Abstract
The contribution of nuclear factor of activated T cells 5 (NFAT5) to the regulation of tumor necrosis factor-α (TNF) production in medullary thick ascending limb (mTAL) cells is unclear. RT-PCR analysis was performed on primary cultures of mouse mTAL cells and freshly isolated mTAL tubules to determine which NFAT isoforms are present in this nephron segment. Primer pairs were designed, based on published sequences for mouse NFAT1-5, to produce fragments of ∼200 bp. Analysis of PCR products by gel electrophoresis and subsequent DNA sequencing indicated that cells and tubules contained mRNA for all five NFAT isoforms. The relative expression of NFAT isoforms was then determined using quantitative real-time RT-PCR. The data indicate that NFAT isoforms 5 ≥ 1 are the predominant isoforms present in mTAL cells and tubules. Western blot analysis demonstrated constitutive expression of NFAT5 in nuclear extracts from mTAL tubules and primary culture cells; expression in mTAL cells also was detected by immunofluorescence. Expression of NFAT5 was increased in mTAL cells transiently transfected with an NFAT5 overexpression vector (pcDNA3.1-NFAT5), resulting in increased basal and calcium-sensing receptor (CaR)-mediated TNF production. Transient transfection of mTAL cells with a small hairpin RNA vector that targeted exon 8 of NFAT5 (U6-N5 ex8) significantly inhibited TNF promoter activity. Transient transfection with U6-N5 ex8 also reduced nuclear expression of NFAT5, TNF mRNA accumulation, and attenuated CaR-mediated activation of Cl− entry into polarized mTAL cells. Collectively, these data suggest that activation of NFAT5 is part of a TNF-dependent pathway that inhibits apical Cl− influx in the mTAL after activation of CaR.
Keywords: NFAT isoforms, TNF, calcium-sensing receptor, Ton/EBP
the thick ascending limb (TAL) is responsible for reabsorption of ∼25% of filtered Na+ as well as Ca2+ (21) and contributes to the generation of the osmotic gradient that drives vasopressin-dependent water reabsorption by the collecting duct. The transport of NaCl across the apical membrane of the TAL occurs mainly via a bumetanide-sensitive Na+-K+-2Cl− cotransporter (NKCC2 or BSC1) (23). Reabsorption of NaCl is then completed as Na+ exits the cell via the basolateral Na+-K+-ATPase while Cl− diffuses along its electrochemical gradient through basolateral KCl cotransporters or Cl− channels. The apical renal outer medullary K+ channel (ROMK) is a major contributor to K+ recycling, which is essential for maintenance of Na+-K+-2Cl− cotransporter activity (25, 26). Activation of the calcium-sensing receptor (CaR) regulates Na+, K+, Cl−, and divalent cation reabsorption in the TAL; however, the mechanisms that contribute to these effects have not been fully determined (28). We recently showed that CaR activation increases TNF production in mTAL cells and proposed that this cytokine may contribute to CaR function in this segment of the nephron (47).
The nuclear factor of activated T cells (NFAT) family of transcription factors includes five proteins evolutionarily related to the Rel/NF-κB family (10). The classic members of this family comprise NFAT1–NFAT4, and the primordial member is NFAT5 (36). Although originally described as transcription factors critical to signaling via the T cell receptor for antigen, NFAT proteins are expressed in many nonlymphoid tissues where they contribute to diverse cellular functions (5, 11, 34). NFAT5 (also called Ton/EBP for tonicity element binding protein or OREBP for osmotic response element binding protein, a transcription factor crucial for cellular responses to hypertonic stress) (35, 39) is expressed in the kidney and contributes to induction of genes that increase the accumulation of organic osmolytes that protect cells against damage in a hypertonic environment (8). Accordingly, current concepts of NFAT5 function clearly define its role in countering the deleterious effects of the hypertonic environment found within the renal medulla (8, 20, 50, 51). The mechanism for accumulation of osmolytes in response to high concentrations of NaCl and urea in renal medullary cells includes increased transcription of the genes for aldose reductase and neuropathy target esterase, which encode proteins that increase sorbitol and glycerophosphocholine synthesis, respectively. The promoter regions of betaine, sodium myo-inositol, and taurine transporters also contain at least one osmotic response element, and transcriptional activity of these genes is stimulated by NFAT5 in response to hypertonicity (8).
Our recent study demonstrated activation of NFAT via stimulation of CaR (1). However, no information is available regarding the presence and function of NFAT isoforms in medullary TAL (mTAL) cells. We were prompted to determine the contribution of NFAT5 to TNF production and CaR-mediated transport function in the mTAL as NFAT5 has been linked with regulatory mechanisms in association with NKCC2 and ROMK activity (22, 32). In the present study, we identified five NFAT isoforms in mTAL tubules and primary cultures from mice. We demonstrate that mTAL cells have high constitutive levels of NFAT5 mRNA and protein and that this isoform contributes to CaR-mediated regulation of apical Cl− entry in these cells. The present studies will contribute to an understanding of the molecular mechanisms by which CaR activation increases TNF production and function in mTAL cells and suggest that NFAT5 plays an important role in salt and water balance as part of the CaR/TNF/cyclooxygenase 2 (COX-2) system that regulates ion transport pathways in these cells.
MATERIALS AND METHODS
Chemicals and reagents.
The anti-NFAT5 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used at 1:1,000 for immunoblot analysis and 1:500 for immunofluorescence and laser scanning cytometry (LSC). Tissue culture media was obtained from Life Technologies (Grand Island, NY). Collagenase (type 1A) was from Sigma (St. Louis, MO), and polyvinylidene difluoride (PVDF) membranes were obtained from Amersham (Arlington Heights, IL). Reagents for preparation of the TNF ELISA were purchased from Pharmingen (San Diego, CA), and the luciferase assay kit was from Promega (Madison, WI). All other chemicals were of the highest grade commercially available.
Animals.
Male C57BL6/J mice (8–12 wk) purchased from Jackson Laboratory were maintained on a standard diet and given tap water ad libitum. Experimental procedures were conducted in accordance with institutional and international guidelines for the welfare of animals (animal welfare assurance number A3362-01, Office of Laboratory Animal Welfare, PHS, NIH).
Isolation of mTAL tubules and cells.
mTAL tubules and cells (90–95% purity) were isolated from mice in a manner similar to that established in our laboratory for rats, with the exception that a lower concentration of collagenase (0.01%) was used (9, 31). Briefly, mice were anesthetized with an intraperitoneal injection of pentobarbital sodium (0.065 mg/10 g body wt). The kidneys were perfused with sterile 0.9% saline via retrograde perfusion of the aorta and cut along the corticopapillary axis. The inner stripe of the outer medulla was excised, minced with a sterile blade, and incubated for 10 min at 37°C in a 0.01% collagenase solution gassed with 95% oxygen. The suspension was sedimented on ice, mixed with Hanks’ balanced salt solution (HBSS) containing 2% BSA, and the supernatant containing the crude suspension of tubules was collected. The collagenase digestion was repeated three times with the remaining undigested tissue, and the combined supernatants were centrifuged for 10 min, resuspended in HBSS, and filtered through a 52-μm nylon mesh membrane (Fisher Scientific, Springfield, NJ). The filtered solution was discarded, and the tubules retained on the mesh were resuspended in HBSS and centrifuged at 500 rpm for 10 min; pelleted tubules were used in experiments or to establish primary cultures of mouse mTAL cells. Cells were grown in six-well plates using Renal Epithelial Cell Basal Medium (REBM, Cambrex), containing Renal Epithelial Cell Growth Medium (REGM, Cambrex) consisting of rhEGF, insulin, hydrocortisone, GA-1,000 (gentamicin sulfate and amphotericin B), FBS, epinephrine, T3 (triiodothronine), and transferrin. After 6–7 days, monolayers of cells were 70–80% confluent. The cells were quiesced for 24 h in RPMI containing 0.42 mM CaCl2 and 0.5% FBS, l-glutamine (2 mM), 100 U/ml streptomycin/penicillin (GIBCO), MEM nonessential amino acids (GIBCO), MEM sodium pyruvate, and β-mercaptoethanol before their use. In all experiments, “control conditions” (i.e., no addition of CaCl2) reflects that cells were incubated in media containing 0.42 mM Ca2+. This amount of calcium should be added to the amounts used to challenge the cells to calculate total Ca2+ present. These control conditions were selected based on previous work showing that the CaR is functionally insensitive when Ca2+ concentrations are <1 mM (48).
Isolation of total RNA and amplification of cDNA fragments.
Total RNA was isolated from mouse mTAL tubules and primary cultures of mTAL cells by adding 1 ml TRIzol reagent and incubating at room temperature for 10 min. Chloroform (0.2 ml) was then added at room temperature for 2–3 min followed by centrifugation at 4°C at 12,000 rpm for 15 min. Isopropanol (3 vol) was added to the recovered supernatant, and the mixture was incubated at room temperature for 10 min, then centrifuged at 4°C at 12,000 rpm for 15 min. The supernatant was discarded, the pellet was washed in 1 ml of 75% EtOH, mixed gently, and centrifuged for 5 min at 7,500 rpm at 4°C; the supernatant was removed and the pellet was dried for 5–10 min. Finally, the RNA pellet was resuspended in 50 μl of RNase-free dH2O and stored at −70°C. After total RNA was treated with deoxyribonuclease I for 30 min, a 3-μg aliquot was used for cDNA synthesis using the Superscript Preamplification system (Life Technologies) in a 20-μl reaction mixture containing Superscript II reverse transcriptase (200 U/μl) and random hexamers (50 ng/μl). The reaction was incubated at room temperature for 10 min to allow extension of the primers by reverse transcriptase, then at 42°C for 50 min, 70°C for 15 min, and 4°C for 5 min. cDNA fragments were separated on a 1% agarose gel and stained with ethidium bromide.
Detection of target gene expression.
A 0.5-μg aliquot of total RNA was converted to cDNA using random primers and PowerScript RT (Clontech) according to the manufacturer's protocol. The cDNA from each RNA sample was put in a 20-μl RT-PCR mixture using a FastStart DNA Master SYBR Green I kit (Roche) supplemented with 3 mM MgCl2 and Platinum Taq polymerase (Invitrogen). Quantitative real-time PCR (qRT-PCR) was used to determine the accumulation of mRNA for NFAT isoforms and TNF. The specific primer pairs for murine NFAT isoforms are provided in Table 1; murine TNF primers (forward: 5-GCCACCACGCTCTTCTGT-3; reverse: 5-TTGAGATCCATGCCGTTGG-3) were designed based on accession no. NM_013693. Input cDNAs were normalized using the housekeeping gene β-actin, and the efficiency of primer pair amplification was determined using a standard curve generated using serially diluted plasmid DNA for NFAT isoforms (40, 41). Relative TNF mRNA accumulation was calculated by the 2(−ΔΔCT) method (33).
Table 1.
Oligonucleotide primers for polymerase chain reaction of murine isoforms of NFAT
| Primer | Sequence | PCR Size, bp | Position | Reference |
|---|---|---|---|---|
| F1-NFAT1m | 5-CTACATGGAGAACAAGCCT-3 | 1439–1457 | (mice)(cDNA) NM_010899 | |
| R1-NFAT1m | 5-GTTTCGGAGCTTCAGGATGC-3 | 228 | 1667–1648 | (mice)(cDNA) NM_010899 |
| F1-NFAT2m | 5-CCTTCGGAAGGGTGCCTTTT-3 | 1317–1297 | (mice)(cDNA) NM_016791 | |
| R1-NFAT2m | 5-CGGCTGCCTTCCGTCTCATAG-3 | 215 | 1638–1618 | (mice)(cDNA) NM_016791 |
| F1-NFAT3m | 5-CATTGGCACTGCAGATGAG-3 | 1430–1448 | (mice)(cDNA) NM_023699 | |
| R1-NFAT3m | 5-CGTAGCTCAATGTCTGAAT-3 | 209 | 1639–1621 | (mice)(cDNA) NM_023699 |
| F1-NFAT4m | 5-CTACTGGTGGCCATCCTGTTGT-3 | 1383–1404 | (mice)(cDNA) NM_010901 | |
| R1-NFAT4m | 5-ACTTTTGTGCTGGCGATTAT-3 | 183 | 1566–1547 | (mice)(cDNA) NM_010901 |
| F1-NFAT5m | 5-AACATTGGACAGCCAAAAGG-3 | 804–823 | (mice)(cDNA) NM_018823 | |
| R1-NFAT5m | 5-GCAACACCACTGGTTCATTA-3 | 223 | 1027–1008 | (mice)(cDNA) NM_018823 |
NFAT, nuclear factor of activated T cells.
NFAT5-dominant negative, small hairpin RNA, pTonE_Luc, and TNF-Luc constructs.
A NFAT5-dominant negative (NFAT5-DN) expression plasmid was generated by cloning a NFAT5 cDNA fragment containing the NFAT5 DNA binding domain (DBD) into the pcDNA3.1 vector (Invitrogen); a cDNA (nucleotides 1074-1694 encoding amino acids 175-471, NM_018823.1) was used in this study (34). The pcDNA3.1-NFAT5 or NFAT5-DN cDNA was amplified from the pBluescript SK+ vector containing full-length NFAT5 using restriction enzymes BamHI and XhoI and ligated into the pcDNA3.1 vector.
Small hairpin (sh)RNA-expressing constructs were made by cloning the first 315 bp of the murine U6 promoter up to the base before the G marking the transcriptional start site. The fragment was cloned between EcoRI and BamHI in pBluescript (Stratagene), and the construct was used as a template for PCR of a U6-shRNA construct whose hairpin targeted exon 8 of the NFAT5 gene (U6-N5 ex8). A U6-shRNA construct with a scrambled hairpin was designed (U6-N5). These PCR reactions used a common primer for the 5′-end, sense: gcagaattcGATCCGACGCCGCCATCTCT. The antisense primers for the 3′-ends were as follows: gcagctagcCTCGAGAAAAAAGCAATGTCAGAGTAGAGCCCTACACAAAGGCTCTACTCT GACATTGCAAACAAGGCTTTTCTCCAAGGGATA (shRNA targeting NFAT5 exon 8); and gcagctagcCTCGAGAAAAAAGAACGTTCGATAATGGATCCTACACAAAGATCCATTATC. A U6-shRNA construct with a scrambled hairpin was also designed (U6-N1) for hairpin targeted exon 4 of the NFAT1 gene (U6-N1 ex4). The antisense primers for the 3′-ends were as follows: gcagctagcCTCGAGAAAAAAGACTGATTGGAGAGTGGCCCTACACAAAGGCCACTCTCCAATCAGTCAAACAAGGCTTTTCTCCAAGGGATA (shRNA targeting NFAT1 exon 4); GAACGTTCAAACAAGGCTTTTCTCCAAGGGATA (scrambled nonsilencing shRNA) (12, 18). The pTonE_Luc reporter (43) (originally from Dr. Steffan N. Ho), was provided by Dr. Feng Cheng (Washington University, St. Louis, MO).
Using PCR, we amplified the TNF promoter region from −200 to +68 nt relative to the transcription start site, and the NheI-BglII restriction fragment was subcloned into firefly luciferase reporter plasmid pGL3 Basic (Promega) (18); plasmids were isolated with EndoFree plasmid kits (Qiagen) for use in the mTAL cells.
Transient transfection.
After murine mTAL cells were cultured to 70–80% confluence, the medium was removed and cells were placed in 1 ml of serum-free OPTI-MEM medium containing different plasmid DNA constructs and 10 μl lipofectamine reagent (Life Technologies) or Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions, for 4 h at 37°C/5% CO2. After the transfection period, 1 ml of DMEM/F12 containing 20% FBS was added and cells were incubated overnight at 37°C/5% CO2. The medium was then removed, and cells were cultured for an additional 12 h in DMEM/F12 containing 10% FBS. The different assays were performed after transfection. Transfection efficiency was evaluated by flow cytometry of cells transfected with pcDNA3.1-EGFP. Stimulation was performed with calcium for 6–9 h after cells were quiesced overnight in RPMI containing 0.5% FBS. In the TNF promoter experiments, cells were transfected by DEAE-dextran with 1 μg of the firefly luciferase reporter plasmids, 0.6 μg of pRL-TK plasmids (Promega) containing a Renilla reniformis luciferase gene as a control plasmid, and 7 μg of the dominant negative version of NFAT5 (NFAT5-DN) expressing or the corresponding empty plasmid vector (pcDNA3.1). The luciferase activities of cell extracts were determined using the Dual-Luciferase Reporter Assay System according to the manufacturer's instructions (Promega). Luciferase activity was calculated as relative light units from firefly luciferase normalized to R. reniformis luciferase values.
Nuclear extracts and protein assay.
Nuclear extracts were prepared by a modification of the method of Dignam et al. (1, 14). One day after transfection, cells were quiesced overnight in RPMI medium containing 0.5% FBS. After treatment, cells were harvested with RIPA buffer into a 1.5-ml Eppendorf tube and spun for 5 min at 4,000 rpm and 4°C. The cell pellet was lysed in CE buffer (10 mM Tris, pH 8.0, 60 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 0.5 mM PMSF, 10 μg/ml aprotinin, 25 μM leupeptin, 2 μM pepstatin A, and 0.3% Nonidet P-40) at 4°C and centrifuged for 5 min at 4,000 rpm and 4°C. The nuclei were kept on ice and washed in 0.5 ml CE buffer without Nonidet P-40 for 5 min at 4,000 rpm, and nuclear proteins were extracted under high-salt conditions in a solution containing 20 mM Tris, pH 7.8, 0.42 M NaCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.2 mM EDTA, 0.5 mM PMSF, 10 μg aprotinin/ml, 25 μM leupeptin, 2 μM pepstatin A, and 25% (vol/vol) glycerol for 30 min at 4°C. After centrifugation at 12,000 rpm for 30 min, protein concentration in the supernatant was determined with a Bio-Rad protein assay kit.
Immunoblot analysis.
Equal amounts of cellular protein lysates were separated by SDS-PAGE and electrophoretically transferred to nitrocellulose membranes. Following overnight treatment with 5% skim milk at 4°C, membranes were probed with appropriated antibodies for 1 h followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia Biotech). Membranes were washed, and proteins were detected by the enhanced chemiluminescence system (Amersham, Arlington Heights, IL).
Measurement of TNF.
Primary cultured murine mTAL cells were quiesced overnight and then challenged with CaCl2 for different times at 37°C/5% CO2. TNF levels in cell-free supernatants were determined by ELISA (Pharmingen), according to the protocol provided by the manufacturer and as previously described (47).
Immunofluorescence and nuclear staining.
Cells were seeded onto two chamber tissue culture-treated glass slides (BD Bioscience), washed several times with PBS, fixed with freshly prepared 4% paraformaldehyde in PBS for 1 h, rinsed several times with fresh PBS, and stored in the cell culture plates at 4°C. For staining, cells were permeabilized with 0.1% Triton X-100 in PBS for 1 h at room temperature. After each sequence with either a primary (rabbit) or secondary antibody (donkey anti-rabbit-conjugated with Alexa Fluor 488; Invitrogen), slides were washed five times with a high-salt solution containing 1% BSA and 2.3% sodium chloride in PBS, followed by a single wash with PBS. Cells were then washed three times and stained with 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) for 5 min followed by a single wash with PBS. The slides were examined using a Nikon Microphot FXA microscope equipped for epifluorescence illumination. Laser-scanning cytometry (LSC; iCys; CompuCyte, Cambridge, MA) was used to measure expression of nuclear NFAT5. In brief, nuclear and cytoplasmic fluorescence was measured by LSC using UV and 488-nm wavelength argon ion lasers to excite the fluorescence of DAPI and Alexa Fluor 488, respectively. Nuclear contouring was based on blue fluorescence of DAPI. The intensity of blue (DAPI) and green (Alexa Fluor 488) fluorescence emission was measured by separate photomultipliers. The integrated value of green fluorescence representing NFAT5 immunofluorescence was measured in the nucleus and cytoplasm, which was defined by integration and peripheral contour settings, respectively, as described before (13).
Cl− measurements.
Cl− levels in cells were determined using 6-methoxy-N-ethylquinolinium (MEQ), a Cl−-sensitive fluorescent dye (4, 17, 37). Cells were grown to 70–80% confluence on six-well plates and then transfected with either scrambled control U6-N5 or U6-N5 ex8, harvested, and seeded onto six-well cell culture inserts (Becton-Dickinson) to obtain polarized mTAL cells. Cells were quiesced overnight and then incubated in the absence or presence of CaCl2 for 6 h at 37°C/5% CO2. After treatment, cells were preincubated for 90 min with cholera toxin (CTX; 100 nM) and then washed twice in Cl−-free buffer containing 140 mM NaNO3, 5 mM KNO3, 5 mM HEPES, 1 mM Mg(NO3)2, and 5 mM glucose, pH 7.4 (15). Cells were then exposed for 1 min to a Cl− -containing buffer (same composition as the Cl−-free buffer except that NO3− was replaced with Cl−) to initiate Cl− influx, collected by scraping in 1.0 ml Cl−-free buffer and homogenized twice for 3 min. Lysates were centrifuged for 10 min at 1,400 rpm, and a 100-μl supernatant was put into the wells of 96-well plates containing 100 μl of 100 μM MEQ dye. A FLx 800 microplate fluorescence reader (Bio-Tek) was used to measure the fluorescence with excitation at 360/40 nm and emission at 460 nm. Protein concentration in the supernatant was determined with a Bio-Rad protein assay kit. Cl− influx was monitored by determining the difference in quenching of dye fluorescence upon exposure of cells to Cl−-containing and Cl−-free buffers and is expressed as arbitrary MEQ fluorescent units (ΔAFU min/mg protein).
Statistics.
All data are presented as means ± SD and were subjected to one-way ANOVA. Multiple means were compared using Tukey's test. Student's t-test or an analysis of variance with multiple comparisons was used to determine the statistical significance.
RESULTS
Identification of NFAT isoforms in mTAL tubules and cells.
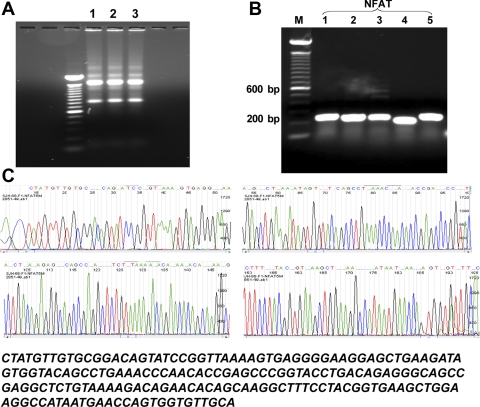
Total RNA was isolated from mouse mTAL tubules and primary mTAL cells using TRIzol reagent and assessed by separation on a 1% agarose gel stained with ethidium bromide (Fig. 1A). Then cDNA was synthesized and amplified using Taq DNA polymerase (2.5 U) in the presence of sense and antisense primers (1 μM) for murine NFAT isoforms 1–5 (NFAT1-5), or β-actin. Primers were prepared using a DNA synthesizer according to published murine NFAT1-5 cDNA sequences [Table 1; National Center for Biotechnology Information (NCBI) nos, NM_010899, NM_016791, NM_023699, NM_010901, and NM_018823]. Five pairs of specific primers were designed using Epicentre software according to cDNA fragment size and primer condition requirements. Analysis of PCR products, separated on a 1% agarose gel and stained with ethidium bromide, indicates that primary cultures of mouse mTAL cells contain mRNA for all five NFAT isoforms (Fig. 1B). Freshly isolated mouse mTAL tubules also contained mRNA for all five NFAT isoforms (not shown), demonstrating that the expression pattern in mTAL cells is not a function of culture conditions. The positions and sequences of these primer pairs were confirmed using NCBI BLAST, and cDNA amplification yielded fragments sizes consistent with those predicted by the primers sets used for each isoform (Table 1 and Fig. 1B). In control experiments, total RNA was amplified before cDNA synthesis to exclude the possibility of contamination with genomic DNA (not shown). RT-PCR products shown in Fig. 1B were sequenced using ABI Big Dye V3.0 sequencing chemistry; purified sequencing products were analyzed using an ABI 3730 DNA Sequencer. The DNA sequences for each of the PCR fragments were identical to those previously published for the corresponding isoforms, confirming the specificity of the primers; results for NFAT5 are shown in Fig. 1C.
Fig. 1.
Identification of nuclear factor of activated T cells (NFAT) isoforms in the medullary thick ascending limb of Henle's loop (mTAL). A: total RNA quality and size were assessed by separation on a 1% agarose gel stained with ethidium bromide. Two distinct 28S and 18S ribosomal RNA bands were observed, and an intensity of 28S was twice that of 18S band with no degradation. Lanes 1–3: total RNA from mouse primary mTAL cells, mTAL tubules, and outer medulla. B: cDNA fragments for NFAT1-5 were generated by RT-PCR using total RNA from mouse mTAL cells. Possible contamination was ruled out by including PCR control samples with no DNA as a template. M, markers; 1–5, NFAT isoforms. C: DNA sequence analysis of NFAT5 cDNA fragment from mTAL cells.
Relative expression of NFAT isoforms in mTAL tubules and cells.
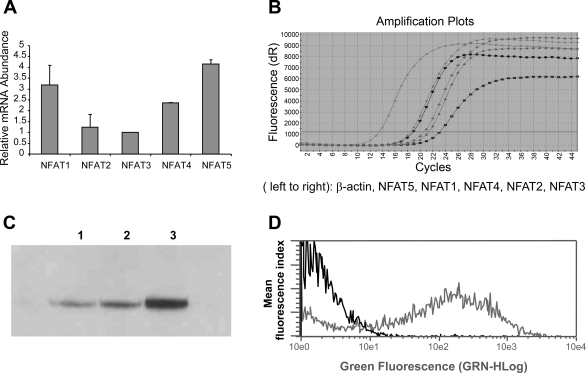
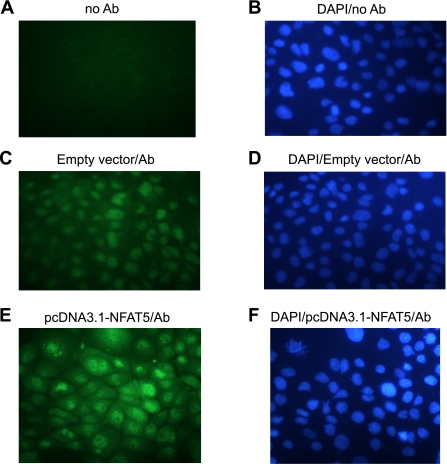
The relative abundance of mRNA levels for NFAT isoforms in mTAL tubules and cells was determined using quantitative real-time RT-PCR. While mRNA for all NFAT isoforms was readily detected, NFAT isoforms 1 and 5 were the predominant isoforms observed in mTAL cells, followed by NFAT4, -2, and -3 (Figs. 2, A and B); similar results were obtained for freshly isolated tubules (not shown). The complete absence of any detectable signal in the negative control samples (no input cDNA) indicated that the observed signals were specific. Moreover, Northern blot analysis using the same probe identified a single NFAT5 mRNA transcript with no additional high-stringency hybridization signals (data not shown). Verification of NFAT5 protein expression was provided by Western blot analysis of nuclear protein from mTAL tubules and cells (Fig. 2C). Transfection efficiency was ∼60%, as demonstrated by flow cytometry with pcDNA3.1-EGFP (Fig. 2D). Endogenous NFAT5, observed using fluorescence microscopy, was constitutively expressed in the nucleus and cytoplasm of mTAL cells (Fig. 3C); nuclear staining (DAPI) of the corresponding cells in Fig. 3, A, C, and E is illustrated in Fig. 3, B, D, and F, respectively. Transient transfection of mTAL cells with overexpression plasmid pcDNA3.1-NFAT5 increased NFAT5 protein expression, with a similar intracellular distribution, as the level of expression was higher than that observed in cells transfected with empty vector (Fig. 3, E vs. C); no staining was detected when the primary antibody was omitted (Fig. 3A). Accordingly, the immunohistochemical data support Western blot analysis showing that NFAT5 protein is expressed in unstimulated primary mouse mTAL cells.
Fig. 2.
Relative expression of NFAT isoforms in the mTAL. Relative expression (A) and amplification plots (B) of NFAT isoforms in mTAL cells. Values are means ± SD of measurements from 3 independent experiments. C: expression of NFAT5 protein was measured in nuclear protein extracts from mTAL tubules (lane 1), primary mTAL cells (lane 2), and mTAL cells transfected with pcDNA3.1-NFAT5 (lane 3). D: flow cytometric assessment of transfection efficiency (>60%) in mTAL cells.
Fig. 3.
Expresssion of NFAT5 in mTAL cells. Immunofluorescence analysis is shown of cells stained with anti-NFAT5-polyclonal antibody (Alexa 488; green) and DAPI (blue) and excited sequentially at different wavelengths. Primary antibody was not present in A and B, and mTAL cells were transfected with either empty pcDNA3.1 plasmid DNA (C and D) or pcDNA3.1-NFAT5 (E and F). The slides were examined using a Nikon Microphot FXA microscope equipped for epifluorescence illumination. Images were obtained at a magnification of ×40. A representative figure from 4 similar experiments is shown.
NFAT5 regulates CaR-mediated TNF production.
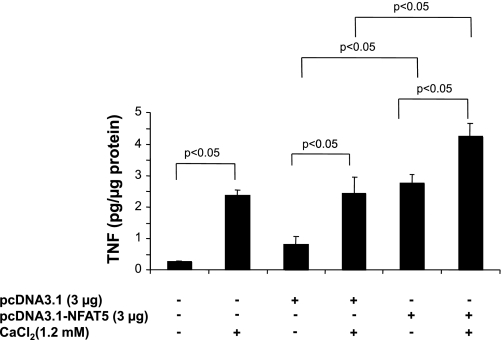
In a previous study, we demonstrated that TNF production in response to exogenous CaCl2 was mediated by activation of CaR (1). As NFAT5 contributes importantly to TNF gene transcription (18, 34), an overexpression vector for NFAT5 was used to assess the contribution of this transcription factor to TNF production in mTAL cells. TNF production by mTAL cells incubated in the absence or presence of CaCl2 (1.2 mM) was determined after transient transfection with either pcDNA3.1-NFAT5 or the empty vector control (pcDNA3.1). CaR activation increased TNF production by mTAL cells, an effect that was similar in untransfected cells and cells transfected with the empty plasmid (Fig. 4). Overexpression of NFAT5 increased TNF production by about threefold compared with the empty vector in the absence of CaR activation and also significantly increased production in cells challenged with Ca2+ (Fig. 4). These data support the notion that NFAT5 contributes to enhanced TNF production after CaR activation and suggests a role for this transcription factor in basal TNF production by cultured mTAL cells.
Fig. 4.
NFAT5 contributes to TNF production in mTAL cells. mTAL cells were incubated in the absence or presence of 1.2 mM CaCl2 for 9 h following transfection with pcDNA3.1-NFAT5 or the empty plasmid vector without the NFAT5 cDNA (pcDNA3.1). Supernatants were harvested, and TNF concentrations were determined by ELISA. Values are means ± SD. P < 0.05; n = 3.
Inhibition of NFAT5 attenuates CaR-mediated TNF reporter activity and mRNA accumulation.
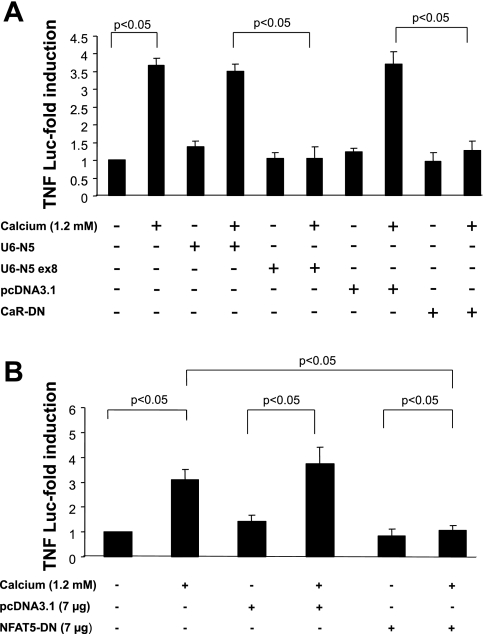
To further establish a link between NFAT5 and TNF gene transcription in response to CaR activation, mTAL cells were cotransfected with a shRNA construct under the control of the murine U6 small nuclear (sn)RNA promoter targeting exon 8 of the murine NFAT5 gene (U6-N5 ex8), and a TNF promoter-luciferase reporter gene construct (18). Activity of the TNF promoter construct increased after untransfected cells or cells transfected with scrambled U6-N5 were challenged with calcium (Fig. 5A). In contrast, activation was not observed in cells transfected with U6-N5 ex8. The increase in TNF promoter activity in response to CaCl2 required activation of CaR as the effect was prevented in cells transfected with a dominant negative (R796W) CaR construct (Fig. 5A).
Fig. 5.
Inhibition of NFAT5 blocks TNF promoter activity in mTAL cells. mTAL cells (5 × 105) were incubated in the absence or presence of 1.2 mM CaCl2 for 9 h after transfection with a TNF-luciferase promoter construct and either a small hairpin (sh)RNA vector targeting NFAT5 at exon 8 (U6-N5 ex8) or dominant negative calcium-sensing receptor (CaR; CaR-DN; A) or dominant negative NFAT5 (NFAT5-DN; B). Cells were harvested after stimulation, and firefly luciferase and Renilla activities were assayed; corresponding empty or scrambled vectors were included as controls. Values are means ± SD. P < 0.05; n = 3.
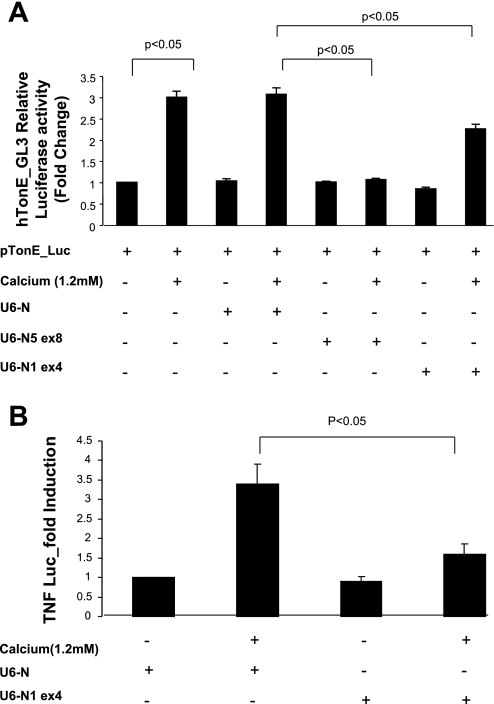
TNF promoter activity also was inhibited in cells transfected with a dominant negative NFAT5 construct (NFAT5-DN, 7 μg), but not the empty vector (Fig. 5B). Additional evidence for a role of NFAT5 is evident from experiments using hTonE-GL3, a reporter construct driven exclusively by a consensus NFAT5 binding site (43). Activity of hTonE-GL3 increased after untransfected cells or cells transfected with scrambled U6-N5 were challenged with calcium (Fig. 6A). In contrast, activation was not observed in cells transfected with U6-N5 ex8 and was partially inhibited when NFAT1 was blocked (U6-N1 ex4), suggesting an interaction between these isoforms. Moreover, inhibition of NFAT1 abolished TNF promoter activity (Fig. 6B) since TNF gene transcription is regulated by both NFAT5 and NFAT1 (18, 38).
Fig. 6.
shRNA inhibition of NFAT5 transcriptional and TNF promoter activities. mTAL cells were incubated in the absence or presence of 1.2 mM CaCl2 for 6 h after transfection with pTonE_Luc (A) or a TNF-luciferase promoter construct in combination with constructs to inhibit NFAT5 (U6-N5 ex8) or NFAT1 (U6-N1 ex4; B), as indicated. Cells were harvested after stimulation, and firefly luciferase and Renilla activities were assayed; scrambled U6-N was included as a control. Values are means ± SD. P < 0.05; n = 3.
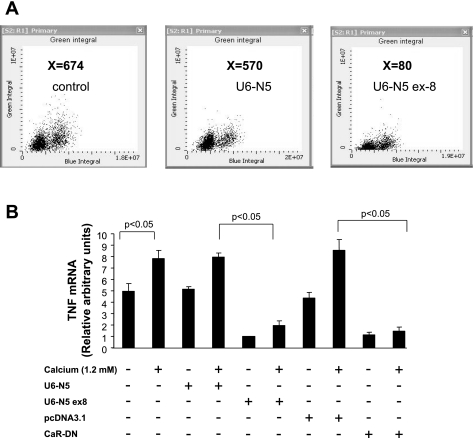
Effects of U6-N5 ex8 on endogenous NFAT5 protein expression and TNF mRNA accumulation in mTAL cells were evaluated by LSC and quantitative real-time RT-PCR, respectively. Analysis by LSC demonstrated that the level of NFAT5 protein was inhibited dramatically in mTAL cells transfected with U6-N5 ex8, but not a construct expressing scrambled U6-N5 (Fig. 7A). Accumulation of TNF mRNA also decreased significantly in mTAL cells challenged with CaCl2 and transfected with either U6-N5 ex8 or dominant negative CaR but not the respective control constructs (Fig. 7B). Collectively, these data demonstrate that NFAT5 contributes to TNF gene transcription in mTAL cells in response to CaR activation.
Fig. 7.
shRNA inhibition of NFAT5 protein and TNF mRNA accumulation in mTAL cells. A: mTAL cells were cultured on glass slides (∼90% confluent) and transfected with 3 μg of either U6-N5 ex8 or U6-N5. LSC analysis showed that U6-N5 ex8, but not scrambled U6-N, lowered NFAT5 protein levels in mTAL cells. Values (X, shown in bold) correspond to relative fluorescence intensity; a representative figure from 3 similar experiments is shown. B: mTAL cells (5 × 105) were incubated in the absence or presence of 1.2 mM CaCl2 after transfection with U6-N5, U6-N5 ex8, pcDNA3.1, or CaR-DN. TNF mRNA was determined using quantitative real-time RT-PCR and specific primers for TNF and β-actin. Values are means ± SD of measurements from 3 independent experiments.
Inhibition of NFAT5 attenuates CaR-mediated effects on Cl− influx.
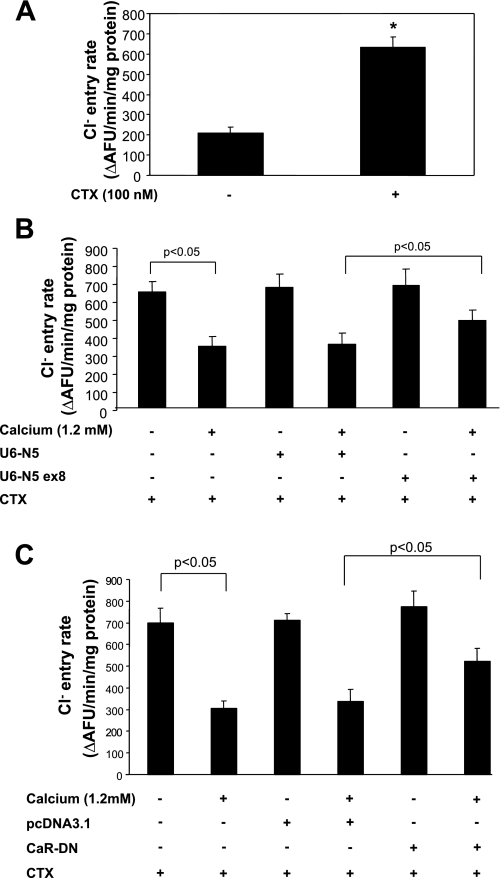
Transient transfection experiments were performed to assess the contribution of NFAT5 to CaR-mediated inhibition of apical Cl− entry in mTAL cells. Inhibition of NFAT5 was accomplished by transfecting cells with a shRNA-expressing construct targeting exon 8 of the murine NFAT5 gene (U6-N5 ex8). Cells were incubated with CTX, an irreversible direct activator of adenylyl cyclase, to increase the amount of NKCC2 protein at the plasma membrane (24). Entry of Cl− increased in cells incubated with CTX but was inhibited when cells were exposed to 1.2 mM CaCl2 for 6 h (Fig. 8, A and B, respectively). The inhibitory effect on Cl− influx was partially reversed when cells were transfected with U6-N5 ex8, but not the control U6-N5 construct (Fig. 8B). Transfection of mTAL cells with dominant negative CaR also attenuated the inhibitory effect on Cl− influx (Fig. 8C). These data indicate that NFAT5 is part of the mechanism whereby CaR activation inhibits Cl− uptake by mTAL cells.
Fig. 8.
Inhibition of NFAT5 prevents apical Cl− entry in mTAL cells. A: polarized mTAL cells were quiesced overnight and then incubated with CTX (100 nM) to increase Cl− influx. B: inhibition of NFAT5 with U6-N5 ex8 attenuates CaCl2-mediated inhibition of Cl− uptake; scrambled U6-N had no effect; n = 3. C: transfection with CaR-DN, but not empty plasmid (pcDNA3.1) attenuated CaCl2-mediated inhibitory effect on Cl− influx. Values are means ± SD. P < 0.05; n = 3.
DISCUSSION
We demonstrated that NFAT isoforms 1–5 are differentially expressed in mouse mTAL tubules and primary cultures of mTAL cells. Experiments using quantitative real-time RT-PCR revealed greater accumulation of NFAT5 (TonEBP/OREBP) and NFAT1 mRNA compared with NFAT4, -2, and -3, respectively. NFAT5 protein was constitutively expressed in mTAL tubules and cells and present in the nucleus of untreated mTAL cells, as well as cells transiently transfected with an NFAT5 overexpression vector. CaR-mediated TNF production was NFAT5 dependent and induced via a transcriptional mechanism in mTAL cells as TNF promoter activity was inhibited following transfection with dnNFAT5 and shRNA (U6-N5 ex8) constructs, which suppress NFAT5 activity by two different mechanisms. U6-N5 ex8, but not scrambled U6-N5, also inhibited CaR-mediated accumulation of TNF mRNA. The ability of CaR to inhibit apical Cl− entry was attenuated when NFAT5 activity was blocked, indicating that this transcription factor contributes to a mechanism that regulates NaCl reabsorption in the mTAL.
NFAT family members are expressed in cell types participating in immunity and inflammation as well as in many organs including the kidney (39, 42). They play a pivotal role in the transcription of cytokine and other genes critical for the immune response and tissue-specific function. NFAT5 is a member of the Rel family of transcriptional activators, as are NF-κB and the NFAT1-4 isoforms. Like other Rel proteins, the DBD of NFAT5 is in an NH2-terminal rel-homology region highly similar (up to 43% sequence identity) to the DBD in NFAT1-4 (35). NFAT1-4 isoforms and NF-κB interact with AP-1 transcription factors to facilitate transactivation of downstream genes, and this interaction at multiple rel-homology region residues results in stabilization of the ternary complex on DNA (36). Although NFAT5 is a distant member of the NFAT family because it lacks the calcineurin binding domain (35), it requires AP-1 for full expression of high NaCl-dependent increases in transactivation of genes encoding organic osmolytes, and can form a protein complex with c-Fos and c-Jun at specific binding sites (29). The molecular interactions by which NFAT5 regulates CaR-mediated TNF production in TAL cells are not yet clear but may also require an interaction with AP-1. The predominant NFAT site in the TNF promoter is the quasi-palindromic κ3 site, which resembles an NF-κB/Rel binding site (36, 38). Although strict physical cooperativity between NFAT and AP-1 is not required for TNF gene transcription, AP-1 family members do cooperate functionally with NFAT to promote TNF gene transcription (19, 44). Moreover, immediately adjacent to the κ3 site of the TNF promoter is a binding site for ATF2-Jun heterodimers, factors which cooperate functionally with NFAT dimers bound to the κ3 site (36). Interestingly, NFAT5 is the only member of the NFAT family to bind the −76 NFAT and κ3 sites in the TNF promoter, suggesting that the DNA-binding specificity of NFAT5 is different from that of other NFAT isoforms (18, 34). Consequently, NFAT5 also may act through unspecified alternative DNA sequences different from the NFAT sites that are currently known in cytokine gene promoters. Therefore, depending on the cell type and stimulation conditions, the NFAT5 homodimer, like other NFAT isoform dimers, may cooperate with transcription factors bound to the proximal TNF promoter, to form enhanceosome complexes that drive TNF gene transcription. Accordingly, it is of interest that TNF promoter activity in mTAL cells was blocked when NFAT1 activity was inhibited. The precise interaction of NFAT5 with other transcription factors, including NFAT1, that increase CaR-mediated TNF production in mTAL cells is currently being determined.
There is little information regarding activation of transcription factors or production of cytokines subsequent to CaR stimulation. However, activation of several signal transduction pathways upon CaR activation suggests that autocrine and paracrine effects of cytokines and eicosanoids could contribute to functions mediated via activation of this receptor (1, 3). The inhibitory effects of CaR activation on ouabain-sensitive O2 consumption via COX-2, in the absence of a direct effect on Na+-K+-ATPase activity in mTAL cells, suggest that CaR activation inhibits Na+ entry in these cells (2). The transport of NaCl across the apical membrane of the TAL occurs mainly via NKCC2 (23). Although NFAT5 is regulated by tonicity in T cells and the kidney (22, 34), our data demonstrate that CaR activation promotes induction of TNF gene transcription via NFAT5 and subsequently inhibits Cl− reabsorption in mTAL cells. The CaR is part of a mechanism that maintains tight control over calcium homeostasis, and renal expression of this receptor may be important for the regulation of salt and water balance (7). For instance, raising the serum-ionized Ca2+ level by 25% increased the urinary excretion of Na+ by 150% (16). Although the CaR is known to increase Ca2+ excretion and affect NaCl reabsorption, the mechanisms for these effects are not well understood.
Activation of the CaR expressed on basolateral membranes of the TAL initiates a mechanism that contributes to regulation of sodium and chloride movement in a furosemide-like manner (6, 27). The CaR expressed by primary cultures of mTAL cells regulates TNF production, COX-2 expression, and PGE2 synthesis, thereby contributing to a mechanism that regulates NaCl reabsorption in this nephron segment (45–47). Accordingly, CaR regulates TAL function by 1) a “short-term” inhibitory effect of 20- HETE on apical K+ channels, inhibition of cAMP, and an increase in phosphodiesterase activity; and 2) a “long-term” mechanism involving TNF/COX-2/PGE2 (47, 49). CaR stimulation initiates Gq- and Gi-dependent signaling pathways that activate NFAT to increase PGE2 synthesis via a TNF-dependent mechanism (3). The finding that the CaR activates NFAT5 in mTAL cells is consistent with a recent report on the analysis of NFAT family members in the kidney showing abundance of NFAT5 in the TAL (22). It also reveals a role for NFAT5 in CaR-mediated regulation of Cl− influx in this segment of the nephron, in addition to the role played by this transcription factor in the regulation of genes involved in protecting cells from hypertonic stress. It is interesting to note that a relationship between NFAT5 and two major apical transport molecules, namely, NKCC2 and ROMK, that are critical to mTAL function has recently been demonstrated (30, 32).
In summary, the present study describes the presence of NFAT5, the most primordial NFAT family member, and other NFAT isoforms in mTAL tubules and mTAL cells. We have shown that NFAT5 is integral to the activation of TNF gene transcription in response to CaR activation. The data demonstrate that inhibition of NFAT5 by two different mechanisms blocks CaR-mediated TNF production in mTAL cells. Although NFAT5 is critically important to cellular protective mechanisms in response to osmotic stress, the physiological function of NFAT5 remains to be elucidated in tissues where osmolality is maintained at fairly constant levels as well as in the kidney. The present study illustrates that NFAT5-dependent induction of TNF produced locally may subserve physiological regulation of ion transport pathways and extends the scope of NFAT5 function in the kidney by identifying a role for this transcription factor as a determinant of TNF expression and function in response to CaR activation in the mTAL.
GRANTS
This work was supported by National Institutes of Health Grants HL-085439 and R01-CA-28704.
Acknowledgments
The authors thank Drs. Jun Chen and Michael S. Goligorsky for help with immunofluorescence microscopy and Dr. Huda Abdullah for critically evaluating the manuscript. We also thank Dr. Hisashi Koga (Kazusa DNA Research Institute) for kindly supplying the mouse cDNA clone for NFAT5 and Dr. Karin Rodland (Pacific Northwest National Laboratory) for the CaR (DN) construct.
REFERENCES
- 1.Abdullah HI, Pedraza PL, Hao S, Rodland KD, McGiff JC, Ferreri NR. NFAT regulates calcium-sensing receptor-mediated TNF production. Am J Physiol Renal Physiol 290: F1110–F1117, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Abdullah HI, Pedraza PL, McGiff JC, Ferreri NR. Calcium-sensing receptor signaling pathways in medullary thick ascending limb cells mediate COX-2-derived PGE2 production: functional significance. Am J Physiol Renal Physiol 295: F1082–F1089, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdullah HI, Pedraza PL, McGiff JC, Ferreri NR. CaR activation increases TNF production by mTAL cells via a Gi-dependent mechanism. Am J Physiol Renal Physiol 294: F345–F354, 2008. [DOI] [PubMed] [Google Scholar]
- 4.Biwersi J, Verkman AS. Cell-permeable fluorescent indicator for cytosolic chloride. Biochemistry 30: 7879–7883, 1991. [DOI] [PubMed] [Google Scholar]
- 5.Boss V, Abbott KL, Wang XF, Pavlath GK, Murphy TJ. The cyclosporin A-sensitive nuclear factor of activated T cells (NFAT) proteins are expressed in vascular smooth muscle cells. Differential localization of NFAT isoforms and induction of NFAT-mediated transcription by phospholipase C-coupled cell surface receptors. J Biol Chem 273: 19664–19671, 1998. [DOI] [PubMed] [Google Scholar]
- 6.Brown EM, Chattopadhyay N, Vassilev PM, Hebert SC. The calcium-sensing receptor (CaR) permits Ca2+ to function as a versatile extracellular first messenger. Recent Prog Horm Res 53: 257–280, 1998. [PubMed] [Google Scholar]
- 7.Brown EM, Macleod RJ. Extracellular calcium sensing and extracellular calcium signaling. Physiol Rev 81: 239–297, 2001. [DOI] [PubMed] [Google Scholar]
- 8.Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiol Rev 87: 1441–1474, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Carroll MA, McGiff JC, Ferreri NR. Products of arachidonic acid metabolism. Methods Mol Med 86: 385–397, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Chytil M, Verdine GL. The Rel family of eukaryotic transcription factors. Curr Opin Struct Biol 6: 91–100, 1996. [DOI] [PubMed] [Google Scholar]
- 11.Cockerill GW, Bert AG, Ryan GR, Gamble JR, Vadas MA, Cockerill PN. Regulation of granulocyte-macrophage colony-stimulating factor and E-selectin expression in endothelial cells by cyclosporin A and the T-cell transcription factor NFAT. Blood 86: 2689–2698, 1995. [PubMed] [Google Scholar]
- 12.Colla E, Lee SD, Sheen MR, Woo SK, Kwon HM. TonEBP is inhibited by RNA helicase A via interaction involving the E'F loop. Biochem J 393: 411–419, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deptala A, Bedner E, Gorczyca W, Darzynkiewicz Z. Activation of nuclear factor kappa B (NF-kappaB) assayed by laser scanning cytometry (LSC). Cytometry 33: 376–382, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragomir A, Roomans GM. Increased chloride efflux in colchicine-resistant airway epithelial cell lines. Biochem Pharmacol 68: 253–261, 2004. [DOI] [PubMed] [Google Scholar]
- 16.El Hajj FG, Seifter J, Scott J, Brown EM. Calcium-regulated renal calcium handling in healthy men: relationship to sodium handling. J Clin Endocrinol Metab 83: 2366–2372, 1998. [DOI] [PubMed] [Google Scholar]
- 17.Eng B, Mukhopadhyay S, Vio CP, Pedraza PL, Hao S, Battula S, Sehgal PB, McGiff JC, Ferreri NR. Characterization of a long-term rat mTAL cell line. Am J Physiol Renal Physiol 293: F1413–F1422, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Esensten JH, Tsytsykova AV, Lopez-Rodriguez C, Ligeiro FA, Rao A, Goldfeld AE. NFAT5 binds to the TNF promoter distinctly from NFATp, c, 3 and 4, and activates TNF transcription during hypertonic stress alone. Nucleic Acids Res 33: 3845–3854, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falvo JV, Uglialoro AM, Brinkman BM, Merika M, Parekh BS, Tsai EY, King HC, Morielli AD, Peralta EG, Maniatis T, Thanos D, Goldfeld AE. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol Cell Biol 20: 2239–2247, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferraris JD, Burg MB. Tonicity-regulated gene expression. Methods Enzymol 428: 279–296, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Friedman PA, Gesek FA. Cellular calcium transport in renal epithelia: measurement, mechanisms, and regulation. Physiol Rev 75: 429–471, 1995. [DOI] [PubMed] [Google Scholar]
- 22.Gallazzini M, Karim Z, Bichara M. Regulation of ROMK (Kir 1.1) channel expression in kidney thick ascending limb by hypertonicity: role of TonEBP and MAPK pathways. Nephron Physiol 104: 126–135, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Gamba G, Miyanoshita A, Lombardi M, Lytton J, Lee WS, Hediger MA, Hebert SC. Molecular cloning, primary structure, and characterization of two members of the mammalian electroneutral sodium-(potassium)-chloride cotransporter family expressed in kidney. J Biol Chem 269: 17713–17722, 1994. [PubMed] [Google Scholar]
- 24.Geibel J, Sritharan K, Geibel R, Geibel P, Persing JS, Seeger A, Roepke TK, Deichstetter M, Prinz C, Cheng SX, Martin D, Hebert SC. Calcium-sensing receptor abrogates secretagogue- induced increases in intestinal net fluid secretion by enhancing cyclic nucleotide destruction. Proc Natl Acad Sci USA 103: 9390–9397, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giebisch G Renal potassium transport: mechanisms and regulation. Am J Physiol Renal Physiol 274: F817–F833, 1998. [DOI] [PubMed] [Google Scholar]
- 26.Greger R Ion transport mechanisms in thick ascending limb of Henle's loop of mammalian nephron. Physiol Rev 65: 760–797, 1985. [DOI] [PubMed] [Google Scholar]
- 27.Hebert SC Extracellular calcium-sensing receptor: implications for calcium and magnesium handling in the kidney. Kidney Int 50: 2129–2139, 1996. [DOI] [PubMed] [Google Scholar]
- 28.Hebert SC, Cheng S, Geibel J. Functions and roles of the extracellular Ca2+-sensing receptor in the gastrointestinal tract. Cell Calcium 35: 239–247, 2004. [DOI] [PubMed] [Google Scholar]
- 29.Irarrazabal CE, Williams CK, Ely MA, Birrer MJ, Garcia-Perez A, Burg MB, Ferraris JD. Activator protein-1 contributes to high NaCl-induced increase in tonicity-responsive enhancer/osmotic response element-binding protein transactivating activity. J Biol Chem 283: 2554–2563, 2008. [DOI] [PubMed] [Google Scholar]
- 30.Jeon US, Han KH, Park SH, Lee SD, Sheen MR, Jung JY, Kim WY, Sands JM, Kim J, Kwon HM. Downregulation of renal TonEBP in hypokalemic rats. Am J Physiol Renal Physiol 293: F408–F415, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Kikeri D, Azar S, Sun A, Zeidel ML, Hebert SC. Na+-H+ antiporter and Na+-(HCO3−)n symporter regulate intracellular pH in mouse medullary thick limbs of Henle. Am J Physiol Renal Fluid Electrolyte Physiol 258: F445–F456, 1990. [DOI] [PubMed] [Google Scholar]
- 32.Lee HW, Kim WY, Song HK, Yang CW, Han KH, Kwon HM, Kim J. Sequential expression of NKCC2, TonEBP, aldose reductase, and urea transporter-A in developing mouse kidney. Am J Physiol Renal Physiol 292: F269–F277, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Rodriguez C, Aramburu J, Jin L, Rakeman AS, Michino M, Rao A. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 15: 47–58, 2001. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Rodriguez C, Aramburu J, Rakeman AS, Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA 96: 7214–7219, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene 20: 2476–2489, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Mahlangu DA, Dix JA. Halide fluxes in epithelial cells measured with an automated cell plate reader. Anal Biochem 325: 28–34, 2004. [DOI] [PubMed] [Google Scholar]
- 38.McCaffrey PG, Goldfeld AE, Rao A. The role of NFATp in cyclosporin A-sensitive tumor necrosis factor-alpha gene transcription. J Biol Chem 269: 30445–30450, 1994. [PubMed] [Google Scholar]
- 39.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA 96: 2538–2542, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbergh L, Valckx D, Waer M, Mathieu C. Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 11: 305–312, 1999. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl MW A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 15: 707–747, 1997. [DOI] [PubMed] [Google Scholar]
- 43.Trama J, Lu Q, Hawley RG, Ho SN. The NFAT-related protein NFATL1 (TonEBP/NFAT5) is induced upon T cell activation in a calcineurin-dependent manner. J Immunol 165: 4884–4894, 2000. [DOI] [PubMed] [Google Scholar]
- 44.Tsai EY, Jain J, Pesavento PA, Rao A, Goldfeld AE. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol Cell Biol 16: 459–467, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, An SJ, Wang WH, McGiff JC, Ferreri NR. CaR-mediated COX-2 expression in primary cultured mTAL cells. Am J Physiol Renal Physiol 281: F658–F664, 2001. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, McGiff JC, Ferreri NR. Regulation of cyclooxygenase isoforms in the renal thick ascending limb: effects of extracellular calcium. J Physiol Pharmacol 51: 587–595, 2000. [PubMed] [Google Scholar]
- 47.Wang D, Pedraza PL, Abdullah HI, McGiff JC, Ferreri NR. Calcium-sensing receptor-mediated TNF production in medullary thick ascending limb cells. Am J Physiol Renal Physiol 283: F963–F970, 2002. [DOI] [PubMed] [Google Scholar]
- 48.Wang W, Lu M, Balazy M, Hebert SC. Phospholipase A2 is involved in mediating the effect of extracellular Ca2+ on apical K+ channels in rat TAL. Am J Physiol Renal Physiol 273: F421–F429, 1997. [DOI] [PubMed] [Google Scholar]
- 49.Wang WH, Lu M, Hebert SC. Cytochrome P-450 metabolites mediate extracellular Ca2+-induced inhibition of apical K+ channels in the TAL. Am J Physiol Cell Physiol 271: C103–C111, 1996. [DOI] [PubMed] [Google Scholar]
- 50.Woo SK, Lee SD, Na KY, Park WK, Kwon HM. TonEBP/NFAT5 stimulates transcription of HSP70 in response to hypertonicity. Mol Cell Biol 22: 5753–5760, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo SK, Nahm O, Kwon HM. How salt regulates genes: function of a Rel-like transcription factor TonEBP. Biochem Biophys Res Commun 278: 269–271, 2000. [DOI] [PubMed] [Google Scholar]