Abstract
Artemis is a multifunctional phospho-protein with roles in V(D)J recombination, repair of double-strand breaks by nonhomologous end-joining, and regulation of cell cycle checkpoints after DNA damage. Here, we describe a novel function of Artemis as a negative regulator of p53 in response to oxidative stress in both primary cells and cancer cell lines. We show that depletion of Artemis under typical culture conditions (21% oxygen) leads to a spontaneous phosphorylation and stabilization of p53, and resulting cellular G1 arrest and apoptosis. These effects are suppressed by co-depletion of DNA-PKcs, but not ATM, indicating that Artemis is an inhibitor of DNA-PKcs-mediated stabilization of p53. Culturing of cells at 3% oxygen or treatment with an antioxidant abrogated p53 stabilization indicating that oxidative stress is the responsible cellular stimulus. Treatment with IR or hydrogen peroxide did not cause activation of this signaling pathway, while inhibitors of mitochondrial electron transport were effective in reducing its activation. In addition, we show that p53-inducible genes involved in reducing reactive oxygen species (ROS) are upregulated by Artemis depletion. These findings indicate that Artemis and DNA-PKcs participate in a novel, signaling pathway to modulate p53 function in response to oxidative stress produced by mitochondrial respiration.
Keywords: Artemis, p53, DNA-PKcs, oxidative stress
Introduction
Artemis is a phospho-protein that appears to play multiple roles in mammalian cells. It is know to be required for V(D)J recombination and its functional loss causes a severe combined immunodeficiency (Ma et al., 2002; Moshous et al., 2001; Moshous et al., 2000; Rooney et al., 2002). In addition Artemis-deficient cells are radiosensitive and are defective in the repair of a minor fraction of double-strand breaks created by ionizing radiation (IR) (Riballo et al., 2004). Artemis also has roles in cell cycle regulation in response to DNA damage as it has been proven to be a substrate of the phosphatidylinositol-3-OH kinase-like kinases (PIKKs) ATM (Ataxia telangiectasia mutated), ATR (ataxia-telangiectasia and Rad3-related) and DNA-PKcs (DNA-dependent protein kinase catalytic subunit) both in vitro and in vivo (Chen et al., 2005; Geng et al., 2007; Ma et al., 2005; Ma et al., 2002; Poinsignon et al., 2004; Riballo et al., 2004; Wang et al., 2005; Zhang et al., 2004). Our previous findings have also shown that it is involved in the recovery from the G2/M cell cycle checkpoint by regulating the activation of Cdk2-cyclin B at the centrosome (Geng et al., 2007; Zhang et al., 2004). In our studies reported here we now show that Artemis also has a role in regulating the G1 phase of the cell cycle by acting as a negative regulator of p53.
The tumor suppressor p53 has well established roles in mediating cell cycle arrest, senescence, and apoptosis in response to DNA damage; however, more recent findings indicate that it also has a role in regulating energy metabolism and oxidative stress (Bensaad and Vousden, 2007; Karawajew et al., 2005). For example p53 regulates a number of genes that are involved in either mitochondrial respiration (SCO2) or glycolysis (phosphoglycerate mutase) (Kondoh et al., 2005; Matoba et al., 2006). In fact, disruption of p53 function may account for the Warburg effect in which cancer cells switch toward glycolysis for energy production. The major source of ROS within cells is mitochondrial respiration, thus, the ability of p53 to regulate this process as well as glycolysis allows it to modulate the intracellular levels of ROS, and thus to control oxidative stress. However, the control of oxidative stress by p53 is complex as it positively regulates both antioxidant and pro-oxidant genes. Under low stress conditions antioxidant genes such as the sestrins, glutathione peroxidase, and the aldehyde 4 family are all upregulated by p53 in order to reduce levels of ROS (Bensaad and Vousden, 2007). In addition, p53 regulates the gene TIGAR (TP53-induced glycolysis and apoptosis regulator) that switches glucose consumption from glycolysis to an alternate pathway that results in production of NADPH which is required for ROS scavenging by reduced glutathione (Bensaad et al., 2006). Interestingly, p53 also positively regulates pro-oxidant genes such as PIG-3 and proline oxidase, and the aforementioned SCO2 gene, all of which act to increase intracellular ROS levels under high stress conditions. Although many of the downstream targets of p53 in the regulation of oxidative stress have been characterized, the upstream regulation of p53 in response to oxidative stress is poorly understood. Our findings reported here help to clarify these pathways by showing that Artemis and DNA-PKcs function antagonistically to regulate the activation of p53 in response to oxidative stress derived from oxidative phosphorylation.
Materials and Methods
Plasmid construction and siRNA
Human wild type p53 cDNA was inserted into pcDNA3.1 at the BamH I and Apa I sites. p53 mutants were prepared using the QuickChange site-directed mutagenesis kit (Stratagene). Artemis plasmids were constructed as described previously (Geng et al., 2007). Lipofectamine 2000 (Invitrogen) was used for DNA transfections.
Artemis siRNAs were: A1 (CUGAAGAGAGCUAGAACAG); A2 (5′-UUAGGAGUCCAGGUUCAUG); A3 (GCUGCAAGUUAGUGAAACA); A4 (GCAUGGAUGUGAUUCAACA); A5 (CGAGUAACCAGCUCAUAAA). Control, DNA-PKcs and ATM siRNA sequences have been described previously (Geng et al., 2007; Zhang et al., 2004). Ku80 siRNA (NM-021141) and Ku70 siRNA (SC-29383) were purchased from Dharmacon and Santa Cruz Biotechnology, Inc., respectively. siRNAs were transfected into cells using Oligofectamine or Lipofectamine 2000 reagents (Invitrogen).
Cell culture
U2OS (osteosarcoma), HeLa (cervical carcinoma), H1299 (nonsmall cell lung carcinoma), HCT-116 (colon carcinoma) and primary fibroblast MRC-5 cells were cultured in DMEM with 10% fetal calf serum. Mouse embryonic fibroblast cells (MEFs) were cultured in DMEM with 10% fetal calf serum plus nonessential amino acids. HCT-116 cells (p53+/+, p53−/−, p21+/+ and p21−/−) were provided by B. Vogelstein.
Immunoprecipitation, BrdU labeling, and cell cycle analysis
Immunoprecipitations, cell cycle analysis, and BrdU labeling were performed as described previously (Zhang et al., 2004).
Antibodies
Antibodies against Artemis, GAPDH, DNA-PKcs, ATM, γ-H2AX, Chk1, and Chk2 have been described previously (Geng et al., 2007; Zhang et al., 2004). Rabbit polyclonal antibodies against p53 (FL393), Bax (SC-493), PARP (SC-7150) and Goat polyclonal antibodies against p21(SC-397), Ku70 (C19), Ku86 (M20) were from Santa Cruz Biotech. Rabbit polyclonal antibody (CM-5) and Mouse monoclonal antibody against p53 (DO-1) were from Novocastra Laboratories Ltd. and EMD CALBIOCHEM, respectively. p53-pS15, p53-pS37 antibodies were from Cell Signaling Technology. Rabbit polyclonal antibodies against TIGAR and Sestrin 2 were from ProSci, Inc. and ProteinTec Group, Inc., respectively.
Artemis mice and MEFs
Artemis mice were provided by Frederick Alt. Preparation of MEF cells and genotyping was performed as described previously (Rooney et al., 2002).
Assay for Annexin V
p21−/− and p21+/+ HCT116 cells were harvested 48 hrs after transfection with Artemis siRNA. Annexin V was labeled using Annexin V-FITC Apoptosis Detection Kit 1 (BD Pharmingen).
Results
Depletion of Artemis Induces p53 Accumulation, Cell Cycle Arrest, and Apoptosis
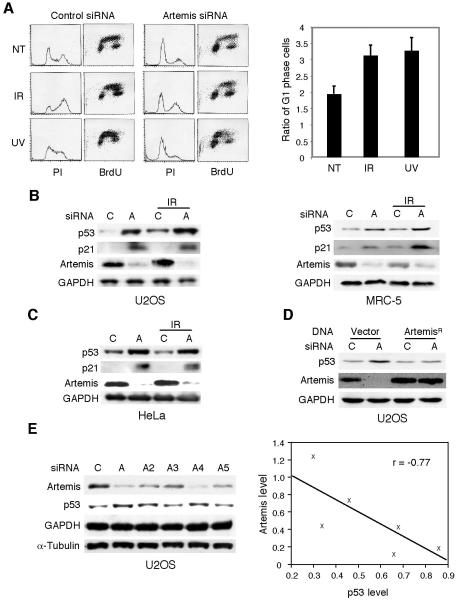
Our prior studies of Artemis have shown that it plays a role in cell cycle checkpoint regulation after exposure of cells to DNA damage (Geng et al., 2007; Zhang et al., 2004). During the course of these studies we noted that depletion of Artemis by small interfering RNA (siRNA) caused an accumulation of U2OS cells in the G1 phase of the cell cycle whether or not cells were exposed to DNA damage (Fig. 1A). A similar result was also observed in HeLa cells (data not shown). Since p53 is a well-established mediator of G1 arrest, we assessed whether p53 levels were altered by Artemis depletion. In both cancer cell lines and primary fibroblast cells (MRC-5) Artemis depletion induced upregulation and activation of p53 as indicated by the concomitant upregulation of p21 (Fig. 1B). Surprisingly, this accumulation of p53 also occurred in HeLa cells (Fig. 1C), which are typically unable to upregulate p53 due to the presence of the human papilloma virus (HPV) E6 protein which mediates degradation of p53 through an associated E3 ubiquitin ligase (Scheffner et al., 1993). A second cell line termed SiHa (Seedorf et al., 1987), which expresses the HPV E6 protein also showed upregulation of p53 upon Artemis depletion (data not shown). Thus, Artemis depletion appears to stabilize p53 through a mechanism distinct from that induced by DNA damage. To demonstrate that the depletion of Artemis by this siRNA was specific, an Artemis construct refractory to the siRNA was shown to suppress the upregulation of p53 (Fig. 1D). To further validate this approach, we used five different siRNAs to target Artemis and monitored the changes in p53 levels. As shown (Fig. 1E), depletion of Artemis strongly statistically correlated with accumulation of p53 as indicated by regression analysis.
Figure 1. Depletion of Artemis causes accumulation and activation of p53.
(A) FACS analysis of U2OS cells subjected to the indicated siRNAs and either not treated (NT) or treated with IR (3 Gy) or UV (3 J/m2) ( (left panel). Right panel shows quantitation of results shown in left panel. In each experiment the ratio of the fraction of G1 cells for the Artemis siRNA relative to the control siRNA was calculated. Error bars represent SEM. (B,C) Immunoblots showing depletion of Artemis by siRNA results in accumulation of p53 and upregulation of p21 in normal and cancer cell lines without exposure to exogenous DNA damage. “A” and “C” indicate Artemis and control siRNAs, respectively. IR dose was 6 Gy. (D) Accumulation of p53 is prevented by transfection of an Artemis construct (ArtemisR) refractory to the Artemis siRNA. (E) Immunoblot showing depletion of Artemis by different siRNAs results in accumulation of p53 (left panel). GAPDH and α-Tubulin were used as loading controls. Right panel shows the regression analysis of results shown in left panel. “r” indicates the correlation coefficient.
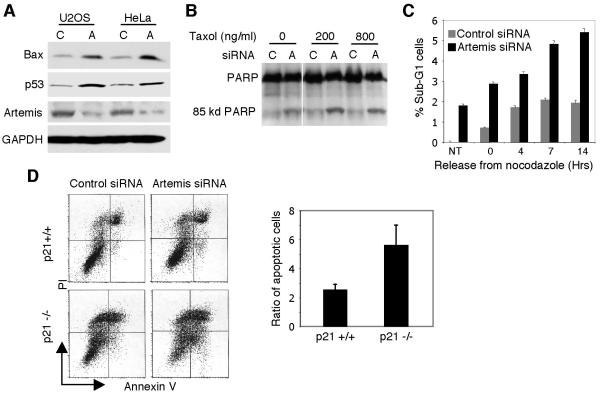
In addition to cell cycle arrest, activated p53 also induces programmed cell death (Vogelstein et al., 2000). We, therefore, examined markers of apoptosis after Artemis depletion. As shown, the pro-apoptotic gene Bax (Zhang et al., 2000) was upregulated (Fig. 2A), and both PARP cleavage and sub-G1 cells were increased (Fig. 2B,C). Furthermore, simultaneous treatment with either one of the spindle poisons Taxol or nocadazole significantly enhanced the level of apoptosis observed with Artemis depletion. To further confirm this phenotype, we examined p21 null cells (Waldman et al., 1995) for apoptosis mediated by Artemis depletion. Others have shown that in the absence of p21 apoptosis due to cellular stress is enhanced (Sohn et al., 2006; Tian et al., 2000). In the absence of p21, Artemis-depleted HCT116 cells exhibited a significantly enhanced level of apoptosis (Fig. 2D) indicating that reduction of Artemis levels strongly induces programmed cell death.
Figure 2. Depletion of Artemis causes apoptosis in cancer cell lines.
(A) Immunoblot showing depletion of Artemis causes upregulation of the apoptotic marker Bax. (B,C) Taxol and nocodazole enhance apoptosis induced by Artemis depletion as measured by PARP degradation and cellular sub-G1 content in U2OS cells. (D) FACS analysis (left panel) and quantitation (right panel) of annexin V staining in HCT116 cells shows absence of p21 enhances apoptosis induced by Artemis depletion. The ratio of apoptotic cells was quantitated as described in Fig. 1A.
Artemis Depletion Stabilizes p53 by DNA-PKcs-Mediated Phosphorylation
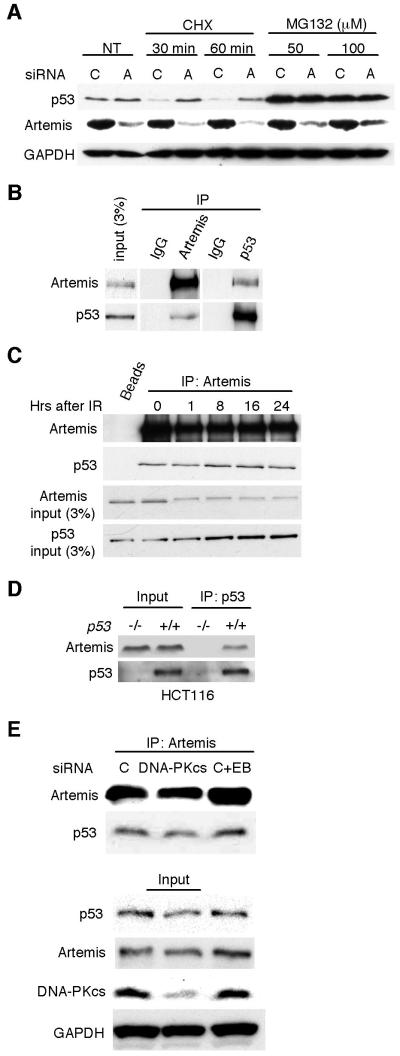
We next examined whether Artemis depletion upregulated p53 by a transcriptional or posttranslational mechanism. Knockdown of Artemis in HeLa cells did not alter the levels of p53 transcripts compared to control cells as determined by quantitative RT-PCR (Fig. S1). However, in the presence of the translational inhibitor cycloheximide stabilization of p53 was observed with Artemis depletion by siRNA (Fig. 3A). In the presence of the 26S proteosome inhibitor MG132, stabilization of p53 was observed after treatment with either control or Artemis siRNA (Fig. 3A). In addition, the half-life of p53 was increased in U2OS cells treated with Artemis siRNA compared to control siRNA (Fig. S2). Taken together, these results indicate that Artemis depletion causes stabilization of p53 by a posttranslational mechanism. This latter finding suggested that Artemis and p53 might physically interact. We, therefore, performed reciprocal co-immunoprecipitation experiments, and as shown (Fig. 3B-D), the results indicated that Artemis and p53 reside in a common complex. Furthermore, depletion of DNA-PKcs did not affect the interaction between Artemis and p53, nor did the inclusion of ethidium bromide indicating that the interaction is not mediated by DNA (Fig. 3E). Taken together these results suggest that Artemis may directly regulate p53.
Figure 3. Artemis acts at the posttranslational level and interacts with p53.
(A) Immunoblot showing that accumulation of p53 induced by Artemis depletion is not inhibited by the translation inhibitor cycloheximide (CHX) in U2OS cells. P53 accumulation occurs in the presence of the 26S proteosome inhibitor MG132 with or without Artemis depletion. (B) Reciprocal co-IP assays performed in HCT-116 cells between Artemis and p53. “IgG” indicates a nonspecific immunoglobulin. (C) Co-IP assay showing that Artemis interacts with p53 before and after IR treatment in HCT-116 cells. IR dose was 6 Gy, and the “0” lane indicates cell that were not irradiated. “Beads” indicates assay performed without Artemis antibody. (D) Co-IP between p53 and Artemis is not observed in p53-deficient HCT-116 cells. (E) Co-IP between p53 and Artemis occurs after depletion of DNA-PKcs. “C+EB” indicates control siRNA and that the co-IP was performed in the presence of ethidium bromide.
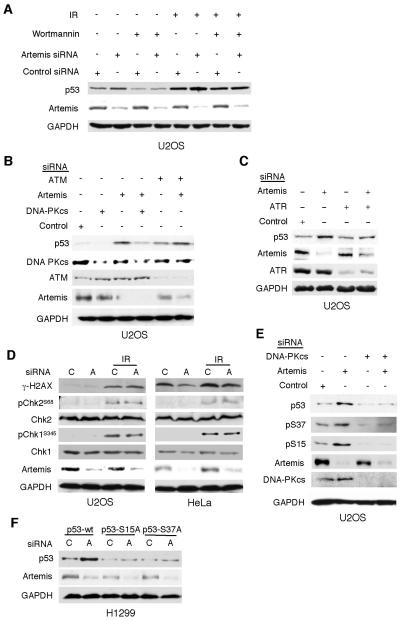
Previous studies from our laboratory and that of others have shown that Artemis is an in vivo substrate of the phosphatidylinositol-3-OH kinase-like kinases (PIKKs) DNA-PKcs, ATM and ATR (Chen et al., 2005; Riballo et al., 2004; Wang et al., 2005; Zhang et al., 2004). All three of these kinases are also known to phosphorylate p53 in order to mediate its stabilization and activation. In the presence of wortmannin, an inhibitor of all three of these PIKKs (Sarkaria et al., 1998), stabilization of p53 by Artemis depletion was suppressed (Fig. 4A). We next used siRNA to deplete each of these kinases to determine which one was responsible for the stabilization of p53 upon Artemis depletion. Depletion of ATM did not suppress p53 stabilization after knockdown of Artemis, while depletion of DNA-PKcs strongly suppressed this effect (Fig. 4B). Use of a second siRNA directed against DNA-PKcs confirmed this result (data not shown). Simultaneous knockdown of Artemis and ATR led to only a small increase in p53 levels compared to knockdown of ATR alone (Fig. 4C), thus, the role of this kinase, if any, is unclear. Since Artemis has been implicated in repair of DSBs via nonhomologous end-joining (NHEJ) (Ma et al., 2002; Riballo et al., 2004; Wang et al., 2005), depletion of Artemis could result in unrepaired DNA damage, and the activation of DNA damage response pathways. However, depletion of Artemis caused neither the phosphorylation of H2AX nor Chk2 or Chk1 (Fig. 4D), indicating that unrepaired DNA damage was not the cause of the observed p53 stabilization. This result also affirmed our conclusion that ATM and ATR were not activated by Artemis depletion. Because DNA-PKcs was implicated, we next examined whether the heterodimer Ku, the DNA end-binding component of DNA-PK (Meek et al., 2004), was required for p53 stabilization. Neither Ku70 nor Ku80 suppressed p53 stabilization upon Artemis depletion indicating that only the catalytic subunit of DNA-PK is involved (Fig. S3). PIKKs are known to phosphorylate p53 at serine residues S15 and S37 to prevent its degradation by E3 ubiquitin ligases (Shieh et al., 1997). Examination of these two sites showed that both were phosphorylated upon Artemis depletion, and that the concurrent depletion of DNA-PKcs suppressed this modification (Fig. 4E). Concurrent knockdown of ATM, but not ATR, failed to suppress these modifications (Fig. S4), thus, as indicated above, we cannot exclude a role for ATR in this pathway, although, it is clearly not activated for the phosphorylation of Chk1. Finally, mutation of these sites to alanine abrogated the stabilization of p53 upon Artemis depletion (Fig. 4F). Taken together, these findings indicate that Artemis depletion results in the spontaneous phosphorylation and stabilization of p53, and that this process is mediated by DNA-PKcs without a requirement for the Ku heterodimer. Interestingly, phosphorylation at these sites also occurs in HeLa cells after IR (data not shown), although stabilization of p53 is not observed (Fig. 1C), suggesting that Artemis depletion results in a separate p53 modification that prevents degradation by the HPV E6 associated ubiquitin ligase.
Figure 4. Stabilization of p53 by Artemis depletion is mediated by DNA-PKcs.
(A) Immunoblots showing that wortmannin (30 μM) inhibits stabilization of p53 by depletion of Artemis. For IR experiments the dose was 3 Gy. (B,C) Immunoblots showing co-depletion of DNA-PKcs, but not ATM, suppresses the stabilization of p53 by Artemis depletion. (D) Chk1, Chk2, and H2AX are not phosphorylated by Artemis depletion in the absence of exogenous DNA damage. (E) Artemis depletion causes phosphorylation of S15 and S37 sites of p53, and these modifications are suppressed by co-depletion of DNA-PKcs. (F) Mutation of S15 or S37 to alanine (S15A, S37A) suppresses the stabilization of p53 by Artemis depletion. Wild-type and mutant p53s were stably expressed in H1299 cells prior to siRNA-mediated depletion of Artemis.
Mitochondrial Oxidative Stress Induces Stabilization of p53 in the Absence of Artemis
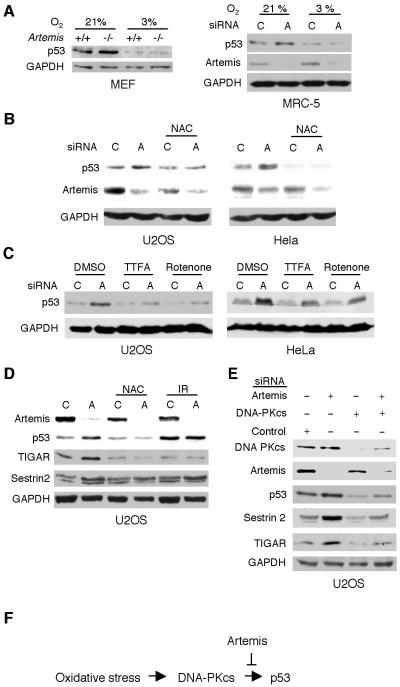
High levels of p53 are known to result in an embryonic lethal phenotype such as is observed in Mdm2 nullizygous mice (Montes de Oca Luna et al., 1995). However, Artemis nullizygous mice develop normally, except for impaired lymphocyte maturation (Rooney et al., 2002), suggesting that p53 levels are not abnormally high in these animals. Consistent with this supposition, we examined four tissues from wild-type and Artemis−/− mice and found no differences in p53 expression levels (Fig. S5A). Interestingly, however, culturing of Artemis mouse embryonic fibroblasts (MEFs) showed that with increasing passage number the levels of p53 increased to a much greater extent in nullizygous MEFs compared to wild-type MEFs (Fig. S5B). In addition, the nullizygous MEFs exhibited a greater fraction of cells in the G1 phase than did wild-type MEFs, and moreover, stable expression of Artemis in the nullizygous cells greatly reduced the G1 population (Fig. S6). Taken together, these findings suggested that culture stress might be the stimulus that induces stabilization of p53 upon Artemis depletion. To examine this hypothesis, we cultured both MEF and MRC5 cells at 3% O2, and found that depletion of Artemis no longer induced a strong stabilization of p53 (Fig. 5A). Furthermore, exposing MEFs cultured at 3% O2 to increasing doses of IR did not cause higher stabilization of p53 in Artemis−/− cells further validating our conclusion that DNA damage is not the stimulus that activates DNA-PKcs in the absence of Artemis (Fig. S7A). Hyperoxic conditions produce high levels of intracellular ROS which could provide the signal for the activation of this pathway, however, treatment with hydrogen peroxide (H2O2) of cells cultured at 3% O2 did not result in differential stabilization of p53 (Fig. S7B). Nevertheless, incubation with the antioxidant N-acetyl-l-cysteine (NAC) abrogated the stabilization of p53 induced by Artemis depletion in HeLa and U2OS cells cultured at 21% O2 indicating that ROS is, in fact, the stimulus for p53 accumulation (Fig. 5B). Recently, it has been shown that mitochondrial respiration plays a critical role in the activation of p53 (Karawajew et al., 2005). This finding combined with our results suggested that ROS produced by mitochondrial respiration might be the source of the signaling stimulus. As a test of this premise, two inhibitors of oxidative phosphorylation, rotenone and thenoyltrifluoroacetone (TTFA), were shown to reduce the stabilization of p53 mediated by Artemis depletion (Fig. 5C). Finally, p53 positively regulates TIGAR and Sestrin 2, two genes involved in reducing ROS (Bensaad et al., 2006; Budanov et al., 2004). As shown (Fig. 5D), Artemis depletion, but not IR treatment, caused ROS-dependent upregulation of these two genes. This effect was suppressed by co-depletion of DNA-PKcs (Fig. 5E) indicating that Artemis and DNA-PKcs regulate p53 by a mechanism that is distinct from DNA damage-mediated activation of p53.
Figure 5. Stabilization of p53 by Artemis depletion is induced by oxidative stress derived from mitochondrial respiration.
(A) Immunoblots showing that reduction of oxygen tension from 21% to 3% abrogates stabilization of p53 by Artemis depletion in primary cell lines. (B) The antioxidant NAC (10 mM) inhibits p53 stabilization induced by Artemis depletion. (C) Rotenone (0.04 mM) and TTFA (0.2 mM), inhibitors of mitochondrial electron transport, reduce the stabilization of p53 induced by Artemis depletion. (D,E) p53 responsive oxidative stress genes, TIGAR and Sestrin 2, are upregulated by Artemis depletion, and this effect is suppressed by co-depletion of DNA-PKcs.
Discussion
Our findings indicate that ROS, produced by mitochondrial respiration, provide a signal that activates DNA-PKcs to phosphorylate and thereby stabilize p53, which in turn broadly activates genes involved in cellular senescence, apoptosis, and regulation of oxidative stress. This pathway is negatively regulated by Artemis, presumably directly since Artemis independently interacts physically with both p53 and DNA-PKcs (Fig. 5F). Thus Artemis may act as a rheostat to control the degree of activation of p53 in response to oxidative stress. The pathway by which ROS produced by mitochondrial respiration signals to DNA-PKcs is unclear at present. However, ROS have been implicated as second messengers in multiple signaling pathways (Mayer and Noble, 1994; Punj and Chakrabarty, 2003). Furthermore, redox-regulating oxidoreductases have been shown to function as interacting partners of p53, which indicates the importance of intracellular redox-regulating factors in p53-activation. For example, WOX1, an oxidoreductase, is a proapoptotic protein, which becomes phosphorylated in response to stress or apoptotic stimuli and forms a complex with p53, which subsequently translocates to the mitochondria and further to nuclei to induce apoptosis (Chang et al., 2003). In addition, the NQO1 oxidoreductase regulates p53 in an MDM2-independent manner, and is required for the stabilization of p53 (Anwar et al., 2003; Asher et al., 2001; Asher et al., 2002). Finally ROS-induced stress, created by mitochondrial respiration, has been shown to be directly responsible for the activation of p53 (Karawajew et al., 2005). Clearly, the full extent of the in vivo implications of this Artemis-regulated p53 pathway will require further investigation.
DNA damage does not appear to contribute to the signaling mechanism since phosphorylation of Chk1, Chk2, and γH2AX was not observed, and there is no requirement for the Ku heterodimer. Recently, it has been shown that direct tethering of DNA-PKcs to chromatin causes activation of its kinase activity in the absence of Ku (Soutoglou and Misteli, 2008). This finding indicates, as we have shown here, that in certain contexts Ku is not an essential co-factor for DNA-PKcs. We were also able to exclude ATM as a component of this pathway, but, were unable to rigorously exclude ATR, although, if it is involved it is clearly not redundant with DNA-PKcs. Taken together, our findings reveal a novel pathway for the regulation of oxidative stress by DNA-PKcs and p53, and have implications for the processes of aging and tumor suppression. Interestingly, both hyperoxic and hypoxic conditions, commonly found in tumor microenvironments, lead to the formation of excess ROS from mitochondria (Guzy and Schumacker, 2006). Artemis is, therefore, a potential target for anticancer therapy since its inhibition in tumors may activate p53 which has been shown to lead to tumor regression (Ventura et al., 2007; Xue et al., 2007). In fact, observations in mice support such a possibility, since Artemis null mice only exhibit accelerated tumorigenesis in the absence of p53 even though lack of Artemis results in genomic instability (Rooney et al., 2004; Rooney et al., 2002). Thus, in the absence of Artemis, p53 may be more readily activated in response to cellular stress, thereby preventing tumor formation. However, in the absence of both Artemis and p53 increased cellular stress leads to an increased incidence of cancer. These considerations also suggest the possibility that Artemis may function both as a tumor suppressor and an oncogene. In an overexpressed state Artemis may suppress the activation of p53 and thus promote tumorigenesis, alternatiavely, its role in the maintenance of genomic stability via its functions in DNA repair and cell cycle control indicate that it can act as a tumor suppressor.
Supplementary Material
Acknowledgements
We thank G. Lozano for helpful discussions of the manuscript. This work was supported by National Cancer Institute grant CA096574 (R.J.L.).
References
- Anwar A, Dehn D, Siegel D, Kepa JK, Tang LJ, Pietenpol JA, et al. Interaction of human NAD(P)H:quinone oxidoreductase 1 (NQO1) with the tumor suppressor protein p53 in cells and cell-free systems. J Biol Chem. 2003;278:10368–73. doi: 10.1074/jbc.M211981200. [DOI] [PubMed] [Google Scholar]
- Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci U S A. 2001;98:1188–93. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A. 2002;99:3099–104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–91. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- Chang NS, Doherty J, Ensign A, Lewis J, Heath J, Schultz L, et al. Molecular mechanisms underlying WOX1 activation during apoptotic and stress responses. Biochem Pharmacol. 2003;66:1347–54. doi: 10.1016/s0006-2952(03)00484-2. [DOI] [PubMed] [Google Scholar]
- Chen L, Morio T, Minegishi Y, Nakada S, Nagasawa M, Komatsu K, et al. Ataxia-telangiectasia-mutated dependent phosphorylation of Artemis in response to DNA damage. Cancer Sci. 2005;96:134–41. doi: 10.1111/j.1349-7006.2005.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Zhang X, Zheng S, Legerski RJ. Artemis links ATM to G2/M checkpoint recovery via regulation of Cdk1-cyclin B. Mol Cell Biol. 2007;27:2625–35. doi: 10.1128/MCB.02072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91:807–19. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- Karawajew L, Rhein P, Czerwony G, Ludwig WD. Stress-induced activation of the p53 tumor suppressor in leukemia cells and normal lymphocytes requires mitochondrial activity and reactive oxygen species. Blood. 2005;105:4767–75. doi: 10.1182/blood-2004-09-3428. [DOI] [PubMed] [Google Scholar]
- Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–85. [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J Biol Chem. 2005;280:33839–46. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–94. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mayer M, Noble M. N-acetyl-L-cysteine is a pluripotent protector against cell death and enhancer of trophic factor-mediated cell survival in vitro. Proc Natl Acad Sci U S A. 1994;91:7496–500. doi: 10.1073/pnas.91.16.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–41. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–86. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- Moshous D, Li L, Chasseval R, Philippe N, Jabado N, Cowan MJ, et al. A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum Mol Genet. 2000;9:583–8. doi: 10.1093/hmg/9.4.583. [DOI] [PubMed] [Google Scholar]
- Poinsignon C, de Chasseval R, Soubeyrand S, Moshous D, Fischer A, Hache RJ, et al. Phosphorylation of Artemis following irradiation-induced DNA damage. Eur J Immunol. 2004;34:3146–55. doi: 10.1002/eji.200425455. [DOI] [PubMed] [Google Scholar]
- Punj V, Chakrabarty AM. Redox proteins in mammalian cell death: an evolutionarily conserved function in mitochondria and prokaryotes. Cell Microbiol. 2003;5:225–31. doi: 10.1046/j.1462-5822.2003.00269.x. [DOI] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–24. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Whitlow S, Eckersdorff M, Manis JP, Lee C, et al. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci U S A. 2004;101:2410–5. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–90. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–82. [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Seedorf K, Oltersdorf T, Krammer G, Rowekamp W. Identification of early proteins of the human papilloma viruses type 16 (HPV 16) and type 18 (HPV 18) in cervical carcinoma cells. Embo J. 1987;6:139–44. doi: 10.1002/j.1460-2075.1987.tb04731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Sohn D, Essmann F, Schulze-Osthoff K, Janicke RU. p21 blocks irradiation-induced apoptosis downstream of mitochondria by inhibition of cyclin-dependent kinase-mediated caspase-9 activation. Cancer Res. 2006;66:11254–62. doi: 10.1158/0008-5472.CAN-06-1569. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–10. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Wittmack EK, Jorgensen TJ. p21WAF1/CIP1 antisense therapy radiosensitizes human colon cancer by converting growth arrest to apoptosis. Cancer Res. 2000;60:679–84. [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–90. [PubMed] [Google Scholar]
- Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair (Amst) 2005;4:556–70. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu J, Park BH, Kinzler KW, Vogelstein B. Role of BAX in the apoptotic response to anticancer agents. Science. 2000;290:989–92. doi: 10.1126/science.290.5493.989. [DOI] [PubMed] [Google Scholar]
- Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol. 2004;24:9207–20. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.