goepfert F, Loetscher H, Adessi C, Czech C. A 13 kDa carboxy-terminal fragment of ApoE stabilizes Abeta hexamers. J. Neurochem. 2005;94:1351–1360. doi: 10.1111/j.1471-4159.2005.03295.x. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M, Vedhachalam C, Sakamoto T, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4. Biochemistry. 2006;45:4240–4247. doi: 10.1021/bi060023b. [DOI] [PubMed] [Google Scholar]
- 28.Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zannis VI, Chroni A, Kypreos KE, Kan HY, Cesar TB, Zanni EE, Kardassis D. Probing the pathways of chylomicron and HDL metabolism using adenovirus-mediated gene transfer. Curr. Opin. Lipidol. 2004;15:151–166. doi: 10.1097/00041433-200404000-00008. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Kypreos K, Zanni EE, Zannis V. Domains of apoE required for binding to apoE receptor 2 and to phospholipids: Implications for the functions of apoE in the brain. Biochemistry. 2003;42:10406–10417. doi: 10.1021/bi027093c. [DOI] [PubMed] [Google Scholar]
- 31.Chroni A, Nieland TJ, Kypreos KE, Krieger M, Zannis VI. SR-BI mediates cholesterol efflux via its interactions with lipid-bound ApoE. Structural mutations in SR-BI diminish cholesterol efflux. Biochemistry. 2005;44:13132–13143. doi: 10.1021/bi051029o. [DOI] [PubMed] [Google Scholar]
- 32.Zanni EE, Kouvatsi A, Hadzopoulou-Cladaras M, Krieger M, Zannis VI. Expression of ApoE gene in Chinese hamster cells with a reversible defect in O-glycosylation. Glycosylation is not required for apoE secretion. J. Biol. Chem. 1989;264:9137–9140. [PubMed] [Google Scholar]
- 33.Wernette-Hammond ME, Lauer SJ, Corsini A, Walker D, Taylor JM, Rall SC., Jr Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J. Biol. Chem. 1989;264:9094–9101. [PubMed] [Google Scholar]
- 34.Li X, Kan HY, Lavrentiadou S, Krieger M, Zannis V. Reconstituted discoidal apoE-phospholipid particles are ligands for the scavenger receptor BI. The amino-terminal 1–165 domain of apoE suffices for receptor binding. J. Biol. Chem. 2002;277:21149–21157. doi: 10.1074/jbc.M200658200. [DOI] [PubMed] [Google Scholar]
- 35.Greenfield N, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 36.Morrisett JD, David JS, Pownall HJ, Gotto AM., Jr Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry. 1973;12:1290–1299. doi: 10.1021/bi00731a008. [DOI] [PubMed] [Google Scholar]
- 37.Acharya P, Segall ML, Zaiou M, Morrow J, Weisgraber KH, Phillips MC, Lund-Katz S, Snow J. Comparison of the stabilities and unfolding pathways of human apolipoprotein E isoforms by differential scanning calorimetry and circular dichroism. Biochim. Biophys. Acta. 2002;1584:9–19. doi: 10.1016/s1388-1981(02)00263-9. [DOI] [PubMed] [Google Scholar]
- 38.Martel CL, Mackic JB, Matsubara E, Governale S, Miguel C, Miao W, McComb JG, Frangione B, Ghiso J, Zlokovic BV. Isoform-specific effects of apolipoproteins E2, E3, and E4 on cerebral capillary sequestration and blood-brain barrier transport of circulating Alzheimer’s amyloid beta. J. Neurochem. 1997;69:1995–2004. doi: 10.1046/j.1471-4159.1997.69051995.x. [DOI] [PubMed] [Google Scholar]
- 39.Chou CY, Lin YL, Huang YC, Sheu SY, Lin TH, Tsay HJ, Chang GG, Shiao MS. Structural variation in human apolipoprotein E3 and E4: secondary structure, tertiary structure, and size distribution. Biophys. J. 2005;88:455–466. doi: 10.1529/biophysj.104.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 41.Kypreos KE, Morani P, Van Dijk KW, Havekes LM, Zannis VI. The amino-terminal 1–185 domain of apoE promotes the clearance of lipoprotein remnants in vivo. The carboxy-terminal domain is required for induction of hyperlipidemia in normal and apoE-deficient mice. Biochemistry. 2001;40:6027–6035. doi: 10.1021/bi002414a. [DOI] [PubMed] [Google Scholar]
- 42.Kypreos KE, Van Dijk KW, van Der ZA, Havekes LM, Zannis VI. Domains of apolipoprotein E contributing to triglyceride and cholesterol homeostasis in vivo. Carboxyl-terminal region 203–299 promotes hepatic very low density lipoprotein-triglyceride secretion. J. Biol. Chem. 2001;276:19778–19786. doi: 10.1074/jbc.M100418200. [DOI] [PubMed] [Google Scholar]
- 43.Peters-Libeu CA, Newhouse Y, Hall SC, Witkowska HE, Weisgraber KH. Apolipoprotein E*dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J. Lipid Res. 2007;48:1035–1044. doi: 10.1194/jlr.M600545-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto T, Tanaka M, Vedhachalam C, Nickel M, Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Contributions of the Carboxyl-Terminal Helical Segment to the Self-Association and Lipoprotein Preferences of Human Apolipoprotein E3 and E4 Isoforms. Biochemistry. 2008;47:2968–2977. doi: 10.1021/bi701923h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Remaley AT, Stonik JA, Demosky SJ, Neufeld EB, Bocharov AV, Vishnyakova TG, Eggerman TL, Patterson AP, Duverger NJ, Santamarina-Fojo S, Brewer HB., Jr Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem. Biophys. Res. Commun. 2001;280:818–823. doi: 10.1006/bbrc.2000.4219. [DOI] [PubMed] [Google Scholar]
- 46.Vedhachalam C, Narayanaswami V, Neto N, Forte TM, Phillips MC, Lund-Katz S, Bielicki JK. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 2007;46:2583–2593. doi: 10.1021/bi602407r. [DOI] [PubMed] [Google Scholar]