Abstract
Fermented food contains numerous peptides derived from material proteins. Bitter peptides formed during the fermentation process are responsible for the bitter taste of fermented food. We investigated whether human bitter receptors (hTAS2Rs) recognize bitterness of peptides with a heterologous expression system. HEK293 cells expressing hTAS2R1, hTAS2R4, hTAS2R14, and hTAS2R16 responded to bitter casein digests. Among those cells, the hTAS2R1-expressing cell was most strongly activated by the synthesized bitter peptides Gly-Phe and Gly-Leu, and none of the cells was activated by the non-bitter dipeptide Gly-Gly. The results showed that these bitter peptides, as well as many other bitter compounds, activate hTAS2Rs, suggesting that humans utilize these hTAS2Rs to recognize and perceive the structure and bitterness of peptides.
Keywords: Bitter taste, hTAS2Rs, Heterologous expression, Bitter peptide, Casein hydrolysate
Because many toxins are bitter, mammals have developed an instinctive aversion to bitter taste to prevent them from ingesting poisonous substances [1]. Nevertheless, adult humans are not always averse to bitter taste, and some degree of bitterness is preferred in certain foodstuffs and beverages, including chocolate, coffee, and beer. In these instances, bitter taste works as a component of flavor and is indispensable to improve the palatability of the food or beverage, although excessive bitterness decreases their sensory quality. Fermented foods such as cheese [2], soy sauce [3], and miso [4] contain numerous peptides derived from material proteins by proteolysis during the aging process. Taste properties and structures of the peptides in fermented food and protein hydrolysates have been widely investigated. Examples include acidic oligopeptides in fish sauce, which have a savory taste [5] and hydrophobic peptides in cheese and casein hydrolysate, which have a bitter taste [6,7]. In general, tasty oligopeptides elicit a weaker taste sensation than other major tasty compounds. They usually are not the principal taste constituents; instead they contribute to the complexity and balance of taste, thereby enhancing the palatability of foodstuffs using a mixture of peptides [8,9]. Because enzymatic hydrolysis frequently results in bitter taste [10], it is supposed that bitter peptides exist universally in fermented food products. Therefore, elucidating a mechanism for bitter taste perception of peptides would aid in evaluating the sensory quality of fermented food. In addition, this knowledge would be helpful for further research on general food acceptability.
Recent research has revealed that some peptides in food have various health-related physiological activities, such as antihypertensive (angiotensin I-converting enzyme [ACE] inhibition), opioid, immunomodulating, antioxidative, antimicrobial, antithrombotic, antiamnestic, and hypocholesterolemic activities [11,12]. Among them, peptides that inhibit ACE, which regulates peripheral blood pressure [13], have been investigated most extensively. Many ACE inhibitory peptides were isolated and identified from various fermented foods [13,14]. It has been reported that many bitter dipeptides had ACE-inhibitory activity [14,15]. Therefore, bitterness of peptides should be considered for modification of the bioactive effect of fermented food and protein hydrolysate.
Several papers have shown that bitter substances are recognized by the bitter receptor TAS2R, a family of G-protein-coupled receptors expressed in taste tissue [16]. In humans, more than 25 TAS2Rs have been identified; some of these are activated by bitter substances and function as bitter taste receptors. Because many bitter tastants activate hTAS2R [17-20], it has been thought that humans perceive bitterness via those receptors. However, despite the sensory, nutritional, and bioactive importance of peptides, it is unclear whether the bitterness associated with these compounds is detected by TAS2Rs or by another signal transduction pathway.
This report describes functional expression experiments that we conducted in human embryonic kidney cells to investigate whether the human bitter receptors, hTAS2Rs, respond to bitter peptides in casein hydrolysate and synthetic peptides, and how humans perceive bitterness of peptides in fermented food.
Materials and methods
Materials
Quinine hydrochloride dihydrate, denatonium benzoate, cycloheximide, theobromine, casein, trypsin, bromelin, α-chymotrypsin, and carbamylcholine chloride (carbachol) were purchased from Wako Pure Chemical Industries Co., Ltd. (Osaka, Japan). Caffeine, salicin, 6-propyl-2-thiouracil (PROP), and naringin were purchased from Sigma–Aldrich (Tokyo, Japan). Glycyl-l-leucine (Gly-Leu), glycyl-l-phenylalanine (Gly-Phe), and glycylglycine (Gly-Gly) were purchased from Peptide Institute (Osaka, Japan). Isoproterenol hydrochloride was purchased from Tocris Bioscience (Ellisville, MO). Fluo 4-acetoxymethyl ester (Fluo 4-AM) was purchased from Dojindo Laboratories (Kumamoto, Japan).
Molecular cloning of the human Tas2r genes
The human Tas2r genes were amplified by polymerase chain reaction (PCR) using gene-specific primers that spanned the complete coding region of the individual hTas2r genes from human genomic DNA (Takara Bio Inc., Shiga, Japan). The amplicons of hTas2r1 (GeneBank Accession No. BC101729), hTas2r4 (NM_016944), hTas2r14 (NM_023922), and hTas2r16 (NM_016945) were cloned into the multiple cloning sites of a plasmid, pcDNA3.1 (Invitrogen Japan KK, Tokyo, Japan). The cDNA fragment encoding the first 38 amino acids of mouse rhodopsin was placed in frame with the 5’-end of human Tas2r coding cDNA sequences to generate the human Tas2R expression vector pcDNA3.1/mRho/hTAS2Rs, since it is known that the first 39 amino acids of bovine rhodopsin are extremely effective in targeting hTAS2Rs to the plasma membrane of HEK293 cells [17]. The rhodopsin segment was amplified from mouse cDNA, which was made by reverse transcriptase-PCR (RT-PCR) from total RNA extracted from mouse retina. Human G protein α15 cDNA (Guthrie cDNA Resources Center, Sayre, PA) was subcloned into pcDNA5/FRT (Invitrogen Japan KK) to generate the Gα15 expression vector pcDNA5/FRT/G15.
Transfection and screening for stable cell lines
Flp-In293 cells (Invitrogen Japan KK) were grown and maintained in a complete medium, low-glucose Dulbecco’s modified Eagle’s medium with GlutaMax (Invitrogen Japan KK) supplemented with 10% fetal bovine serum (Invitrogen Japan KK) and 100 μg/ml of Zeocin (Invitrogen Japan KK), at 37 °C in a 5% CO2 incubator. The cells were transfected with pcDNA5/FRT/G15 using Lipofectamine 2000 (Invitrogen Japan KK) according to the manufacturer’s instructions. Cell lines stably expressing Gα15 (Flp-In293/G15 cell) were selected based on stable resistance against 100 μg/ml of hygromycin.
To establish stable cell lines expressing hTAS2Rs, Flp-In293/G15 cells were transfected with pcDNA3.1/mRho/hTAS2Rs using Lipofectamine 2000. The cells were grown and selected in a complete medium containing 100 μg/ml of hygromycin and 400 μg/ml of G418 (Invitrogen Japan KK). Stable expression of the genes for hTAS2Rs in the transfected cells was confirmed by RT-PCR using a SuperScript III CellsDirect cDNA Synthesis Kit (Invitrogen Japan KK) and TAS2R-specific PCR primers.
Calcium imaging
Intracellular-calcium of cells stimulated with bitter tastants was measured to study the bitter response using a method described previously [21,22]. Briefly, to load the intracellular-calcium indicator dye, cells at 70–80% confluency were incubated with 2 μM Fluo 4-AM for 30 min at room temperature followed by 30 min at 37 °C. Next, the cells were washed with D-PBS (Invitrogen Japan KK) to remove excess Fluo 4-AM. The fluorescence images were recorded at 5-s intervals using excitation and emission wavelengths of 450–490 nm and >510 nm, respectively, with a DP70 digital camera (Olympus Co., Tokyo, Japan) equipped with an IX-71 inverted microscope (Olympus Co.) and analyzed using DP Controller software and DP Manager software (Olympus Co.). The result was quantitatively represented as the percentage of the highest count of responsive cells to all cells observed in the microscopic field during stimulation.
Preparation of casein hydrolysate
Casein at the concentration of 1% was hydrolyzed with trypsin (enzyme–substrate ratio, 0.05; pH 8.0) at 37 °C for 12 h. The hydrolysate was the boiled for 15 min to inactivate the protease, and any precipitate that formed was removed by centrifugation. After lyophilization, the hydrolysate was dissolved in 10% ethanol and subjected to gel filtration chromatography using a column (2.6 cm × 60 cm) of Sephadex G-25 gel (Amersham Biosciences AB, Uppsala, Sweden) that had been equilibrated with 10% ethanol. The elution of peptides with 10% ethanol was monitored with the biuret reaction by measuring optical density (OD) at 280 and 540 nm.
Results and discussion
Establishment of stable cell lines for hTAS2Rs expression
First, we established Gα15-expressing cells to couple activation of hTAS2Rs to the release of Ca2+, which can be measured using a calcium-sensitive fluorescent dye. In calcium imaging experiments using cells transfected with pcDNA5/FRT/Gα15 and screened for hygromycin resistance, most of the cells showed responses to 5 μM of isoproterenol, indicating that Gα15 was functionally expressed in the cells. The cells stably expressing Gα15 were then transfected with human bitter receptor expression vectors pcDNA3.1/mRho/hTAS2Rs. After transfection and screening for G418 resistance, several cells were obtained and expanded into stable cell lines. Expression of hTAS2R genes and proteins was checked by RT-PCR and immuno-staining, respectively. Because all hTAS2Rs were expressed as fusion proteins with the first 38 amino acids of mouse rhodopsin at their N termini, cell surface expression of hTAS2Rs was confirmed by immunostaining with an anti-rhodopsin antibody.
Responses of hTAS2Rs to casein hydrolysate
To obtain bitter peptides, casein was hydrolyzed with trypsin. It is well known that casein hydrolysate tastes strongly bitter [6], and bitter peptides isolated from a hydrolysate with trypsin are reported [7]. Because casein hydrolysate contains whole protein and free amino acids along with small peptides, it was subjected to gel filtration chromatography using a Sephadex G-25 column to separate the peptide fraction. As shown in Fig. 1, the hydrolysate was separated into three peak-fractions, H, M, and L, based on the biuret reaction measuring OD at 280 and 540 nm. Fraction H was likely to contain unbroken protein, and fraction L was likely to contain mainly amino acids. Therefore, fraction M was collected and used as the bitter casein peptide sample. Many peaks were detected on the HPLC elution profile using the ODS column of the casein peptide (fraction M), which indicated that numerous peptides were included in this fraction. Using 2.5 mg/ml of this casein peptide fraction, which tastes moderately bitter (equivalent to the bitterness of 0.125 mM of quinine), calcium imaging experiments were performed to determine the responses of hTAS2R-expressing cells to bitter peptides.
Fig. 1.
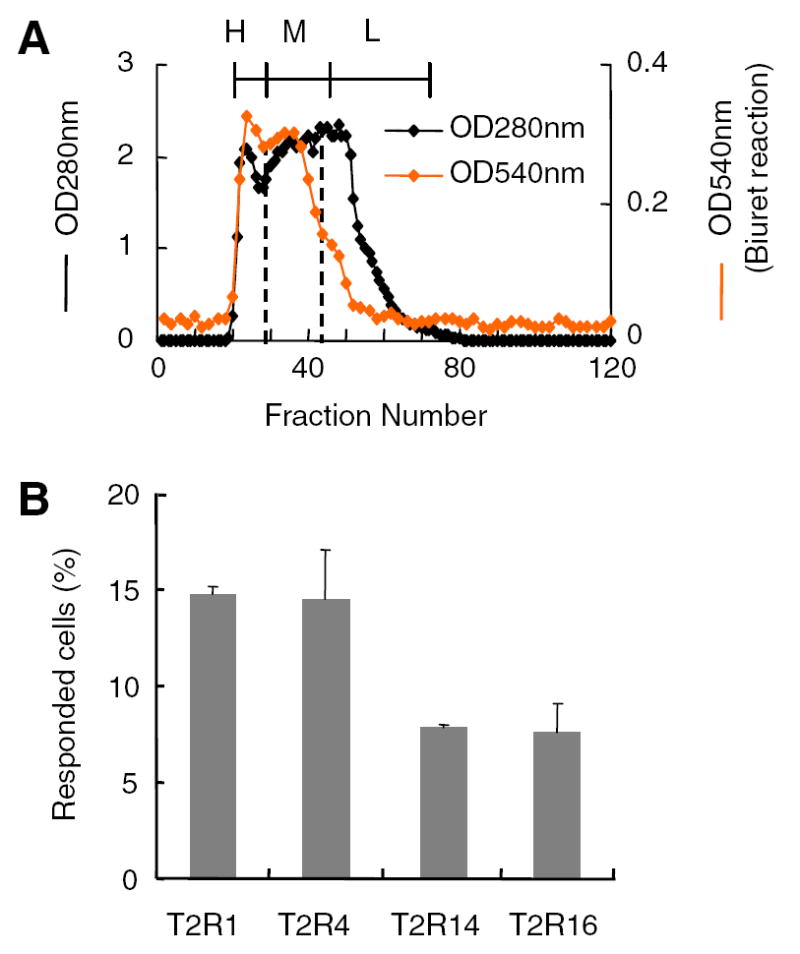
The bitter peptide fraction M from casein hydrolysate stimulates hTAS2Rs. (A) Separation of casein hydrolysate by gel filtration chromatography on Sephadex G-25 column. At 2.5 mg/ml, the tastes of fractions H, M, and L, were weakly bitter, bitter, and bitter, respectively. (B) Responses of hTAS2R-expressing cells to the bitter peptide fraction M (2.5 mg/ml).
When bitter peptide fraction M was applied to the cells expressing hTAS2R1, hTAS2R4, hTAS2R14, and hTAS2R16, all the cells, particularly hTAS2R1 and hTAS2R4, elicited a response to the fraction. It was concluded that structurally diverse peptides contained in this fraction could stimulate all four receptors, although they might have different potency and specificity in structure recognition. Because bitter peptides exist universally in fermented food and are responsible for their bitterness, it was proved that bitterness of fermented food is also perceived by the bitter receptors, hTAS2Rs, in the same way as are other bitter compounds.
Responses of hT2Rs to synthetic peptides
Human TAS2R4 and TAS2R16 are known to respond specifically to denatonium [17] and salicin [18], respectively. Moreover, TAS2R14 is known to respond to a variety of bitter compounds [19]. Therefore, we examined calcium imaging to verify whether our stable cell lines showed response profiles similar to those reported elsewhere. As shown in Fig. 2, application of salicin to the cells expressing hTAS2R16 elicited robust transient elevation of cytosolic Ca2+ concentration as a function of salicin concentration between 0.1 and 10 mM. This result was consistent with that of Bufe et al. [18]. hTAS2R14 is known as a receptor that is broadly tuned for various bitter compounds, including the potent neurotoxins such as (−)-α-thujone, which is the pharmacologically active component of absinthe, and picrotoxinin, which is a poisonous substance found in fishberries [19]. In our dose–response results, hTAS2R14 was activated by picrotin to the same degree as that reported previously [19].
Fig. 2.
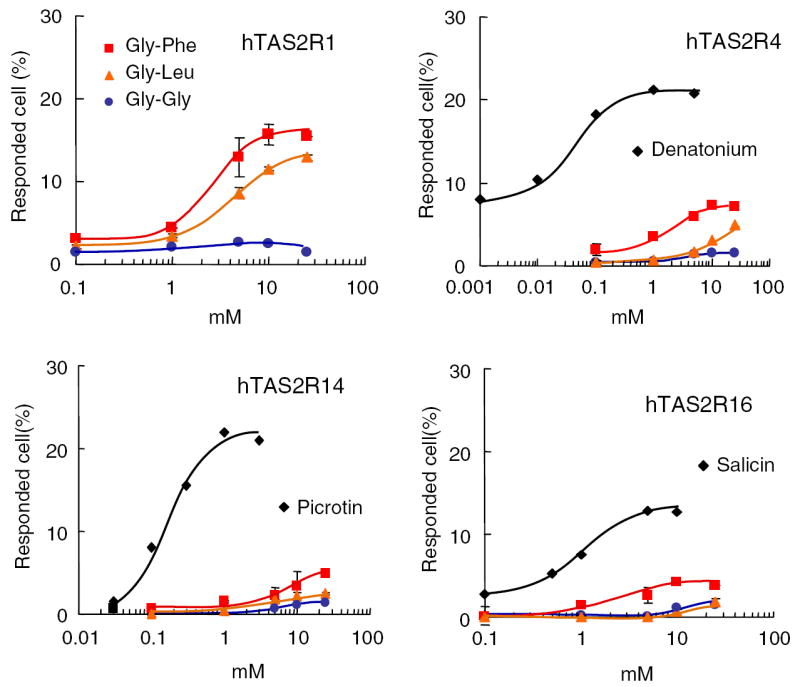
Dose–response curves of hTAS2Rs to synthesized bitter dipeptides.
Bitter peptide fraction M contained numerous peptides with different structures, and probably also included non-bitter peptides; thus, it was not certain whether bitter peptides stimulated the receptors. To obtain a definitive answer, we examined the effect of individual peptides. Three synthetic peptides including two bitter peptides, Gly-Leu and Gly-Phe, and one non-bitter peptide, Gly-Gly, were employed for calcium imaging using hTAS2R-expressing cells. Glycyl dipeptides are of simple structures, which facilitates elucidation of the structure and bitterness relationships, although their tastes are not as strong as some other bitter peptides. The two bitter peptides, Gly-Phe and Gly-Leu, have a hydrophobic amino acid in their structures, and their bitterness threshold values have been determined to be 1.2 mM [23] and 25 mM [24], respectively, by a sensory test. Gly-Gly, which does not have a hydrophobic amino acid, is tasteless in the sensory test. The tastes of those peptides were also evaluated by the sensory test as the bitterness equivalent to 0.125 mM of quinine hydrochloride, which is of medium bitterness. Gly-Gly was tasteless at 100 mM. Because the bitterness equivalent was determined to be 57.5 ± 3.2 and 100 ± 3.9 mM for Gly-Phe and Gly-Leu, respectively, it was confirmed that both Gly-Phe and Gly-Leu were bitter, and that Gly-Phe was more bitter than Gly-Leu, as previously reported [7].
Using these bitter and non-bitter dipeptides, the responses of four hTAS2R-expressing cells were examined. The responses of the cells expressing Gα15 alone were subtracted from those of hTAS2R-expressing cells as a background signal independent of the TAS2R receptor. The responses of cells expressing Gα15 alone to any bitter compounds were less than 5%. As shown in Fig. 2, the tasteless dipeptide, Gly-Gly, did not activate any hTAS2Rs. hTAS2R4, hTAS2R14, and hTAS2R16 were activated robustly by their specific ligands, denatonium, picrotin, and salicin, respectively. Two bitter peptides, Gly-Phe and Gly-Leu, activated hTAS2R1 most strongly, whereas they evoked no or weak responses in other receptors. In the case of hTAS2R4 and hTAS2R14, greater responses were observed to higher concentrations of the bitter peptides. However, the background signal increased as well due to non-specific stimulation of cells. Because hTAS2R4 was more strongly activated by casein hydrolysate as shown in Fig. 1, it indicates that other bitter peptides in the hydrolysate may be able to activate hTAS2R4 more potently. It was found that hTAS2R1 was not activated by a non-bitter peptide, although it was activated by bitter peptides. However, it was unclear whether the response to bitter peptides was specific. Therefore, the response of hTAS2R1 to various bitter compounds was examined. As shown in Fig. 3, it was apparent that hTAS2R1 was more sensitive to bitter peptides than to other bitter compounds such as denatonium, picrotin, salicin, and caffeine. These results indicate that Gly-Phe and Gly-Leu are the two most potent ligands among the ligands that have been tested, thus implicating that hTAS2R1 can be a receptor for bitter peptides.
Fig. 3.

Responses of hTAS2R1-expressing cells to various bitter compounds. G-F, 25 mM Gly-Phe; G-L, 25 mM Gly-Leu; DEN, 1 mM denatonium; PIC, 3 mM picrotin; SAL, 5 mM salicin; CAF, 1 mM caffeine.
Many bitter peptides have been isolated from various sources, and the relationship between bitterness and structure has been studied extensively [25,26]. It is believed that there is an empirical correlation between the hydrophobicity of the peptides and their bitter taste. This correlation is known as the Q rule [27,28], which predicts a bitter taste for peptides with Q values higher than 1400 cal/mol and molecular weights between 100 and 6000 kDa. Matoba and Hata [26] also proposed that hydrophobic amino acid side chains were responsible for the bitter taste. According to their calculation, Gly-Phe and Gly-Leu are hydrophobic and bitter, whereas Gly-Gly is not sufficiently hydrophobic to elicit bitterness [7,29]. Our results, which showed that bitter peptides stimulated the human bitter receptors hTAS2Rs, support this hypothesis based on human sensory tests that have been conducted by several investigators in the past. These results suggest that humans utilize TAS2Rs to recognize and perceive the structure and bitterness of peptides.
Acknowledgments
We thank Dr. A. Ohnishi of Tokyo University of Agriculture for his help in fluorometric imaging experiment. We also thank Ms. M. Ishigaki and Mr. Y. Matsushita for their technical assistance. This work was supported by a grant from Asahi Breweries Foundation. (KM) and National Institutes of Health grants DC007487(LH) and DC007974(HW).
References
- 1.Glendinning JI. Is the bitter rejection response always adaptive? Physiol Behav. 1994;56:1217–1227. doi: 10.1016/0031-9384(94)90369-7. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton JS, Hill RD, Leewen HV. A bitter peptide from cheddar cheese. Agric Biol Chem. 1974;38:375–379. [Google Scholar]
- 3.Oka S, Nagata K. Isolation and characterization of acidic peptides in soy sauce. Agric Biol Chem. 1974;38:1195–1202. [Google Scholar]
- 4.Takeuchi T, Yoshii H. Studies on peptides in miso and soy-sauce. J Ferment Technol. 1969;47:496–501. [Google Scholar]
- 5.Park J-N, Ishida K, Watanabe T, Endoh K, Watanabe K, Murakami M, Abe H. Taste effects of oligopeptides in a Vietnamese fish sauce. Fish Sci. 2002;68:921–928. [Google Scholar]
- 6.Bumberger E, Belitz H-D. Bitter taste of enzymic hydrolysates of casein. Z Lebensm Unters Forsch. 1993;197:14–19. doi: 10.1007/BF01202693. [DOI] [PubMed] [Google Scholar]
- 7.Roudot-Algaron F. The taste of amino acids, peptides and proteins: examples of tasty peptides in casein hydrolysates. Lait. 1996;76:313–348. [Google Scholar]
- 8.Kirimura J, Shimizu A, Kimizuka A, Ninomiya T, Katsuya N. The contribution of peptides and amino acids to the taste of foodstuffs. J Agric Food Chem. 1969;17:689–695. [Google Scholar]
- 9.Maehashi K, Matsuzaki M, Yamamoto Y, Udaka S. Isolation of peptides in enzymatic hydrolysate of food proteins and characterization of their taste properties. Biosci Biotech Biochem. 1999;63:555–559. doi: 10.1271/bbb.63.555. [DOI] [PubMed] [Google Scholar]
- 10.Lovsin-Kukman I, Zelenik-Blatnik M, Abram V. Bitterness intensity of soybean protein hydrolysates—chemical and organoleptic characterization. Z Lebensm Unters Forsch. 1996;203:272–276. [Google Scholar]
- 11.Arai S, Osawa T, Ohigashi H, Yoshikawa M, Kaminogawa S, Watanabe M, Ogawa T, Okubo K, Watanabe S, Nishino H, Shinohara K, Esashi T, Hirahara T. A mainstay of functional food science in Japan—history, present status, and future outlook. Biosci Biotechnol Biochem. 2001;65:1–13. doi: 10.1271/bbb.65.1. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa M, Fujita H, Matoba N, Takenaka Y, Yamamoto T, Yamauchi R, Tsuruki H, Takahata Y. Bioactive peptides derived from food proteins preventing lifestyle-related diseases. Biofactors. 2000;12:143–146. doi: 10.1002/biof.5520120122. [DOI] [PubMed] [Google Scholar]
- 13.Li G-H, Le G-W, Shi Y-H, Shrestha S. Angiotensin I-converting enzyme inhibitory peptides derived from food proteins and their physiological and pharmacological effects. Nutr Res. 2004;24:469–486. [Google Scholar]
- 14.Pripp AH, Ardo Y. Modelling relationship between angiotensin-(I)-converting enzyme inhibition and the bitter taste of peptides. Food Chem. 2007;102:880–888. [Google Scholar]
- 15.Wu J, Aluko R. Quantitative structure–activity relationship study of bitter di- and tri-peptides including relationship with angiotensin I-converting enzyme inhibitory activity. J Pept Sci. 2007;13:63–69. doi: 10.1002/psc.800. [DOI] [PubMed] [Google Scholar]
- 16.Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Rhyba JP. The receptors and coding logic for bitter taste. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 17.Chandrashekar J, Muller KL, Hoonm MA, Adler E, Feng L, Guo W, Zuker CS, Ryba JP. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 18.Bufe B, Hofman T, Krautwurst D, Raguse J-D, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to β-glucopyranosides. Nat genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 19.Behrens M, Brockhoff A, Kuhn C, Bufe B, Winnig M, Meyerhof W. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem Biophys Res Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Pronin AN, Tang H, Connor J, Keung W. Identification of ligands for two human bitter T2R receptor. Chem Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- 21.Ariyasu T, Matsumoto S, Kyono F, Hanaya T, Arai S, Ikeda M, Kurimoto M. Taste receptor T1R3 is an essential molecular for the cellular recognition of the disaccharide trehalose. In Vitro Cell Dev Biol—Animal. 2003;39:80–88. doi: 10.1290/1543-706X(2003)039<0080:TRTIAE>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishibashi N, Sadamori K, Yamamoto O, Kanehisa H, Kouge K, Kikuchi E, Okai H, Fukui S. Bitterness of phenylalanine- and tyrosine-containing peptides. Agric Biol Chem. 1987;51:3309–3313. [Google Scholar]
- 24.Ishibashi N, Arita Y, Kanehisa H, Kouge K, Okai H, Fukui S. Bitterness of leucine-containing peptides. Agric Biol Chem. 1987;51:2389–2394. [Google Scholar]
- 25.Ney KH. Prediction of bitterness of peptides from their amino-acid composition. Z Lebensm Unters Forsch. 1971;147:64–68. [Google Scholar]
- 26.Matoba T, Hata T. Relationship between bitterness of peptides and their chemical structures. Agric Biol Chem. 1972;36:1423–1431. [Google Scholar]
- 27.Guigoz Y, Solms J. Bitter peptides, occurrence and structure. Chem Senses Flavor. 1976;2:71–84. [Google Scholar]
- 28.Lemieux L, Simard RE. Bitter flavour in dairy products. II. A review of bitter peptides from caseins: their formation, isolation and identification, structure masking and inhibition. Lait. 1992;72:335–382. [Google Scholar]
- 29.Ishibashi N, Ono I, Kato K, Shigenaga T, Shinoda I, Okai H, Fukui S. Role of the hydrophobic amino acid residue in the bitterness of peptides. Agric Biol Chem. 1988;52:91–94. [Google Scholar]


