Abstract
Fibroblast growth factors play important roles in inner ear development. Previous studies showed that mouse Fgf16 is expressed asymmetrically during the otic cup and vesicle stages of development, suggesting roles in regulating or responding to anteroposterior axial cues. Here, we studied otic Fgf16 expression throughout embryonic development and found transcripts in the developing cristae and in a few cells in the lateral wall of the cochlear duct. To determine the otic function of Fgf16 and to follow the fate of Fgf16-expressing cells, we generated an Fgf16IRESCre allele. We show that Fgf16 does not have a unique role in inner ear development and that the Fgf16 lineage is found throughout the three cristae, in portions of the semicircular canal ducts, and in the cochlear spiral prominence epithelial cells. This strain will be useful for gene ablations in these tissues.
Keywords: Fibroblast growth factor, mouse mutant, lineage, semicircular canals, vestibular system, cochlear duct, spiral prominence, otic cup and vesicle, embryo
Introduction
The inner ear is a morphologically complex organ specialized for sensing sound and relative head motion. In the mouse, epithelial mechanosensory patches are found in six locations. Sound-responsive cells are located along the length of the coiled and ventrally located cochlear duct. The remaining motion-responsive sensory patches are housed within the dorsally located vestibular system. Two sensory organs, the maculae of the utricle and saccule respond to changes in linear motion and three additional sensory organs, the anterior, lateral and posterior cristae, each one located at the base of a semicircular canal, respond to changes in angular motion.
Remarkably, the entire inner ear epithelium is derived from a patch of head ectoderm that is morphologically evident as a placodal thickening at embryonic day (E)8.0 in the mouse (~8–9 somites). Subsequently, from E8.5–E9.0 (~13–20 somites), the placode invaginates to form the otic cup, which continues to deepen, and by E9.5 (~25 somites) closes and separates from the surface ectoderm, forming the roughly spherical otic vesicle. Morphogenesis of the vesicle initiates at E10–10.5 with a dorsomedially directed outgrowth of the endolymphatic duct/sac (EDS) anlage and a ventrally directed outgrowth of the cochlear duct. During the next few days of development, this relatively simple epithelium acquires its mature and complex morphology and begins to differentiate its six sensory patches. Morphogenesis is essentially complete by E15.5, but sensory differentiation continues even through the early postnatal period (Swanson et al., 1990; Morsli et al., 1998; Kiernan et al., 2002).
The ear is an asymmetric structure with each of its developmental axes having distinct morphologic characteristics. For example, the anterior and lateral cristae are anterior structures and the posterior crista is a posterior structure. Although much remains to be determined regarding the timing and order of otic axis initiation and fixation, particularly in the mouse (Bok et al., 2007), it is clear that patterns of gene expression in the otic placode tend to be relatively uniform, and cell fates, as assayed in the chick, are not compartmentalized at this stage (Streit, 2002). Asymmetric otic gene expression can first be discerned at the otic cup stage, when the anteroposterior (AP) axis (likely the first axis to be fixed) is starting to be established (Wu et al., 1998), and fate maps generated by marking chick otic cup cells at stage 13.5 (~20–21 somites) suggest that a compartment boundary separating anterior and posterior cell fates has already formed (Brigande et al., 2000).
We showed previously that Fgf16 transcripts are regionally restricted and polarized along the AP axis of the otic cup and vesicle. Fgf16 expression initiated at the early otic cup stage (13 somites) and was concentrated in a posterior spot. By the early otic vesicle stage (24 somites), Fgf16 was found in a posterodorsolateral spot (Wright et al., 2003). These observations suggested possible signaling roles for FGF16 in otic axis formation and/or cell fate decisions. In addition, the seemingly transient posterior expression of Fgf16 pointed to the opportunity to map the fate of these cells and compare the results with fate maps established by cell marking techniques in other species. To this end, we studied the Fgf16 otic expression pattern throughout embryonic development and generated a targeted mutant mouse strain in which Fgf16 expression was ablated and CRE recombinase was expressed in its place. We found that Fgf16 transcripts continue to be expressed beyond the early otic vesicle stage and mark all three developing cristae. We also detected cochlear duct expression of Fgf16 that was confined to a few cells in the lateral wall. Mice bearing an IRESCre insertion disrupting Fgf16 expression did not exhibit any structural or functional otic abnormalities, but lineage studies of the Fgf16IRESCre strain revealed that all three cristae, but not the maculae or cochlear sensory tissue, are derived from the early posterior, and a subsequent anterior patch of Fgf16-expressing cells, and that derivatives of these cells also populate a proportion of the non-sensory semicircular canal ducts, prior to any expression of Fgf16 in the ducts. The Fgf16 lineage in the cochlear duct marked precursors of spiral prominence epithelial cells, which may contribute to the regulation of endolymph composition (Slepecky, 1996; Santi and Tsuprun, 2001), and thus auditory function. Our results suggest that the Fgf16IRESCre strain may be a useful tool in conditional ablation experiments designed to isolate the function of genes expressed in the developing cristae and/or spiral prominence epithelial cells from their functions in other inner ear tissues.
Results and Discussion
Fgf16 is initially expressed asymmetrically in the otic cup and vesicle, and subsequently in all three semicircular canal cristae and in the lateral wall of the cochlear duct
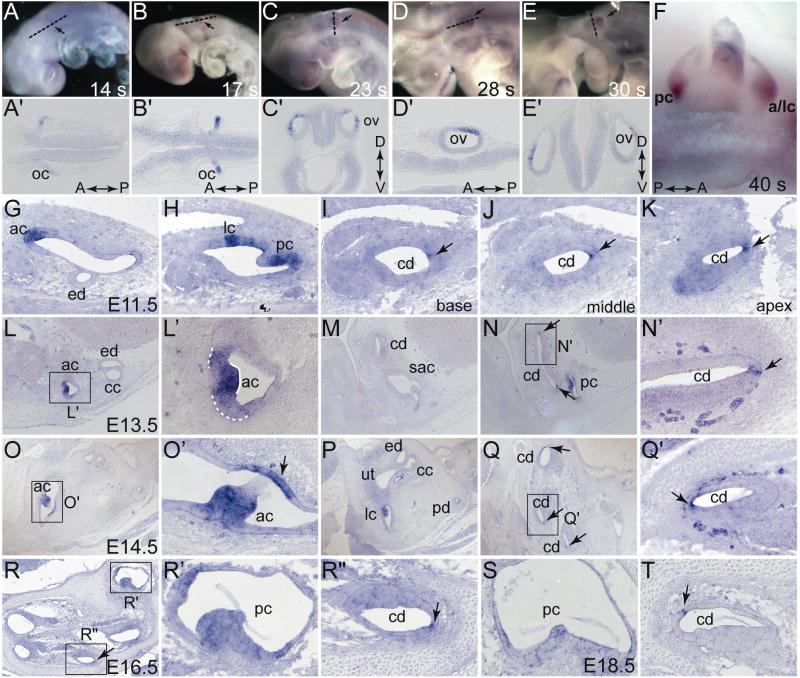
Previous studies showed that both mouse and chick Fgf16 expression is regionally restricted during the early phases of inner ear development (Wright et al., 2003; Chapman et al., 2006). To obtain a more detailed picture of mouse otic Fgf16 expression, we studied closely staged embryos throughout development (Fig. 1). Otic expression of Fgf16 was first detected weakly in the posterior otic cup of 14-somite embryos (E9.0, Figs. 1A,A′). By 17 somites (E9.0), as the cup started to close to form the otic vesicle, Fgf16 transcripts were more strongly expressed and still localized to the posterior cup (Figs. 1B,B′). Starting at 21 somites (E9.5), when the vesicle formed (data not shown) and subsequently at 23 somites (E9.5), Fgf16 was detected in the posterior otic vesicle, marking the dorsolateral region (Figs. 1C,C′). By 28–30 somites (E10.0), Fgf16 expression was further restricted to a posterolateral domain, located midway between the dorsal and ventral poles of the vesicle (Figs. 1D,D′,E,E′). Finally, at 40 somites (E11.0), Fgf16 as found in two spots in the developing cristae (Fig. 1F; a/lc, anterior/lateral cristae; pc posterior crista). Extra-otic sites of embryonic Fgf16 expression at these stages included the developing posterior pharyngeal pouch endoderm and olfactory placode/epithelium (supplementary Figs. S1A-D″), however, unlike in zebrafish embryos (Nomura et al., 2006), mouse Fgf16 expression was not detected in the limb apical ectodermal ridge (data not shown).
Figure 1. Fgf16 mRNA expression from E9.0–E18.5.
Whole-mount 14-somite (A), 17-somite (B), 23-somite (C), and 28-somite (D), 30-somite (E), and 40-somite (F) embryos were probed with labeled antisense Fgf16 cRNA. Anterior is to left (A,B,C,D,E) or right (F). White lines indicate the plane of coronal (A′,B′,D′) or transverse (D′,E′) sections shown in panels below the whole mounts, black arrows indicate the otic vesicle. Fgf16 expression in transverse (G-R″) or sagittal (S,T) paraffin sections. (L′) Solid white line indicates strong expression in anterior crista, dotted white line indicates weaker expression in cells adjacent to the crista. Abbreviations: A, anterior; ac, anterior crista; a/lc, anterior and lateral criate; cc, common crus; cd, cochlear duct; D, dorsal; ed, endolymphatic duct; lc, lateral crista; oc, otic cup; ov, otic vesicle; P, posterior: pc, posterior crista; sac, saccule; ut, utricle; V, ventral.
To determine whether Fgf16 expression persisted in the otic region later in development, we detected Fgf16 transcripts at E11.5, E13.5, E14.5 and E16.5 in transverse sections and at E18.5 in sagittal sections taken through the head. At E11.5, Fgf16 expression was found in all three developing cristae, but not in other regions of the developing semicircular canal ducts (Figs. 1G,H; ac, anterior crista; lc, lateral crista; pc, posterior crista). In addition, several cells at the lateral (non-sensory) edge of the cochlear duct (cd) expressed Fgf16 (Figs. 1I–K, arrow). Expression was stronger at the apex (Fig. IK) than at the base of the duct (Fig. 1I). At E13.5, Fgf16 transcripts continued to be expressed in the developing cristae (Figs. 1L,L′,N), and weak expression extended into adjacent patches of presumptive non-sensory epithelium of the semicircular canals (see for example the anterior semicircular canal in Fig. 1L′), but was completely absent from the developing saccular and utricular sensory patches (Fig. 1M). In addition, Fgf16 continued to be expressed in a few cells of the lateral wall of the cochlear duct (Fig. 1N,N′, arrows). At E14.5, Fgf16 expression in the cristae persisted, but began to weaken (Figs. 1O,O′,P). A discrete patch of expression in the thin canal wall opposite the cristae was also apparent (Figs. 1O,O′; arrow in O′). This expression in non-sensory canal tissue extended a short distance away from the cristae. Fgf16 expression in the lateral cochlear duct continued through E14.5 (Figs. 1Q,Q′). Although Fgf16 could still be detected weakly in the cristae at E16.5 and E18.5 (Figs. 1R,R′,S), its expression was markedly reduced relative to earlier stages. In contrast, expression in the lateral cochlear duct remained strong (Figs. 1R,R″,T). Extra-otic sites of Fgf16 expression detected in this series of sections through a portion of the head included the anterior pituitary and olfactory epithelium (supplementary Figs. S1E-F′).
These results show that Fgf16 expression in the mouse inner ear persists beyond the stages originally analyzed (Wright et al., 2003). As in the chick embryo (Chapman et al., 2006), mouse Fgf16 is expressed initially in a pattern similar to that of Bmp4, a gene that marks the developing cristae and is required for their development and that of their associated semicircular canals (Morsli et al., 1998; Chang et al., 2008). Zebrafish Fgf16 is also expressed in the otic vesicle, but whether it is regionalized is unknown (Nomura et al., 2006). The early asymmetric expression of Fgf16 in the posterior and then posterolateral regions of the mouse otic vesicle suggests possible roles in or responses to signaling for otic axis formation or cell type specification, particularly in the prospective vestibular system. The subsequent restriction of Fgf16 expression to the developing cristae suggests roles in distinguishing this type of sensory development from that of the maculae. It is interesting to note that unlike Bmp4, which ultimately marks the supporting cells of the cristae (Morsli et al., 1998), Fgf16 expression in the cristae does not resolve to distinguish the sensory vs. supporting cell populations, but rather is strongly reduced by E16.5 as this distinction develops.
Expression of Fgf16 in the developing cochlear duct starting at E11.5 is remarkably restricted and persistent in a few cells that appear to lie at the base of the developing stria vascularis, which is required for generation and maintenance of the endocochlear potential needed for hearing function. Although Bmp4 is also expressed from E11.5 in the developing cochlear duct, its domain is likely non-overlapping with that of Fgf16 in this tissue, as Bmp4 marks the precursors of Hensen and Claudius cells (Morsli et al., 1998); whereas Fgf16 marks more laterally located cells. The functional significance of these particular Fgf16-expressing cells, however, is unknown.
Gene targeting at the Fgf16 locus
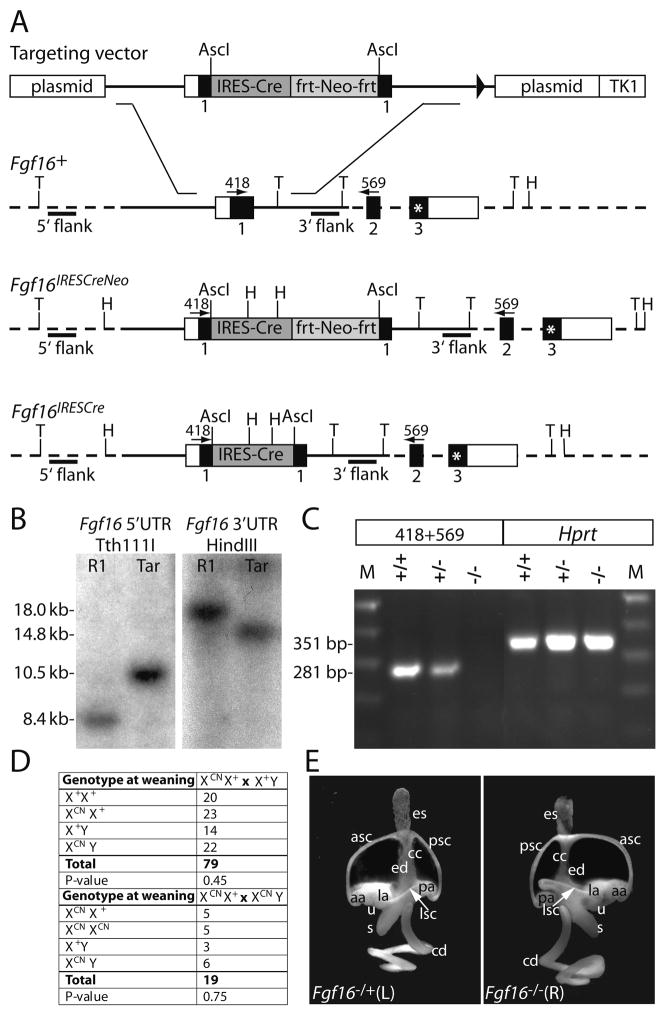
To observe the consequences of Fgf16 inactivation and to follow the fate of Fgf16-expressing cells originating in the posterior otic epithelium and later in the cochlear duct, we targeted the Fgf16 locus using a vector containing an internal ribosomal entry site (IRES) preceding a Cre gene, which was followed by an frt-flanked Neo selection cassette (Fig. 2A) (Arenkiel et al., 2003). To generate a null allele, the IRES-Cre-Neo cassette was placed in the first coding exon of Fgf16 (Fig. 2A), preventing production of the majority of the protein, in particular, the predicted FGFR-binding sequences located between two conserved cysteines.
Figure 2. Gene targeting at the Fgf16 locus.
(A) Structure of the linearized Fgf16 targeting vector and depiction of the wild type Fgf16 allele (Fgf16+), the correctly targeted mutant allele in ES cells and offspring of chimeric mice (Fgf16IRESCreNeo) and the targeted allele after exposure to FLP recombinase (Fgf16IRESCre). Mouse genomic Fgf16 DNA is depicted with solid lines; dotted lines indicate Fgf16 genomic DNA that is not present in the targeting vector; untranslated regions are shown as open boxes, protein coding regions as solid boxes. The IRES-Cre cassette is shown as a dark grey box; the Neo cassette flanked by frt sites as a light grey box; the stop codon in the Fgf16 frame in exon 3 as an asterisk. The plasmid backbone and the thymidine kinase (TK1) gene are depicted as open boxes. Recognition sites for Tth111I and HindIII, used in Southern analysis are indicated by “T” and “H” respectively. Probes used for Southern analysis are shown as black bars. Numbered arrows indicate the identity, position and directionality of primers used for the RT-PCR assay. (B) Southern blot hybridization assay demonstrating correct targeting of Fgf16 in ES cells. Tth111I-digested genomic DNA from either the R1–45 ES cell line (R1) or a correctly targeted cell line (Tar) probed with the external 5′ probe. (C) RT-PCR assay used to detect Fgf16 mRNA in wild type and mutant animals. Total E9.0 RNA isolated from female wild type (+/+), female heterozygous (+/−) and male hemizygous mutant (−/Y) embryos was PCR-amplified using primers 418 and 569. A 351 bp Hprt cDNA fragment was amplified from all samples as a positive control. (D) Fgf16 backcross and intercross genotypes. Offspring of the indicated crosses were genotyped at weaning. P-values were determined using the method of χ2. The Fgf16IRESCreNeo allele is defined as XNeo. (E) Fgf16-deficient inner ears were morphologically normal. Representative lateral views of paint-filled inner ears from E15.5 female heterozygous control and Fgf16IRESCreNeo homozygous mutants. Labeled structures: aa, anterior ampulla; asc, anterior semicircular canal; cc, common crus; cd, cochlear duct; ed, endolymphatic duct; es, endolymphatic sac; la, lateral ampulla; lsc, lateral semicircular canal; pa, posterior ampulla; psc, posterior semicircular canal; s, saccule; u, utricle.
The insertion mutation was introduced into X-linked Fgf16 in male ES cells through standard gene targeting protocols. Correctly targeted cell clones, hemizygous for the introduced mutation, were identified by Southern blot hybridization analysis of Tth111I-digested DNA using a 5′ flanking external probe and of HindIII-digested DNA using a 3′ internal probe, which caused the predicted shifts of wild type Fgf16 DNA from 8.4 kb to 10.5 kb and 18 kb and 14.8 kb, respectively (Figs. 2A,B). Mice bearing the targeted allele, Fgf16IRESCreNeo (MGI accession number 3790778, symbolized Fgf16tm1(Cre)Sms), were generated by standard procedures.
To determine the efficacy of the disruption strategy, an RT-RCR reaction was performed to detect Fgf16 mRNA in wild type and mutant animals (Figs. 2A,C). Using primers that flank the IRES-Cre-Neo insertion site (Fig. 2B), the expected DNA fragment of 281 bp was evident in wild type and heterozygous female samples, but was absent from a male hemizygous mutant sample. In contrast, all samples expressed the X-linked Hprt gene, showing that the targeting strategy specifically disrupted production of normal Fgf16 mRNA.
Fgf16 is not required for mouse development
To determine whether Fgf16 is required for any aspect of embryonic or postnatal development, the F1 female Fgf16IRESCreNeo/+ offspring of germline chimeras were mated to wild type males or F2 Fgf16IRESCreNeo/Y males, and adult progeny were genotyped. The numbers of each genotype were consistent with a normal Mendelian distribution of wild type and mutant alleles (Fig. 2D). No obvious anatomic or behavioral abnormalities, such as circling or head tossing, indicative of vestibular dysfunction, were apparent in homozygous mutant females or hemizygous mutant males, and the Fgf16-deficient animals were fertile, indicating that Fgf16 does not have an obvious and unique role during mouse development. Since zebrafish Fgf16 is expressed in the pectoral fin apical ectodermal ridge and Fgf16 morpholino-treated (knockdown) zebrafish embryos have defects of pectoral fin (limb) bud outgrowth that are evident as early as 32 hrs post-fertilization (Nomura et al., 2006), our results show that limb/fin AER expression and function of Fgf16 has diverged in the teleost and mammalian lineages.
The Fgf16 otic expression pattern suggested possible roles in otic axis formation and/or cell fate decisions. To visualize the consequences of loss of Fgf16 to inner ear morphogenesis, we filled Fgf16IRESCreNeo wild type or heterozygous control and homozygous or hemizygous mutant inner ear epithelia with latex paint at E15.5, a stage when the normal mouse ear has attained its mature morphology. At the gross anatomical level, a total of 12 mutant ears had a normal morphology and showed no obvious differences with wild type or heterozygous controls (Fig. 2E).
To test for cochlear sensory dysfunction, we measured auditory brainstem response (ABR) thresholds in each ear individually at approximately 6 weeks of age in animals of all three Fgf16IRESCreNeo genotypes. No significant differences in auditory thresholds were observed between wild type (n=3), heterozygous female (n=3) and hemizygous male mutant (n=7) animals, which showed thresholds averaging 29 dB, 28 dB and 29 dB, respectively.
Taken together, these results suggest that Fgf16-deficient inner ears are structurally and functionally normal. Thus, if Fgf16 has a function in the ear, it must be redundant with other genes. The most closely related FGFs, which might be expected to share similar receptor specificity (FGFR3c>FGFR2c>FGFR1c, FGFR1b≫GFR4), are FGF9 and FGF20 (Zhang et al., 2006). Fgf9 is expressed primarily in non-sensory regions of the otic epithelium, in the developing semicircular canal fusion plates and then canal ducts, and in Reissner’s membrane in the cochlea. Fgf9 is required directly for otic capsule morphogenesis and therefore indirectly for epithlelial morphogenesis (Pirvola et al., 2004). The finding that Fgf16 is expressed adjacent to and possibly overlapping with some of the Fgf9-expressing vestibular tissues suggests the testable possibility that the Fgf9 otic malformation phenotype could be exacerbated by loss of Fgf16. Redundancy between Fgf16 and Fgf20, which is expressed between E13.5 and E15.5 in the developing sensory cells of the cochlear duct (Hayashi et al., 2008), but not at all in the vestibular system (O. Bermingham-McDonogh, pers. comm.) seems less likely. The other Fgfs expressed in the developing cristae include Fgf3 and Fgf10 (Wilkinson et al., 1989; Pirvola et al., 2000; Pauley et al., 2003, L. Urness and S. Mansour, unpublished). Both are required for normal morphogenesis of the semicircular canals (Mansour et al., 1993; Pauley et al., 2003; Ohuchi et al., 2005; Hatch et al., 2007) and it is possible that the additional loss of Fgf16 might exacerbate their otic phenotypes, though the difference in receptor specificity (FGF3 and FGF10 signal primarily through FGFR2b) makes redundancy less likely.
The Fgf16 lineage is found in both sensory and non-sensory inner ear lineages
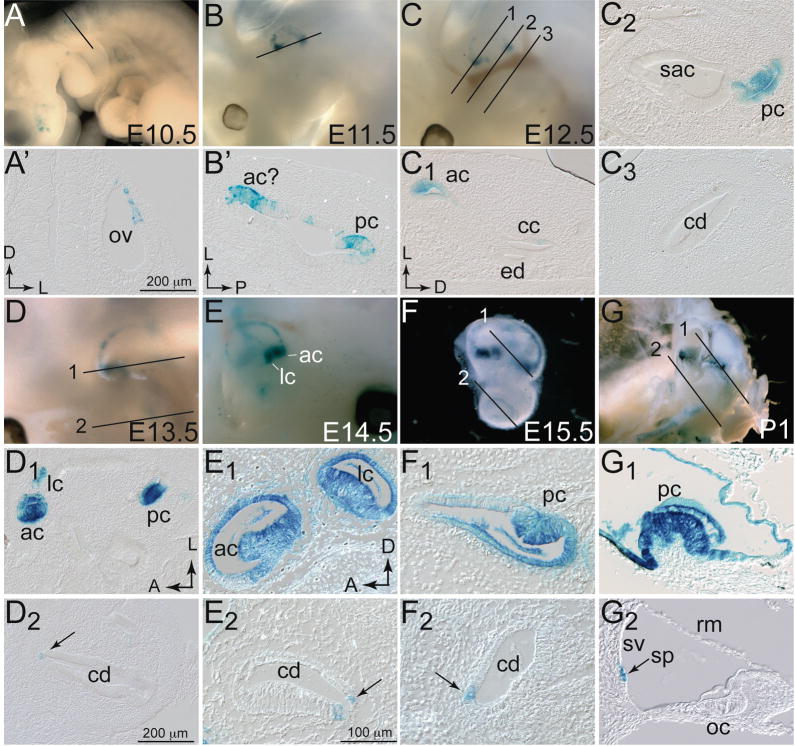
Fgf16IRESCreNeo mice did not express CRE (data not shown), presumably due to the presence of the Neo cassette. To enable analysis of the Fgf16 lineage, the Neo gene was removed by crossing Fgf16IRESCreNeo mice to a FLPe deleter strain (Rodriguez et al., 2000). Mice harboring the Neo-deleted allele (Fgf16IRESCre/Y, MGI accession number 3790784, symbolized Fgf16tm1.1(Cre)Sms) were crossed to Rosa26LacZ/LacZ reporter mice (Soriano, 1999) for lineage studies. Embryos were collected at daily intervals between E9.5 and E15.5, and at P1, and stained with X-gal to detect β-galactosidase (βgal) activity in whole embryos (E9.5–14.5) or after dissection of the inner ears (E15.5, P1). βgal-positive (blue) cells were first apparent at E10.5, in the dorsolateral otic vesicle, although some cells in this region were reporter-negative (Fig. 3A,A′). The delay in the initial detection of βgal relative to the initiation of Fgf16 mRNA expression at E9.0 is likely due to the additive effects of inefficient translation initiation at the IRES element, the time required for CRE protein synthesis and accumulation, LoxP excision, and finally, βgal synthesis and accumulation. Such delays have been noted for other IRESCre alleles (Ohyama and Groves, 2004; Vincent and Robertson, 2004; Lan et al., 2007). The patchiness of the expression could be caused by random inactivation of the Fgf16IRESCre allele in heterozygous female embryos. At E11.5, βgal-positive cells were seen in the developing canal cristae and in scattered cells extending distally from the cristae at the margins of the vertical canal plate (vcp; Figs. 3B,B′). The same pattern of Fgf16 lineage was seen at E12.5, after the canals started forming by fusion of the canal plates (Fig. 3C). βgal positive cells were found in the cristae and at the bases of the canal ducts extending distally (Figs. 3C,C1,C2), but not yet in the cochlear duct (Fig. 3C3). At E13.5, βgal-positive cells were detected in all three cristae and in scattered cells distributed throughout the non-sensory ducts of the anterior and posterior semicircular canals (a/psc), but these cells were more highly concentrated near the cristae (Fig. 3D,D1). At E14.5, the pattern of βgal expression had not changed, but there were increasing numbers of positive cells in the semicircular canal ducts and expression in the cristae appeared to include cells throughout the thickness of the cristae, in precursors to both sensory and supporting cell lineages (Figs. 3E,E1). Fgf16 lineage was apparent in the lateral margin of the cochlear duct, particularly in the most apical regions (Figs. 3E,E2). At E15.5 and P1, reporter-positive cells continued to be detected throughout the cristae, in both sensory and supporting domains, and in many cells of the anterior and posterior non-sensory semicircular canal ducts (Figs. 3F,F1,G,G1). Cochlear Fgf16 lineage was maintained (Figs. 3F2,G2), and by P1, it was clear that these βgal positive cells were located at the base of the stria vascularis (Fig. 3G2) and appeared to comprise the spiral prominence epithelium, the specific function of which is not understood in detail, but is likely to contribute to the maintenance of the endocochlear potential (Slepecky, 1996; Santi and Tsuprun, 2001).
Figure 3. Lineage of Fgf16-expressing cells at E10.5-P1.
Embryos (A–D), inner ears (E,F) or heads (G) harboring both the Rosa26LacZ reporter and Fgf16CreNeo alleles were stained with X-gal at E10.5 (A), E11.5 (B), E12.5 (C), E13.5 (D), E14.5, (E) E15.5 (F), and P1 (G). Anterior is to left in all whole mount panels except E, in which anterior is to the right. Black lines (numbered as appropriate) indicate the plane of transverse (A,C,F,G), coronal (B,D) sections shown in panels below the whole mounts. Sagittal sections of E14.5 left ear (E) were taken in the plane of view, with E1 more lateral than E2. Scale bar in A′ and D1 applies to B′-D2. Scale bar in E1 applies to E2–G2 Abbreviations: ac, anterior crista; cd, cochlear duct; ed, endolymphatic duct; lc, lateral crista; oc, organ of Corti; ov, otic vesicle; pc, posterior crista; rm; Reissner’s membrane; sac, saccule; sp, spiral prominence; sv, stria vascularis.
Consistent with the in situ hybridization study, extra-otic sites of Fgf16 lineage detected in our samples included the olfactory placode and epithelium, the endodermal pouch-derived parathyroid glands and some thymus cells, as well as the developing pituitary. We also noted Fgf16 lineage in E14.5 limbs and lung, and P1 thyroid gland, which is derived from the dorsal oropharynx (foramen cecum, supplemental Fig. S2). We looked for Fgf16 lineage in brown fat and heart, two sites of reported embryonic Fgf16 expression (Miyake et al., 1998; Lavine et al., 2005). Fgf16-lineage was absent from brown fat (P1 and P15, data not shown), but could be detected in the sinoatrial node (the heart’s pacemaker, supplemental Fig. S2). Further investigation will be required to correlate extra-otic Fgf16 expression and lineage data and determine whether Fgf16-deficient mice have subtle or stress-activated phenotypes related to any of these lineages.
The vestibular cells marked by βgal activity from E11.5 and onwards corresponded precisely to the domains of Fgf16 expression in the developing cristae marked by RNA in situ hybridization (Fig. 1G–R). Additional lineage-positive cells were located distally from the cristae in the non-sensory semicircular canal ducts, which did not themselves express significant levels of Fgf16 mRNA until about E14.5 (Figs. 1O,O′), indicating that the Fgf16IRESCre driver recapitulates endogenous ongoing Fgf16 expression in the developing cristae, and in addition, between E11.5 and E14.5, marks duct cells that are likely derived from Fgf16-expressing cells in and/or immediately adjacent to the developing sensory patches. The cells adjacent to the developing cristae have been proposed as a “canal genesis zone” based on DiI fate-mapping of the developing chick canal pouch and the phenotypes of SU5402- or Noggin-treated chick ears (Chang et al., 1999; Gerlach et al., 2000; Chang et al., 2004) and of Fgf10 and Bmp4 mutant mouse ears, in which defective sensory patch development leads to semicircular canal agenesis (Pauley et al., 2003; Ohuchi et al., 2005; Chang et al., 2008). Our expression and Fgf16 lineage tracing data strongly support the existence of a canal genesis zone in the mouse, as Fgf16-expressing cells apparently contribute to the semicircular canals as well as to the developing cristae. By E14.5, Fgf16 expression was detected in the canals, so subsequent lineage observed in the canals may reflect a mixture of cristae- and canal-derived cells.
The cochlear expression of Fgf16 mRNA at the lateral edges of the duct and the βgal-positive ROSA lineage corresponded precisely, except for a 2-day delay in detecting the lineage, suggesting that these Fgf16-expressing cells do not migrate or proliferate significantly.
Although our studies have not yet revealed a function for Fgf16 in the inner ear, Fgf16IRESCre mice could be used to inactivate floxed genes specifically in the developing cristae and spiral prominence epithelial precursors. This might be useful in teasing apart the roles of cristae-expressed genes that are also expressed the other types of otic sensory epithelia. For example, Bmp4 is expressed in developing cristae and at low levels in the maculae (Morsli et al., 1998; Chang et al., 2008), and Fgf10 is expressed in all three types of sensory patches (Pirvola et al., 2000; Pauley et al., 2003), but whether the function of each gene is the same in each sensory patch is unknown. In addition, expression of Fgf16IRESCre in the progenitors of the spiral prominence epithelium provides an opportunity to probe gene function in that tissue. For example, Pendrin, encoded by Slc26a4, is expressed in the epithelial cells of both the stria vascularis and spiral prominence (Wangemann et al., 2004), but it is not known whether its role in endolymph homeostasis requires expression in both locations. Finally, by combining Fgf16IRESCre with a Rosa26stopDTA allele (Ivanova et al., 2005; Wu et al., 2006), it will be possible to ablate the Fgf16 lineage and observe the consequences to inner ear development.
Methods
All work with mice complied with protocols approved by the University of Utah Institutional Animal Care and Use Committee. Embryo ages were determined by counting somite pairs or by considering noon on the day of vaginal plug detection to be E0.5.
RNA in situ hybridization
Fgf16 expression was analyzed at E8.5–10.5 by whole mount RNA in situ hybridization of CD-1 (Charles River Laboratories) embryos using a digoxigenin-labeled Fgf16 RNA probe followed by cryosectioning as described previously (Miyake et al., 1998; Wright et al., 2003). For expression analysis at later stages, whole heads were fixed, embedded in paraffin, sectioned at 10 μm, and stained as described (Urness et al., 2008) using the same probe as for the whole mount expression analysis.
Generation and genotyping of the Fgf16IRESCreNeo and Fgf16IRESCre alleles
Recombination cloning (Zhang et al., 2002) was used to isolate from the λKO2 library (mouse strain129/Sv) a phage bearing a 9.3 kb fragment of Fgf16 genomic DNA with a LoxP site inserted 4.8 kb downstream of exon 1 and flanked by a thymidine kinase gene. After recovery of the plasmid insert, an IRES-Cre-PGKNeo cassette (Arenkiel et al., 2003) was inserted into an artificial AscI site generated in the first exon of Fgf16 and disrupting the 63rd odon. The resulting targeting vector was linearized and electroporated into R1–45 ES cells, which were selected in G-418 and ganciclovir as described (Li et al., 2007). Since Fgf16 is located on the X chromosome, random insertions of the targeting vector were expected to retain a LoxP site, whereas correctly targeted cell lines were expected to lack the LoxP site, which was located near one end of the targeting vector. The presence or absence of the LoxP site was determined by PCR amplification using Fgf16 primers 501 (5′-CCCACTGTTCTTGCCTCTTC-3′) and 502 (5′-GGTTTTGGTGCTGGAGATTG-3′), which flank the LoxP site, and yielded a 376 bp wild type or a 450 bp LoxP-containing DNA fragment. ES clones lacking a LoxP site were analyzed by Southern blot hybridization of Tth111I-digested DNA using a 5′ flanking external probe and of HindIII-digested DNA using a 3′ internal probe was used to confirm correct targeting (Figs. 2A,B). Of 192 drug resistant cell lines, 5 were correctly targeted. Following injection into C57Bl/6 blastocysts, several correctly targeted cell lines generated highly chimeric males that transmitted the targeted allele to offspring. The Fgf16IRESCreNeo allele is officially symbolized as Fgf16tm1(Cre)Sms, MGI accession number 3790778. In subsequent crosses, the wild type and mutant alleles were distinguished by PCR analysis using a three-primer mix containing primers 73 (Neo reverse-5′-TTCAGTGACAACGTCGAGCAC-3′), 418 (Fgf16 exon 1 forward-5′-GTCTTTGCCTCCTTGGACTG-3′) and 419 (Fgf16 exon 1 reverse-5′-CCGTTGGGGAAGATCTCAAG -3′), which produced a wild type band of 218 bp and a mutant band of 691 bp.
To remove the Neo cassette, chimeric males capable of transmitting Fgf16IRESCreNeo were mated to FLP-expressing mice (Rodriguez et al., 2000) and the progeny were screened for loss of the PGK-Neo selection cassette by PCR. Primers 418 and 419 as above and 557 (Cre forward-5′-CAATACCGGAGATCATGCAAG-3′) produced a wild type band of 218 bp and a mutant band of 360 bp. The Neo deleted allele is officially symbolized as Fgf16tm1.1(Cre)Sms, MGI accession number 3790784.
RT-PCR analysis
Total RNA was isolated from female wild type, female heterozygous and male hemizygous mutant embryos at E9.0. Following reverse transcription with an oligo(dT) primer, cDNA fragments were PCR-amplified using Fgf16-specific primers 418 (exon 1, forward) and 569 (exon 2, reverse-5′-GCCAAGCTGATAAATTCCAGA-3′). A 351 bp region within the Hprt positive control gene was PCR-amplified using primers 33 (5′-CCTGCTGGATTACATTAAAGCACTG-3′) and 34 (5′-GTCAAGGGCATATCCAACAACAAAC-3′).
Paint-filling and auditory brainstem response (ABR) threshold measurements
E15.5 embryos were fixed in Modified Carnoy’s solution, cleared in methyl salicylate and the ears were filled through the saccule with white latex paint as described (Morsli et al., 1998). For ABR studies, 3–5 week old mice were anesthetized using 0.02 ml/g Avertin. Thresholds for click stimuli (47 μsec duration, 29.3/sec) presented to each ear individually were determined using high frequency transducers controlled and analyzed by SmartEP software (Intelligent Hearing Systems) (Zheng et al., 1999).
Fgf16 lineage analysis
We crossed Rosa26LacZ/LacZ reporter females (Soriano, 1999) to Fgf16IRESCre/Y males or heterozygous or homozygous Fgf16IRESCre females to homozygous reporter males. Embryos or dissected ears harboring both the Rosa26LacZ and Fgf16IRESCre alleles were stained with X-gal and sectioned as described (Yang et al., 1997).
Supplementary Material
Acknowledgments
We thank Steve Elledge for the recombination screening system reagents, Ben Arenkiel and Mario Capecchi for the IRESCreNeo cassette, and Susan Tamowski of the University of Utah Transgenic/Gene Targeting Facility for chimera generation. We are grateful to Chaoying Li for completing the lineage staining, to Dr. Steven Bleyl for heart expertise, and to Dr. Gary Schoenwolf for developmental expertise and constructive criticism of the manuscript. Supported by NIH grants R01-DC04185 (SLM, LDU and EH) and T32-GM007464 (EH).
References
- Arenkiel BR, Gaufo GO, Capecchi MR. Hoxb1 neural crest preferentially form glia of the PNS. Dev Dyn. 2003;227:379–386. doi: 10.1002/dvdy.10323. [DOI] [PubMed] [Google Scholar]
- Bok J, Chang W, Wu DK. Patterning and morphogenesis of the vertebrate inner ear. Int J Dev Biol. 2007;51:521–533. doi: 10.1387/ijdb.072381jb. [DOI] [PubMed] [Google Scholar]
- Brigande JV, Iten LE, Fekete DM. A fate map of chick otic cup closure reveals lineage boundaries in the dorsal otocyst. Dev Biol. 2000;227:256–270. doi: 10.1006/dbio.2000.9914. [DOI] [PubMed] [Google Scholar]
- Chang W, Brigande JV, Fekete DM, Wu DK. The development of semicircular canals in the inner ear: role of FGFs in sensory cristae. Development. 2004;131:4201–4211. doi: 10.1242/dev.01292. [DOI] [PubMed] [Google Scholar]
- Chang W, Lin Z, Kulessa H, Hebert J, Hogan BL, Wu DK. Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. PLoS Genet. 2008;4:e1000050. doi: 10.1371/journal.pgen.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Nunes FD, De Jesus-Escobar JM, Harland R, Wu DK. Ectopic noggin blocks sensory and nonsensory organ morphogenesis in the chicken inner ear. Dev Biol. 1999;216:369–381. doi: 10.1006/dbio.1999.9457. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Cai Q, Bleyl SB, Schoenwolf GC. Restricted expression of Fgf16 within the developing chick inner ear. Dev Dyn. 2006;235:2276–2281. doi: 10.1002/dvdy.20872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach LM, Hutson MR, Germiller JA, Nguyen-Luu D, Victor JC, Barald KF. Addition of the BMP4 antagonist, noggin, disrupts avian inner ear development. Development. 2000;127:45–54. doi: 10.1242/dev.127.1.45. [DOI] [PubMed] [Google Scholar]
- Hatch EP, Noyes CA, Wang X, Wright TJ, Mansour SL. Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development. 2007 doi: 10.1242/dev.006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci. 2008;28:5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Signore M, Caro N, Greene ND, Copp AJ, Martinez-Barbera JP. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis. 2005;43:129–135. doi: 10.1002/gene.20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Steel KP, Fekete DM. Development of the mouse inner ear. In: Rossant J, Tam PPL, editors. Mouse development: Patterning, morphogenesis and organogenesis. London: Academic Press Inc; 2002. pp. 539–566. [Google Scholar]
- Lan Y, Wang Q, Ovitt CE, Jiang R. A unique mouse strain expressing Cre recombinase for tissue-specific analysis of gene function in palate and kidney development. Genesis. 2007;45:618–624. doi: 10.1002/dvg.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Miyake A, Konishi M, Martin FH, Hernday NA, Ozaki K, Yamamoto S, Mikami T, Arakawa T, Itoh N. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochem Biophys Res Commun. 1998;243:148–152. doi: 10.1006/bbrc.1998.8073. [DOI] [PubMed] [Google Scholar]
- Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura R, Kamei E, Hotta Y, Konishi M, Miyake A, Itoh N. Fgf16 is essential for pectoral fin bud formation in zebrafish. Biochem Biophys Res Commun. 2006;347:340–346. doi: 10.1016/j.bbrc.2006.06.108. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Yasue A, Ono K, Sasaoka S, Tomonari S, Takagi A, Itakura M, Moriyama K, Noji S, Nohno T. Identification of cis-element regulating expression of the mouse Fgf10 gene during inner ear development. Dev Dyn. 2005;233:177–187. doi: 10.1002/dvdy.20319. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, Dickson C, Ylikoski J. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Zhang X, Mantela J, Ornitz DM, Ylikoski J. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Dev Biol. 2004;273:350–360. doi: 10.1016/j.ydbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Santi PA, Tsuprun VL. Physiology of the Ear. Singular; 2001. Cochlear Microanatomy and Ultrastructure. p look this up. [Google Scholar]
- Slepecky NB. Cochlear Structure. In: Dallos P, Popper AN, Fay RR, editors. The Cochlea. New York: Springer-Verlag; 1996. p look this up. [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Streit A. Extensive cell movements accompany formation of the otic placode. Dev Biol. 2002;249:237–254. doi: 10.1006/dbio.2002.0739. [DOI] [PubMed] [Google Scholar]
- Swanson GJ, Howard M, Lewis J. Epithelial autonomy in the development of the inner ear of a bird embryo. Dev Biol. 1990;137:243–257. doi: 10.1016/0012-1606(90)90251-d. [DOI] [PubMed] [Google Scholar]
- Urness LD, Li C, Wang X, Mansour SL. Expression of ERK signaling inhibitors Dusp6, Dusp7, and Dusp9 during mouse ear development. Dev Dyn. 2008;237:163–169. doi: 10.1002/dvdy.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Robertson EJ. Targeted insertion of an IRES Cre into the Hnf4alpha locus: Cre-mediated recombination in the liver, kidney, and gut epithelium. Genesis. 2004;39:206–211. doi: 10.1002/gene.20047. [DOI] [PubMed] [Google Scholar]
- Wangemann P, Itza EM, Albrecht B, Wu T, Jabba SV, Maganti RJ, Lee JH, Everett LA, Wall SM, Royaux IE, Green ED, Marcus DC. Loss of KCNJ10 protein expression abolishes endocochlear potential and causes deafness in Pendred syndrome mouse model. BMC Med. 2004;2:30. doi: 10.1186/1741-7015-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, McMahon AP. Expression pattern of the FGF-related proto-oncogene int-2 suggests multiple roles in fetal development. Development. 1989;105:131–136. doi: 10.1242/dev.105.1.131. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Hatch EP, Karabagli H, Karabagli P, Schoenwolf GC, Mansour SL. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228:267–272. doi: 10.1002/dvdy.10362. [DOI] [PubMed] [Google Scholar]
- Wu DK, Nunes FD, Choo D. Axial specification for sensory organs versus non-sensory structures of the chicken inner ear. Development. 1998;125:11–20. doi: 10.1242/dev.125.1.11. [DOI] [PubMed] [Google Scholar]
- Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Yang W, Musci TS, Mansour SL. Trapping genes expressed in the developing mouse inner ear. Hear Res. 1997;114:53–61. doi: 10.1016/s0378-5955(97)00146-9. [DOI] [PubMed] [Google Scholar]
- Zhang P, Li MZ, Elledge SJ. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nat Genet. 2002;30:31–39. doi: 10.1038/ng797. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.